Abstract
Gemtuzumab ozogamicin (GO) is a conjugate of a monoclonal antibody and calicheamicin, which has been reapproved for the treatment of acute myeloid leukemia (AML). AML patients with the CD33 rs12459419 CC genotype might benefit from the addition of GO to intensive treatment in contrast to patients with CT/TT genotypes. Nevertheless, contradictory results have been reported. We sought to shed light on the prediction of GO response in AML patients with rs12459419 polymorphism who were treated with GO in the consolidation (n = 70) or reinduction (n = 20) phase. The frequency distribution of the rs12459419 polymorphism in the complete cohort of patients was 44.4% (n = 40), 50% (n = 45), and 5.6% (n = 5) for CC, CT, and TT genotypes, respectively. Regarding the patients treated with GO for consolidation, we performed a Kaplan-Meier analysis of overall survival and relapse-free survival according to the rs12459419 polymorphism (CC vs. CT/TT patients) and genetic risk using the European Leukemia Net (ELN) 2010 risk score. We also carried out a Cox regression analysis for the prediction of overall survival, with age and ELN 2010 as covariates. We found no statistical significance in the univariate or multivariate analysis. Additionally, we performed a global Kaplan-Meier analysis for the patients treated with GO for reinduction and did not find significant differences; however, our cohort was too small to draw any conclusion from this analysis. The use of GO in consolidation treatment is included in the approval of the compound; however, evidence regarding its efficacy in this setting is lacking. Rs12459419 polymorphism could help in the selection of patients who might benefit from GO. Regrettably, in our cohort, the rs12459419 polymorphism does not seem to be an adequate tool for the selection of patients who might benefit from the addition of GO in consolidation cycles.
1. Introduction
Acute myeloid leukemia (AML) is a heterogeneous disorder characterized by a clonal expansion of myeloid progenitors [1]. AML remains a difficult-to-treat disease, and novel efficacious therapies are needed. Immunotherapeutic strategies have been proved highly effective in other hematological malignancies [2]. Gemtuzumab ozogamicin (GO) is a humanized anti-CD33 IgG4 mAb conjugated to a cytotoxic agent N-acetyl gamma calicheamicin. GO targets the membrane antigen CD33 that is present on the majority of AML blasts [3]. The efficacy of the drug in three open-label phase II trials resulted in an accelerated approval in 2000 by the US Food and Drug Administration [4]. However, a large follow-up study (SWOG-S0106) showed an increase in early death with the use of GO in AML patients, so the drug manufacturer voluntarily withdrew the US New Drug Application in 2010 [5]. In 2014, a meta-analysis of five randomized controlled trials [6–10] showed that adding GO to induction chemotherapy improved overall survival (OS) and relapse-free survival (RFS) in favorable-risk and, to a lesser degree, intermediate-risk AML patients. Based on these results, GO was reapproved. Nevertheless, three of the trials of the meta-analysis also included GO in consolidation cycles, and the possible clinical benefit of adding GO in the consolidation cycle has not been clearly elucidated.
GO treatment has been associated with hepatotoxicity and hepatic veno-occlusive disease/sinusoidal obstruction syndrome (VOD/SOS), myelosuppression, bleeding/thrombocytopenia, infusion-related reactions, and tumor lysis syndrome. In the ALFHA 701 study, drug-induced neutropenia was more prolonged in the GO group than in the control group after each cycle of consolidation. In addition, the median time to recovery of platelets was longer for patients in the GO arm than in the control arm for each treatment course (persistent grades III-IV thrombocytopenia was observed in 20% of patients in the GO group vs. 2% in the control group) [11].
Because GO is ineffective in many patients and/or may have adverse effects, there is interest in understanding favorable and unfavorable prognostic factors. Therefore, it is important to further refine the selection of patients that might achieve a favorable response from GO administration. Two studies conducted in pediatric and NPM1-mutated AML adult patients treated with GO during the induction phase suggested a potential value of CD33 genotype as a predictor of response [12, 13]. However, the gathered data from several trials did not validate the results described previously [14, 15]. Specifically, subjects with the CD33 rs12459419 CC genotype (about 50% of study entrants) seemed to have a significantly lower risk of relapse and better event-free survival and disease-free survival after GO therapy, whereas this benefit was not seen in patients with the CT or TT genotypes. Rs12459419:c.41C > T; p.Ala14Val is related to the loss of exon 2 in the CD33 transcript, which results in a shorter isoform of the CD33 protein product (D2-CD33) that lacks the IgV domain. This domain, which is encoded by exon 2, is recognized by the antibody that is conjugated to calicheamicin in GO [16–19]. Consequently, the loss of this domain can interfere with the clinical efficacy of GO (Figure 1).
Figure 1.
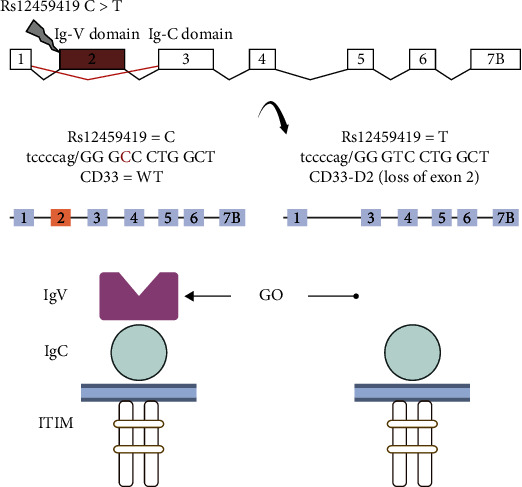
Mechanism of action of GO. Abbreviations: GO: gemtuzumab ozogamicin.
In the present study, we analyzed whether CD33 rs12459419 polymorphism influences the clinical outcome in adult AML patients treated with GO during consolidation or reinduction cycles.
2. Material and Methods
2.1. Patients and Samples
The PETHEMA AML epidemiologic registry (NCT02607059) includes the data of patients diagnosed with AML, regardless of the treatment administered. The primary patient and disease characteristics were collected retrospectively (Table 1). We reviewed the prospectively collected data of 106 adult AML patients treated with GO from the PETHEMA AML epidemiologic registry following the PETHEMA 2007 protocol. Pretreatment samples were available for CD33 polymorphism determination in 90 patients: 48 men and 42 women. The median age was 55.3 years (range 17.4–76.9 years) in these patients with newly diagnosed AML between 2005 and 2013. Seventy patients received GO 3 mg/m (2) in the consolidation cycle and 20 in reinduction treatment. Finally, 19 patients underwent autologous hematopoietic stem-cell transplantation (HSCT), and 33 patients went through allogeneic HSCT.
Table 1.
Baseline characteristics of AML patients treated with GO 3 mg/m(2) during reinduction and consolidation phases based on CD33 rs12459419 polymorphism.
| CD33 rs12459419 genotype in AML patients treated with GO in reinduction and consolidation phases | ||||||
|---|---|---|---|---|---|---|
| Reinduction treatment | Consolidation treatment | |||||
| Patients characteristics | Total; n =20 | CC (n = 12; 60%) | CT/TT (n = 7/1; 35%/5%) | Total; n = 70 | CC (n = 32; 46%) | CT/TT (n = 34/4; 49%/6%) |
| Median age at diagnosis, years | 51 | 49.5 | 53.9 | 57.2 | 57.2 | 57.7 |
| Range | 17.4–64.2 | 17.4-64.1 | 30.4-61.2 | 24.8–76.9 | 24.8-70.0 | 28.2-76.9 |
| Median follow-up, years | 0.58 | 0.58 | 0.25 | 2.13 | 1.96 | 2.17 |
| Sex | ||||||
| Female | 10 (50%) | 8 (67%) | 2 (25%) | 36 (51%) | 18 (56%) | 18 (47%) |
| Male | 10 (10%) | 4 (33%) | 6 (75%) | 34 (49%) | 14 (44%) | 20 (53%) |
| Cytogenetic risk (ELN 2010) | ||||||
| Favorable risk | 5 (25%) | 4 (33%) | 1 (13%) | 28 (40%) | 15 (47%) | 13 (34%) |
| Intermediate-I risk | 2 (10%) | 2 (17%) | ─ | 20 (29%) | 8 (25%) | 12 (32%) |
| Intermediate-II risk | 6 (30%) | 3 (25%) | 3 (38%) | 7 (10%) | 3 (9%) | 4 (11%) |
| Adverse risk | 6 (30%) | 3 (25%) | 3 (38%) | 9 (13%) | 4 (13%) | 5 (13%) |
| Median leucocytes at diagnosis, 109/L | 14.7 | 16.0 | 22.8 | 17.5 | 17.5 | 17.5 |
| Range | 1.6-206.6 | 1.6-100.5 | 1.9-206.6 | 0,7-379 | 1,1-379 | 0,7-324 |
| Median hemoglobin at diagnosis, g/dl | 8.7 | 8.7 | 8.8 | 9.5 | 9,5 | 9,5 |
| Range | 4.6-11.2 | 4.6-11.2 | 5.5-10.9 | 5-13,9 | 5-13,9 | 5,3-13,5 |
| Median platelets at diagnosis, 109/L | 53 | 53 | 49 | 65 | 65 | 62 |
| Range | 15-261 | 30-261 | 15-97 | 14-336 | 14-317 | 13-336 |
| Treatment | ||||||
| Induction therapy | ||||||
| Idarubicin+cytarabine (3 + 7) | 19 (95%) | 11 (92%) | 8 (100%) | 59 (84%) | 27 (84%) | 32 (84%) |
| Idarubicin+cytarabine (2 + 5) | 1 (5%) | 1 (8%) | ─ | 9 (13%) | 5 (16%) | 4 (11%) |
| Other treatments | ─ | ─ | ─ | 2 (3%) | ─ | 2 (5%%) |
| Reinduction | ||||||
| 3 + 7 with GO | 1 (5%) | ─ | 1 (12.5%) | ─ | ─ | ─ |
| IDA-FLAGO | 19 (95%) | 12 (100%) | 7 (87.5%) | ─ | ─ | ─ |
| Other treatments | ─ | ─ | ─ | ─ | ─ | ─ |
| Consolidation | ||||||
| HDARAC+GO | ─ | ─ | ─ | 14 (20%) | 6 (19%) | 8 (21%) |
| 3 + 7 + GO | ─ | ─ | ─ | 56 (80%) | 26 (81%) | 30 (79%) |
| HDARAC | 6 (30%) | 4 (33%) | 2 (25%) | ─ | ─ | |
| IDA-FLAG | 2 (10%) | 2 (17%) | ─ | ─ | ─ | |
| Support treatment | 1 (5%) | 1 (8%) | ─ | ─ | ─ | |
| Stem cell transplant | ||||||
| Allogeneic hematopoietic cell transplantation | 11 (55%) | 8 (67%) | 3 (38%) | 22 (31%) | 11 (34%) | 11 (29%) |
| Autologous hematopoietic cell transplantation | ─ | ─ | ─ | 19 (27%) | 6 (19%) | 13 (29%) |
Abbreviations: GO: gemtuzumab ozogamicin; ELN: European Leukemia Net; IDA-FLAG/IDA-FLAGO: idarubicin, fludarabine, cytarabine, and G-CSF ± GO; ARAC (100 mg/m2 during 5 days); HDARAC (1-3 g/m2 during 1, 3, and 5 days of the cycle).
2.2. CD33 SNP Screening
We received DNA (n = 85) and RNA (n = 14) samples from seven centralized PETHEMA laboratories. The samples analyzed in this study were collected at the diagnostic stage. DNA was extracted using automated or manual DNA extraction kits (Qiagen, Hilden, Germany) following the manufacturer's recommendations. DNA quantification was done using NanoDrop (ThermoFisher Scientific, Waltham, MA) or Qubit fluorometer (ThermoFisher Scientific, Waltham, MA) [20]. Samples were screened for the CD33 SNP using HaeIII restriction enzyme digestion of polymerase chain reaction (PCR)-generated amplicons. The cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit (ThermoFisher, Vilnius, Lithuania). PCR products of 266 base pairs (bp) and 204 bp were generated from genomic DNA and cDNA, respectively, using HotStarTaq® DNA polymerase (Qiagen, Germany) using the manufacturer's recommended conditions and primers (CD33 exon 1/F: 5′--CTGGAAGCTGCTTCCTCAGACATG--3′; CD33 exon 2/R: 5′-GAACCAGTAACCATGAACTGGGGAGTT--3′) at an annealing temperature of 66°C. Products were digested overnight with HaeIII (New England Biolabs, Hitchin, UK) and separated on a 3% agarose gel to discriminate between the C allele (94 + 29 + 143 bp) and T allele (94 + 172 bp) using the DNA and C allele (61 + 143 bp) and T allele (204 bp) using the cDNA.
2.3. Statistical Analysis
OS was calculated from the date of diagnosis of AML until death in all included patients. RFS was calculated from the date of achieving complete response (CR) or CR with incomplete hematologic recovery (CRi) until the date of relapse or death due to any cause. CR and CRi were defined according to current 2017 ELN guidelines [21]. OS and RFS were estimated by the Kaplan-Meier method and compared by log-rank test. Group comparisons were defined according to the rs12459419 genotype (i.e., CC vs. CT and TT). Patients treated with GO in the consolidation phase were analyzed globally and in subgroups according to the ELN 2010 genetic risk [22] (intermediate I and II groups were merged). Multivariate Cox regression analysis was used for OS prediction, with age and ELN genetic risk model as covariates. All patients, who received GO as reinduction, were analyzed separately. A separate analysis using the ELN 2010 genetic risk was not performed due to the reduced number of patients in the reinduction group. Mann–Whitney test was used to compare time to platelet (>50.000/μl) and neutrophil recovery (>500/μl) among patients who received 3 + 7 plus GO as a consolidation cycle. A P value below 0.05 was considered statistically significant. SPSS version 19.0 (IBM, Arkmon) was used for the analyses.
3. Results
The frequency distribution of the rs12459419 genotypes in the whole patient population (n = 90) were 44.4%, 50%, and 5.6% for the CC, CT, and TT polymorphism, respectively. From the total of 70 patients treated with GO during the consolidation cycle, there were 32 patients (45.7%) with CC genotype and 38 (54.3%) with CT/TT genotype. The median duration of OS in the CC group was 2.7 years (95% confidence interval: 1.0-4.3) vs. 4.4 years (95% CI: 0.5-8.2) in the CT/TT group (P = 0.9). The median RFS in the CC group was 1.5 years (95% CI: 1.1-1.9) vs. 1.7 years (95% CI: 0.7-2.6) in the CT/TT group (P = 0.5).
Furthermore, a separate analysis based on the ELN 2010 genetic risk [22] was performed, including 64 patients. In the analysis of OS and RFS, the distribution of rs12459419 genotype was as follows: 15 patients (23.4%) with CC genotype and 13 (20.3%) with CT/TT genotype in the favorable-risk group, 11 patients (17.2%) with CC genotype and 16 (25%) with CT/TT genotype in the intermediate-risk group, and four (6.3%) with CC genotype and five (7.8%) with CT/TT genotype in the adverse-risk group. The OS in the favorable group was 4.7 years (95% CI: not calculable [NC]) in CC genotype and 8.6 years (95% CI: 1.4-15.4) in CT/TT genotype (P = 0.9). The OS in the intermediate group was 2.5 years (95% CI: 1.6-3.5) in CC genotype and 1.7 years (95% CI: NC-7.8) in CT/TT genotype (P = 0.9). The OS in the adverse group was 0.8 years (95% CI: NC-3.0) in CC genotype and 0.1 years (95% CI: 0.4-0.9) in CT/TT genotype (P = 0.8). The RFS in the favorable group was 1.7 years (95% CI: 1.3-2.7) in CC genotype and 2.7 years (95% CI: NC-7.5) in CT/TT genotype (P = 0.6). The RFS in the intermediate group was 1.4 years (95% CI: 1.3-1.6) in CC genotype and 1.6 years (95% CI: NC-9.4) in CT/TT genotype (P = 0.6). The RFS in the adverse group was 0.2 years (95% CI: NC-1.2) in CC genotype and 0.6 years (95% CI: 0.3-0.8) in CT/TT genotype (P = 0.4) (Table 2 and Figure 2). Cox regression analysis for the prediction of OS showed that ELN 2010 and age were independent predictor factors (P = 0.03, HR: 1.4, [95% CI: 1.03-1.85] and P = 0.02, HR: 1.04, [95% CI: 1.01-1.07], respectively). Rs12459419 genotype was not an independent predictor factor (P = 0.5, HR: 0.8, [95% CI: 0.4-1.5]). Proportional hazard assumptions were checked by adding the interaction of every variable with time (Table 3).
Table 2.
Kaplan-Meier and log-rank test analyses of overall survival and relapse-free survival in patients treated with GO in the consolidation phase.
(a).
| Overall survival | ||||
|---|---|---|---|---|
| rs12459419 genotypes | Genetic risk (ELN 2010) | N | Median (95% CI) | P |
| CC | Global | 32 | 2.7 y (1.0-4.3) | 0.9 |
| Favorable risk | 15 | 4.7 y (NC) | 0.9 | |
| Intermediate risk | 11 | 2.5 y (1.6-3.5) | 0.9 | |
| Adverse risk | 4 | 0.8 y (NC-3.0) | 0.8 | |
| CT/TT | Global | 38 | 4.4 y (0.5-8.2) | 0.9 |
| Favorable risk | 13 | 8.6 y (1.4-15.4) | 0.9 | |
| Intermediate risk | 16 | 1.7 y (NC-7.8) | 0.9 | |
| Adverse risk | 5 | 0.1 y (0.4-0.9) | 0.8 | |
(b).
| Relapse-free survival | ||||
|---|---|---|---|---|
| rs12459419 genotypes | Genetic risk (ELN 2010) | N | Median (95% CI) | P |
| CC | Global | 32 | 1.5 y (1.1-1.9) | 0.5 |
| Favorable risk | 15 | 1.7 y (1.3-2.7) | 0.6 | |
| Intermediate risk | 11 | 1.4 y (1.3-1.6) | 0.6 | |
| Adverse risk | 4 | 0.2 y (NC-1.2) | 0.4 | |
| CT/TT | Global | 38 | 1.7 y (0.7-2.6) | 0.5 |
| Favorable risk | 13 | 2.7 y (NC-7.5) | 0.6 | |
| Intermediate risk | 16 | 1.6 y (NC-9.4) | 0.6 | |
| Adverse risk | 5 | 0.6 y (0.3-0.8) | 0.4 | |
Abbreviations: ELN: European Leukemia Net; CI: confidence interval; NC: not calculable.
Figure 2.
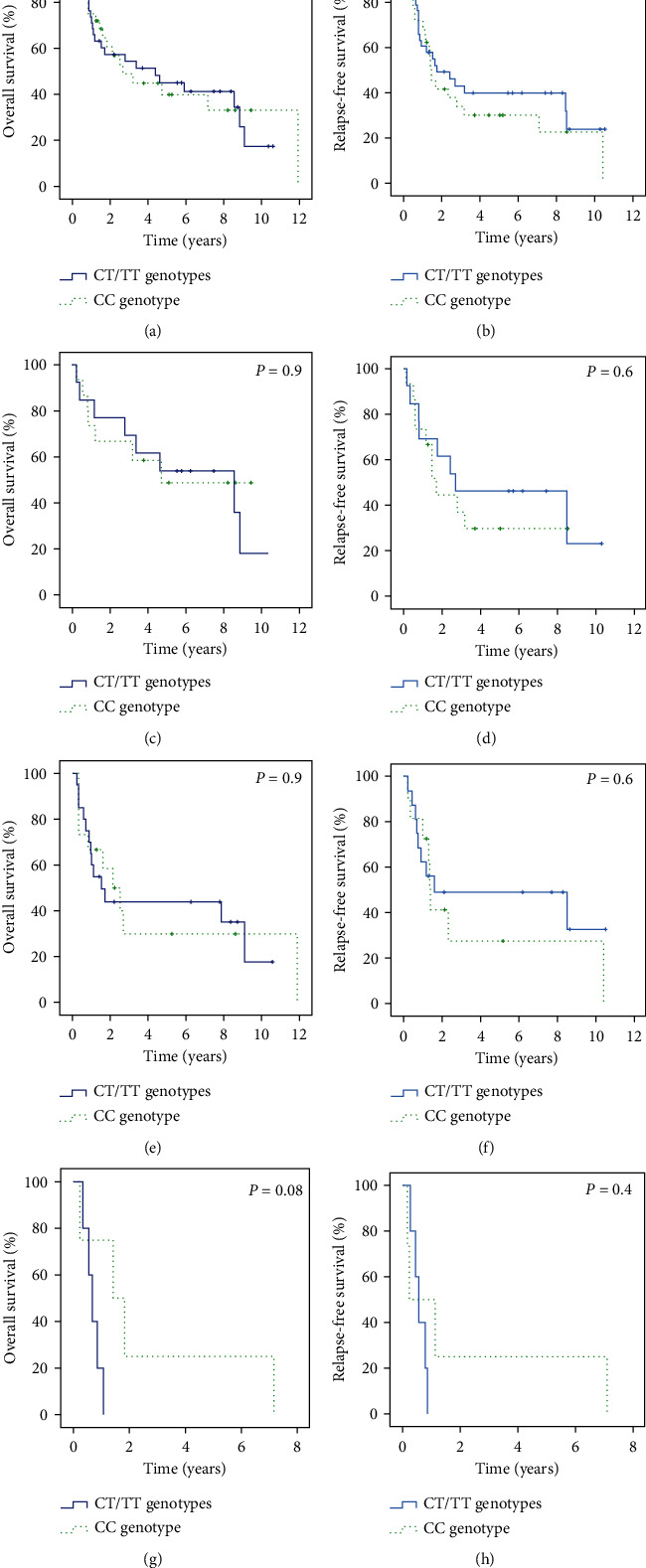
Clinical outcome stratified by the CD33 rs12459419 polymorphism for patients treated with GO during the consolidation phase (a–h). (a) Overall survival (OS). (b) Relapse-free survival (RFS). (c–h) OS and RFS according to CD33 rs12459419 polymorphism and genetic risk. (c and d) OS and RFS in the favorable group; (e and f) OS and RFS in the intermediate group; and (g and h) OS and RFS in the adverse group.
Table 3.
Cox proportional hazard regression model analysis of overall survival.
| Overall survival | ||
|---|---|---|
| Covariate | HR (95% CI) | P |
| Age | 1.04 (1.01-1.07) | 0.02 |
| Genetic risk (ELN 2010) | 1.4(1.03-1.85) | 0.03 |
| CD33 genotypes | 0.8 (0.4-1.5) | 0.5 |
Abbreviations: ELN: European Leukemia; CI: confidence interval.
Finally, in the 20 patients treated with GO during the reinduction phase, the distribution of rs12459419 genotype in OS analysis was 12 patients (60%) with CC genotype and eight (40%) with CT/TT genotype. The median duration of OS in the CC group was 0.6 years (95% CI: 0.5-0.8) vs. 0.3 years (95% CI: 0.3-0.3) in the CT/TT group (P = 0.3). The distribution of rs12459419 genotype in RFS analysis was nine patients (45%) with CC genotype and three (15%) with CT/TT genotype. The median duration of RFS in the CC group was 0.6 years (95% CI: 0.3-0.9) vs. 0.6 years (95% CI: 0.5-2.7) in the CT/TT group (P = 0.8). (Supplementary Material see available here).
The drug-induced neutropenia was similar in the CT/TT and CC genotype groups after the first cycle of consolidation with 3 + 7 plus GO: a median of 23 days (range 19-31) vs. 20 days (range 14-28), respectively (P = 0.3). In addition, we did not find statistical differences regarding the time to recovery of platelets: a median of 25.5 days (range 23-54) in the CT/TT group and 25.5 days (range 20-43) in the CC group (P = 0.8). No patient developed VOD/SOS during cycles of treatment prior to allogeneic HSCT. Regrettably, data on the incidence of VOD/SOS in the postallogeneic HSCT setting were not collected.
4. Discussion
Personalized medicine is based on selecting an adequate treatment for every individual patient [23]. We evaluated the impact of the CD33 rs12459419 genotype on the efficacy of GO treatment in AML patients. Investigating a potential lack of clinical benefit of CT and TT genotypes of the polymorphism, we performed comparisons between the CT/TT and CC groups, who were supposed to benefit from GO treatment. Our real-life cohort is composed of 90 AML patients: none of them were included in clinical trials, and all were treated with GO in reinduction (n = 20) or consolidation treatment (n = 70) following the PETHEMA 2007 protocol. In our cohort, the rs12459419 genotype does not have a prognostic impact in terms of OS or RFS among AML patients treated with GO in either the consolidation or reinduction cycle. Additionally, the time to hematological recovery after the first consolidation cycle with 3 + 7 plus GO cycle was not influenced by the SNP genotype.
The clinical impact of the genotype of this splicing polymorphism in AML patients treated with GO-containing chemotherapy is controversial. In a study in a mainly pediatric population, but also including young adults (COG-AAML0531) with AML (0-29 years old), a benefit of GO was demonstrated in the induction treatment in those with CC genotype of CD33 rs12459419 polymorphism [12]. In contrast to our results, a recent large study in adult AML patients who were eligible for intensive therapy and had an NPM1 mutation showed improved RFS and reduced cumulative incidence of relapse in patients with CC genotype treated with GO during the induction phase [13]. Conversely, the British group analyzed a cohort of mainly adult AML patients (13-69 years old) and found that the patients who received GO in induction treatment did not achieve clinical improvement in terms of OS or RFS regardless of the rs12459419 genotype [14]. Moreover, as in our population, a study in patients treated with decitabine plus GO during the consolidation phase (n = 113) did not reveal a significant impact of the SNP on OS or RFS. However, this study was performed in a patient population unlikely to benefit from GO as it included high-risk myelodysplastic syndrome patients and AML patients with unfavorable risk features [15]. In contrast to the described studies, our cohort was composed of patients who received GO at either consolidation or reinduction cycles. Interestingly, three out of five trials that were included in the meta-analysis of GO efficacy administered GO also in consolidation cycles [10]. Nevertheless, only the MRC AML15 trial performed a new randomization process for consolidation. MRC AML15 trial did not show differences in consolidation depending on GO administration, but this study neither showed differences regarding GO administration in the induction cycle [7]. Indeed, GO approval includes induction and consolidation doses following the ALFA 0701 trial scheme protocol [9]. Therefore, the possible clinical benefit of adding GO only in the consolidation phase has not been clearly elucidated. Nevertheless, a high proportion of minimal residual disease eradication has recently been reported with the addition of GO to consolidation treatment with HIDAC [24]. This finding may support the clinical efficacy of GO in consolidation cycles. Besides, although GO addition to consolidation had not achieved clinical benefit in the overall population, GO could improve outcomes in patients with CC genotype of the CD33 rs12459419 polymorphism. Regrettably, our study has shown that this polymorphism does not have prognostic value in this setting.
Our study has some limitations. First, our patients were selected from an observational registry, which can be interpreted as a limitation given the heterogeneity of treatment or as a strength because our data are more real-life than those derived from a clinical trial. Second, since the number of patients was small, it would be necessary to increase the sample size to validate our results. However, our results support the findings of the previously published study with the largest cohort [14, 15].
In the event that the genotype of the rs12459419 polymorphism played a role in the mechanism of action of GO, it would modify the effect of the drug on leukemia and normal hematopoiesis only in patients with CC genotype. We would expect a longer time to recovery of peripheral blood counts in these patients without an increase in this time in patients with CT or TT genotypes. In our cohort, we did not find differences in the recovery of neutrophils and platelets based on the SNP genotype. These results were similar to those recently published by Short et al., which support the lack of prognostic value of the rs12459419 polymorphism found in our cohort [1].
5. Conclusions
In summary, in our population of 20 AML patients treated with GO in reinduction, we found no prognostic significance of CD33 rs12459419 polymorphism, but no conclusions can be drawn due to the reduced amount of patients analyzed. In the consolidation setting, we analyzed 70 patients and did not observe differences between groups. Therefore, CD33 rs12459419 polymorphism does not seem to be an adequate tool to select patients who might benefit from the addition of GO in consolidation cycles.
Acknowledgments
J.M.A.D. received research funding from Incyte, Celgene, Pfizer, and Astellas Pharma. Thanks to the PETHEMA group for their participation in this study.
Data Availability
Data are available upon request from the authors.
Ethical Approval
The study protocol was conducted following the Declaration of Helsinki and approved by the Ethics Committee for Clinical Research of the Hospital Universitario Fundación Jiménez Díaz (EO160-18_FJD).
Consent
Informed consent was a requisite for patients alive at the time of data lock (January 2019).
Disclosure
This work is submitted in partial fulfillment of the requirement for the PhD.
Conflicts of Interest
T.C.B. is a PhD candidate at Universidad Autónoma de Madrid (UAM). No other conflicts of interest were declared.
Authors' Contributions
Conceptualization was contributed by Tamara Castaño-Bonilla, Juan Manuel Alonso-Domínguez, Eva Barragan, and Pau Montesinos; data curation was contributed by Juan Manuel Alonso-Domínguez and Pau Montesinos; formal analysis was contributed by Juan Manuel Alonso-Domínguez; investigation was performed by Tamara Castaño-Bonilla, Juan Manuel Alonso-Domínguez, and Pau Montesinos; methodology was contributed by Tamara Castaño-Bonilla, Eva Barragán, Claudia Sargas, Alejandro Sanz, Lorenzo Algarra, Pilar Herrera-Puente, Raimundo García-Boyero, Manuel Barrios, David Martinez-Cuadron, Rebeca Rodriguez-Veiga, Blanca Boluda, Cristina Gil, Josefina Serrano-Lopez, Joaquín Martínez-López, Maria Jose Sayas-Lloris, Maria Teresa Olave, Rosalia Riaza-Grau, Teresa Bernal-Del Castillo, Maria Jose Larrayoz, Antonio Jimenez, Raquel Amigo, Joaquin Sanchez, Rosa Ayala, Carlos Blas, Daniel Lainez, Juana Serrano, Miguel A. Sanz, Juan M. Alonso-Dominguez, and Pau Montesinos; project administration was contributed by Juan Manuel Alonso-Domínguez and Pau Montesinos; resources was contributed by Tamara Castaño-Bonilla, Juan Manuel Alonso-Domínguez, and Pau Montesinos; supervision was performed by Juan Manuel Alonso-Domínguez and Pau Montesinos; validation was performed by Tamara Castaño-Bonilla, Juan Manuel Alonso-Domínguez, Eva Barragan, and Pau Montesinos; visualization was performed by Tamara Castaño-Bonilla, Juan Manuel Alonso-Domínguez, Daniel Lainez, Juana Serrano-López, and Pau Montesinos; writing–original draft was performed by Tamara Castaño-Bonilla and Juan Manuel Alonso-Domínguez; writing–review and editing was performed by Tamara Castaño-Bonilla, Eva Barragán, Claudia Sargas, Alejandro Sanz, Lorenzo Algarra, Pilar Herrera-Puente, Raimundo García-Boyero, Manuel Barrios, David Martinez-Cuadron, Rebeca Rodriguez-Veiga, Blanca Boluda, Cristina Gil, Josefina Serrano-Lopez, Joaquín Martínez-López, Maria Jose Sayas-Lloris, Maria Teresa Olave, Rosalia Riaza-Grau, Teresa Bernal-Del Castillo, Maria Jose Larrayoz, Raquel Amigo, Antonio Jimenez, Joaquin Sanchez, Rosa Ayala, Carlos Blas, Daniel Lainez, Juana Serrano, Miguel A. Sanz, Juan M. Alonso-Dominguez, and Pau Montesinos. J.M.A.D. and P.M. contributed equally to this study and should be considered as senior authors.
Supplementary Materials
Supplement Table 1: Kaplan-Meier analysis of overall survival and relapse-free survival in patients treated with GO in the reinduction phase. Supplement Figure 1: clinical outcome stratified according to CD33 rs12459419 polymorphism for patients treated with GO during the reinduction phase (a, b) (a) Overall survival. (b) Relapse-free survival.
References
- 1.Papaemmanuil E., Gerstung M., Bullinger L., et al. Genomic classification and prognosis in acute myeloid leukemia. The New England Journal of Medicine . 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acheampong D. O., Adokoh C. K., Asante D.-B., et al. Immunotherapy for acute myeloid leukemia (AML): a potent alternative therapy. Biomedicine & Pharmacotherapy . 2018;97:225–232. doi: 10.1016/j.biopha.2017.10.100. [DOI] [PubMed] [Google Scholar]
- 3.Godwin C. D., Gale R. P., Walter R. B. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia . 2017;31(9):1855–1868. doi: 10.1038/leu.2017.187. [DOI] [PubMed] [Google Scholar]
- 4.Gottardi M., Sperotto A., Di Rorà A. G. L., et al. Gemtuzumab Ozogamicin in acute myeloid leukemia: past, present and future. Minerva Medica . 2020;111(5):395–410. doi: 10.23736/S0026-4806.20.07019-6. [DOI] [PubMed] [Google Scholar]
- 5.Petersdorf S., Kopecky K., Stuart R. K., et al. Preliminary results of Southwest Oncology Group Study S0106: an international intergroup phase 3 randomized trial comparing the addition of gemtuzumab ozogamicin to standard induction therapy versus standard induction therapy followed by a second randomization to post-consolidation gemtuzumab ozogamicin versus no additional therapy for previously untreated acute myeloid leukemia. Blood . 2009;114(22):790–790. doi: 10.1182/blood.V114.22.790.790. [DOI] [Google Scholar]
- 6.Petersdorf S. H., Kopecky K. J., Slovak M., et al. A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood . 2013;121(24):4854–4860. doi: 10.1182/blood-2013-01-466706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett A. K., Hills R. K., Milligan D., et al. Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: results of the MRC AML15 trial. Journal of Clinical Oncology . 2011;29(4):369–377. doi: 10.1200/JCO.2010.31.4310. [DOI] [PubMed] [Google Scholar]
- 8.Delaunay J., Recher C., Pigneux A., et al. Addition of gemtuzumab ozogamycin to chemotherapy improves event-free survival but not overall survival of AML patients with intermediate cytogenetics not eligible for allogeneic transplantation Results of the GOELAMS AML 2006 IR study. Blood . 2011;118(21):79–79. doi: 10.1182/blood.V118.21.79.79. [DOI] [Google Scholar]
- 9.Castaigne S., Pautas C., Terré C., et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet . 2012;379(9825):1508–1516. doi: 10.1016/S0140-6736(12)60485-1. [DOI] [PubMed] [Google Scholar]
- 10.Hills R. K., Castaigne S., Appelbaum F. R., et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. The Lancet Oncology . 2014;15(9):986–996. doi: 10.1016/S1470-2045(14)70281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert J., Pautas C., Terré C., et al. Gemtuzumab ozogamicin forde novoacute myeloid leukemia: final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica . 2019;104(1):113–119. doi: 10.3324/haematol.2018.188888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamis A. S., Alonzo T. A., Meshinchi S., et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III Children’s Oncology Group Trial AAML0531. Journal of Clinical Oncology . 2014;32(27):3021–3032. doi: 10.1200/JCO.2014.55.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teich K., Krzykalla J., Kapp-Schwoerer S., et al. Cluster of differentiation 33 single nucleotide polymorphism Rs12459419 is a predictive factor in patients with nucleophosmin1 mutated acute myeloid leukemia receiving gemtuzumab ozogamicin. Haematologica . 2021;106(11) doi: 10.3324/haematol.2021.278894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale R. E., Popa T., Wright M., et al. No evidence that CD33 splicing SNP impacts the response to GO in younger adults with AML treated on UK MRC/NCRI trials. Blood . 2018;131(4):468–471. doi: 10.1182/blood-2017-08-802157. [DOI] [PubMed] [Google Scholar]
- 15.Short N. J., Richard-Carpentier G., Kanagal-Shamanna R., et al. Impact of CD33 and ABCB1 single nucleotide polymorphisms in patients with acute myeloid leukemia and advanced myeloid malignancies treated with decitabine plus gemtuzumab ozogamicin. American Journal of Hematology . 2020;95(9):E225–E228. doi: 10.1002/ajh.25854. [DOI] [PubMed] [Google Scholar]
- 16.Cowan A. J., Laszlo G. S., Estey E. H., Walter R. B. Antibody-based therapy of acute myeloid leukemia with gemtuzumab ozogamicin. Frontiers in Bioscience . 2013;18(4):1311–1334. doi: 10.2741/4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gbadamosi M., Meshinchi S., Lamba J. K. Gemtuzumab ozogamicin for treatment of newly diagnosed CD33-positive acute myeloid leukemia. Future Oncology . 2018;14(30):3199–3213. doi: 10.2217/fon-2018-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortland L., Alonzo T. A., Walter R. B., et al. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin–containing chemotherapy. Clinical Cancer Research . 2013;19(6):1620–1627. doi: 10.1158/1078-0432.CCR-12-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laszlo G. S., Beddoe M. E., Godwin C. D., et al. Relationship between CD33 expression, splicing polymorphism, and in vitro cytotoxicity of gemtuzumab ozogamicin and the CD33/CD3 BiTE® AMG 330. 2019;104(2):e59–e62. doi: 10.3324/haematol.2018.202069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castaño-Bonilla T., Alonso-Dominguez J. M., Barragán E., et al. Prognostic significance of FLT3 -ITD length in AML patients treated with intensive regimens. Scientific Reports . 2021;11(1):p. 20745. doi: 10.1038/s41598-021-00050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Döhner H., Estey E., Grimwade D., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood . 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Döhner H., Estey E. H., Amadori S., et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood . 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 23.Lai C., Karp J. E., Hourigan C. S. Precision medicine for acute myeloid leukemia. Expert Review of Hematology . 2016;9(1):1–3. doi: 10.1586/17474086.2016.1107471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inc M. G. Gemtuzumab ozogamicin added to hidac consolidation therapy for by Nurit Horesh. July 2021, https://library.ehaweb.org/eha/2021/eha2021-virtual-congress/325247/nurit.horesh.gemtuzumab.ozogamicin.added.to.hidac.consolidation.therapy.for.html?f=menu%3D6%2Abrowseby%3D8%2Asortby%3D2%2Amedia%3D3%2Ace_id%3D2035%2Alabel%3D21989%2Aot_id%3D25551%2Amarker%3D1286%2Afeatured%3D17286.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table 1: Kaplan-Meier analysis of overall survival and relapse-free survival in patients treated with GO in the reinduction phase. Supplement Figure 1: clinical outcome stratified according to CD33 rs12459419 polymorphism for patients treated with GO during the reinduction phase (a, b) (a) Overall survival. (b) Relapse-free survival.
Data Availability Statement
Data are available upon request from the authors.


