Abstract
Methods
We compared differentially expressed genes (DGEs) in HBV-positive and -negative tumor samples and conducted a Spearman correlation study between the DGEs and HBV titers within The Cancer Genome Atlas (TCGA). Moreover, we validated the results of our in-house samples.
Results
In this study, we discovered a series of genes that correlated statistically with HBV infection based on the TCGA database. These genes were related to increased inflammation and some oncogenic signaling pathways via Gene Set Enrichment Analysis (GSEA). PDZK1 is an ideal gene, which mostly relates positively to HBV infection; moreover, it is overexpressed in human HCC, especially in those HBV-infected HCCs. After analyzing the TCGA data and performing a verification study using our own samples, PDZK1 expression was investigated to be significantly associated with PI3K-Akt signaling and fatty acid metabolism. Further, single-sample GSEA analysis of tumor immune cell infiltration gene sets revealed that high PDZK1expression in HCC tissues was significantly associated with increased tumor-associated macrophages (TAMs) and regulatory T cells(Tregs).
Conclusions
PDZK1 is an HBV infection-related gene, which plays oncogenic roles, possibly due to enhancing PI3K-Akt, fatty acid usage in tumor cells and TAMs, and Treg-induced immunosuppression.
1. Introduction
Chronic hepatitis B virus (HBV) infection is the leading cause of hepatocellular carcinoma (HCC) globally [1]. Chronic carriers have a more than 100-fold increased relative risk of tumor development. Several mechanisms have been proposed to induce HCC using HBV [2]. HBV causes 80% of liver cancer cases and is the second most common carcinogen after smoking. Babies born to e-positive mothers have approximately a 90% risk of becoming persistent carriers after perinatal infection, and preschoolers have a 30% risk [2, 3]. Only 5% to 10% of adults become persistent carriers after infection. In individuals persistently infected with HBV, 10%-30% will develop cirrhosis and HCC. In persistently infected individuals, these highly different results in clearance rates and disease outcomes cannot be fully explained by differences in immune, viral, or environmental factors. Therefore, differences in host genetic factors may influence the natural history of hepatitis B.
The genetic factors that affect the outcome of HBV infection include human leukocyte antigen (HLA), cytokines, chemokines, mannose-binding proteins, vitamin D receptors, and Toll-like receptors (TLRs) [4–8], such as HLA-DPA1 rs3077 and HLA-DPB1 single nucleotide polymorphisms (SNPs) [9] and TNF-alpha promoter SNP and/or haplotypes [10]. However, it is difficult to verify other genes in different populations. Moreover, a single gene or SNP cannot fully explain disease susceptibility because human traits are inherited from multiple genes. The main advantage of genetic association analysis is that it uses distortions in the genetic frequency distribution of comparative populations to detect disease-related genes. However, this advantage has potential drawbacks that can lead to false-positive associations and the deletion of important loci. False-positive associations may result from sampling errors in substructured study populations, variations associated with multiple statistical tests, and linkage imbalances between labeled SNPs and actual disease-related SNPs. Since almost all investigations into HBV-associated HCC have been based on epidemiology and the detailed molecular mechanisms remain unexplored, we can predict whether HBV infection is present in liver cancer samples from the TCGA database based on a report published in Nature Communications [11].
In this study, we compared differentially expressed genes (DGEs) in HBV-positive and -negative tumor samples according to previous reports and conducted a Spearman correlation study between the DGEs and HBV titers within TCGA. We found that PDZK1 may be an ideal gene that is associated with HBV infection and is significantly differentially expressed in cancer and noncancer tissues. Below, we discuss the potential effects of PDZK1 on gene signaling pathways and the tumor microenvironment.
2. Materials and Methods
2.1. Study Subjects
The hospital-based case-control study consisted of 80 study subjects newly diagnosed with HCC, with 50 cases being HBV positive and 30 cases being HBV negative. All subjects were recruited from Sir Run Run Hospital, Nanjing Medical University, and Northern Jiangsu People's Hospital between July 2016 and June 2020. Study subjects with other hematological disorders, previous history of cancer, radiotherapy, immunosuppression/antiviral medication, and chemotherapy were excluded. The cancer-free control subjects from the same geographic area showed no genetic relationship with the cases. Study subjects were classified according to the World Health Organization classification. The Institutional Review Board of Sir Run Run Hospital, Nanjing Medical University, and Northern Jiangsu People's Hospital approved this study, and every patient gave written informed consent.
2.2. Bioinformatics Analysis
Liver hepatocellular carcinoma (LIHC) liver cancer data from the TCGA database were used to extract samples with known HBV infection status according to previous reports. The difference between the two groups of transcripts was calculated using the R software package Limma (v.3.40.6), and the absolute value of fold change (Fc) abs(FC) > 0.5. Adjusted P < 0.05 was used to identify DGEs. clusterProfiler (v.3.12.0) and PHEATMAP (v.1.0.12) were used for cluster analysis and heat map generation Gene Ontology (GO), respectively, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using Cluego25 (v.2.5.5). Data from the TCGA-liver hepatocellular carcinoma (LIHC) dataset were used to extract samples with known HBV infection status according to previous reports. The critical value was a false detection rate < 0.05.
2.3. The Prognosis of Dysregulated Genes Was Analyzed Using Gene Expression Profiling Interactive Analysis (GEPIA) with the Kaplan–Meier Plotter
GEPIA (http://gepia.cancer-pku.cn/) [12] is a new web-based tool containing sequencing expression data from 9736 tumor samples of 33 cancer types and 8587 normal samples. The database includes various analysis modules, such as differential gene expression, survival and prognosis, correlation, and dimensionality reduction analyses.
2.3.1. Real-Time Polymerase Chain Reaction (PCR)
Total RNA in HCC tissues was isolated using the TRIzol reagent. The expression of genes of interest was detected using the cyber-green-based real-time PCR. The primers for genes used in the study are listed in Table 1.
Table 1.
Primer sequence for genes of interest.
| Gene symbol | Sequence | Amplificon (bp) |
|---|---|---|
| PDZK1 | TTCCTGCGAATTGAGAAGGAC | 164 |
| CCCCGAATCGCATTTAAGTGAA | ||
| MET | AGCAATGGGGAGTGTAAAGAGG | 191 |
| CCCAGTCTTGTACTCAGCAAC | ||
| IL6R | CCCCTCAGCAATGTTGTTTGT | 171 |
| CTCCGGGACTGCTAACTGG | ||
| FGFR3 | CCCAAATGGGAGCTGTCTCG | 109 |
| CCCGGTCCTTGTCAATGCC | ||
| BCL2L1 | GAGCTGGTGGTTGACTTTCTC | 119 |
| TCCATCTCCGATTCAGTCCCT | ||
| RAF1 | GGGAGCTTGGAAGACGATCAG | 165 |
| ACACGGATAGTGTTGCTTGTC | ||
| ITGA6 | ATGCACGCGGATCGAGTTT | 160 |
| TTCCTGCTTCGTATTAACATGCT | ||
| ALDH3A2 | AAACCAGTTAAGAAGAACGTGCT | 103 |
| CGAAGGGGTAATTCCAAGCTC | ||
| ADH5 | ATGGCGAACGAGGTTATCAAG | 202 |
| CATGTCCCAAGATCACTGGAAAA | ||
| ADH1B | CCCGGAGAGCAACTACTGC | 224 |
| AACCAGTCGAGAATCCACAGC | ||
| ALDH1B1 | AGGGGGCTGTTTATGGTGG | 167 |
| GGTACGGATACTTTTCCCAGAGT | ||
| HACD2 | GCTGCGAAGTGGAAACCATC | 135 |
| CCTCCTTCTGCACACATTTGAA | ||
| FASN | AAGGACCTGTCTAGGTTTGATGC | 106 |
| TGGCTTCATAGGTGACTTCCA | ||
| HSD17B8 | CCTGGTGTCCAATGGTTGTC | 60 |
| ACCTTTCCTACGATGCTACTGAT | ||
| HACD3 | TGGAGCCCGTGAAAAAGAGC | 75 |
| TCTCCTTCATCTTAGAGGCCAC |
2.3.2. Immunohistochemistry Staining
Sections were stained according to the previous publication. The sections were incubated with primary mouse antihuman Ab for CD68(ab213363), CD163(ab182422), CD4(ab183685), ITGA6 (ab181551), RAF1 (ab181115), FGFR3 (ab133644), ADH5(ab177932), FASN (ab128870), and Foxp3 (ab215206). The sections were stained with 3,3′-diaminobenzidine according to the manufacturer's protocols and then mounted and photographed using a digitalized microscope camera (Nikon, Japan).
2.4. Statistical Analysis
Data are presented as the mean ± SEM. The χ2 test and Student's t-test analysis of variance were used to evaluate differences in demographic and clinical characteristics. Kaplan-Meier survival curves were generated, and the log-rank test was used to investigate the significance of various variables for survival. All expression experiments conducted in vitro were repeated at least thrice with triplicate samples. Pearson's correlation analysis was used to analyze the relationship of associated factors. Statistical analysis was performed using STATA 9.2 and presented with the GraphPad Prism (CA, USA). In all cases, P < 0.05 was considered significant.
3. Results
3.1. HBV Infection-Related Genes Are Associated with Increased Inflammation and Several Oncogenic Signaling Pathways
Based on previous reports [10], we obtained 11 tumor and 6 nontumor HBV-infected samples as well as 22 tumor and 13 nontumor HBV noninfected samples in the TCGA-LIHC database. First, we compared the differential gene expression between HBV-infected and noninfected samples in normal or tumor tissues and obtained DEGs using the criterion with an absolute value of log Fc > 0.5 and adjusted P value of <0.05 (Figures 1(a) and 1(b)). After the DEGs were enriched, GSEA was performed using a typical KEGG pathway subset(CP). The results suggest that HBV infection is associated with an increased inflammatory response due to enrichment of signaling pathways of “leukocyte transendothelial migration” and “cytokine-cytokine receptor interactions” (Figures 1(c) and 1(d)). However, DEGs obtained from tumor tissues were associated with carcinogenic signaling pathways such as focal adhesion, PI3K-Akt, and Ras signaling pathways (Figure 1(c)). These results agreed with previous reports of HBV-induced enhancement of liver inflammation. However, our study also provides new insights that HBV infection in tumor tissues has more oncogenic characteristics than in HBV-negative tumors, which may be mediated by pathways, such as PI3K and Ras signaling pathways.
Figure 1.
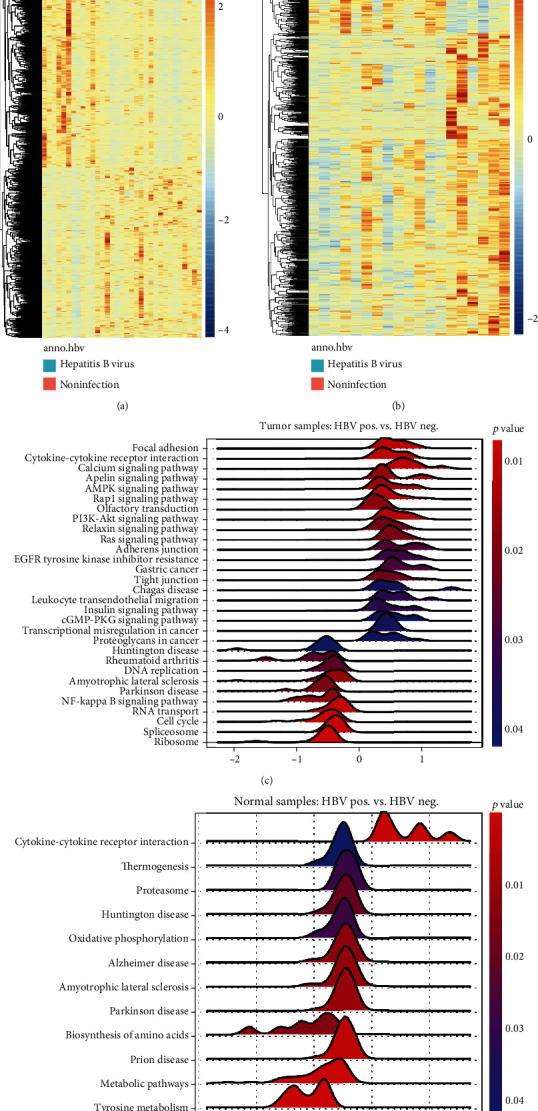
HBV infection-related genes were associated with increased inflammation and several oncogenic signaling pathways. (a, b) Heat map of all significantly differently expression genes (DEGs) (n = 964, HBV Pos. vs. HBV Neg, in tumor tissues, and n = 1098, HBV Pos. vs. HBV Neg. in normal tissues). (c, d) GSEA results obtained by using DEGs by comparing between HBV Pos. and HBV Neg. Samples in tumor (c) and normal (d) tissues.
3.2. PDZK1 Is an HBV-Infection-Related Oncogene in Human HCC
To find the ideal candidate gene for further study, we found commonly up- or downregulated DEGs between tumor and normal samples using the online tool, http://Venny2.1. As presented in the Venny map (Figure 2(a)), we found 27 upregulated and 82 downregulated common genes. Next, we conducted a multiple correlation study on HBV reads provided by previous reports [11] and the expression of each common gene. We found that 63 genes were significantly linearly correlated with HBV reads, with 49 correlating positively and 14 negatively (Figure 2(b)). Among these genes, we found that PDZK1 is an ideal oncogene regulated by HBV. We found that PDZK1 showed a good linear correlation with HBV reads (R = 0.59, P = 0.0013) (Figure 2(c)), and the data in TCGA-LIHC suggested that this gene was significantly overexpressed in HCC tissues (Figure 2(d)).
Figure 2.
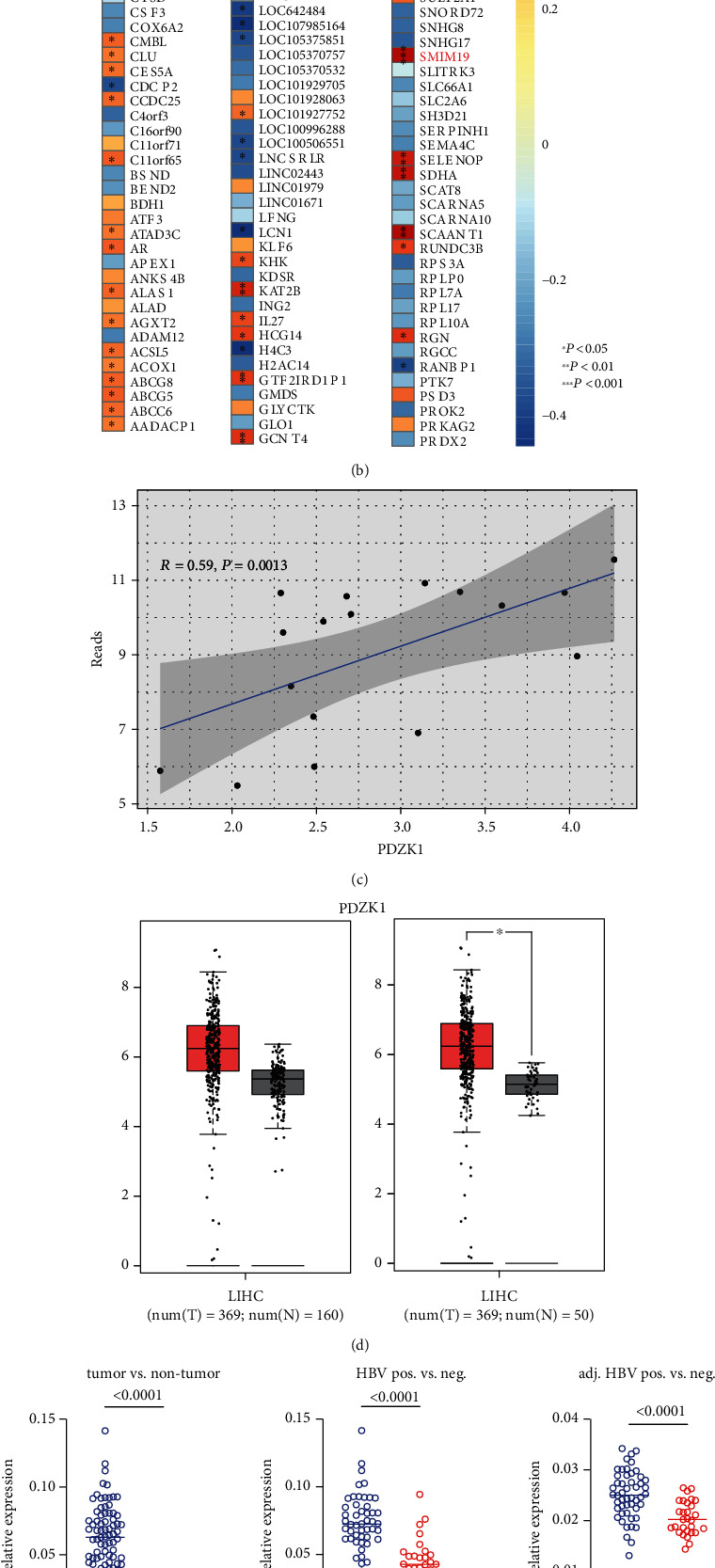
PDZK1 is one of the HBV-infection-related oncogenes in human HCC. (a) Venny map indicating the common up- or downregulated genes both in cancer and normal samples from TCGA-LIHC. (b) Heat map reflating either R or P value obtained from multiple correlation study between HBV reads and gene expression level of common genes. (c) Scatter plot reflecting the correlation between HBV reads and PDZK1 expression in the TCGA samples. (d) The comparison of PDZK1 expression between normal and tumor samples from TCGA-LIHC or TCGA-LIHC and GTEx database. (e) Real-time PCR detection PDZK1 transcription in the sample sets indicated in the figure. (f) Scatter plot reflecting the correlation between HBV reads and PDZK1 expression from clinical samples.
To verify these TCGA results, we collected HCC samples with and without HBV infection (n = 50 and n = 30). First, we confirmed the high expression of the PDZK1 gene in human hepatocellular carcinoma (Figure 2(e)). Additionally, we found that compared with HBV-negative tissues, PDZK1 expression increased significantly in HBV-positive tissues, both in tumors and in adjacent tissues (Figure 2(e)). Finally, we studied the linear correlation between PDZK1 expression and HBV titer, and a good correlation was observed (R = 0.9, P < 0.0001) (Figure 2(f)).
3.3. High PDZK1 Expression in Human HCC Is Associated with Inflammation, Oncogenic Characteristics, and Fatty Acid Metabolism
By comparing the 40 samples with the highest and lowest PDZK1 gene expression in TCGA-LIHC, 4453 DEGs (adjusted P < 0.05) were obtained, among which 2049 genes were downregulated and 2024 genes were upregulated (Figure 3(a)). Furthermore, DEGs were used for GO and KEGG enrichment. For GO (BP) enrichment, we found that PDZK1 is associated with increased inflammatory response, reflected in terms such as “neutrophil activation” and “T-cell activation” (Figure 3(b)). For KEGG enrichment, we found that the PI3K-Akt signaling pathway ranked first among all signaling pathways (Figure 3(c)). Next, we performed GSEA analysis on fold change values of all coding genes (25,717 genes), and we found that PI3K-Akt was also the first significant signaling pathway. The second and third signaling pathways of “fatty acid degradation” and “fatty acid metabolism” were found (Figure 3(d)). Normalized enrichment scores (NES) of the three genomes were 1.765, 1.731, and 1.725 (Figure 3(e)). Some core genes belonging to these three gene sets are listed in Figure 3(f).
Figure 3.
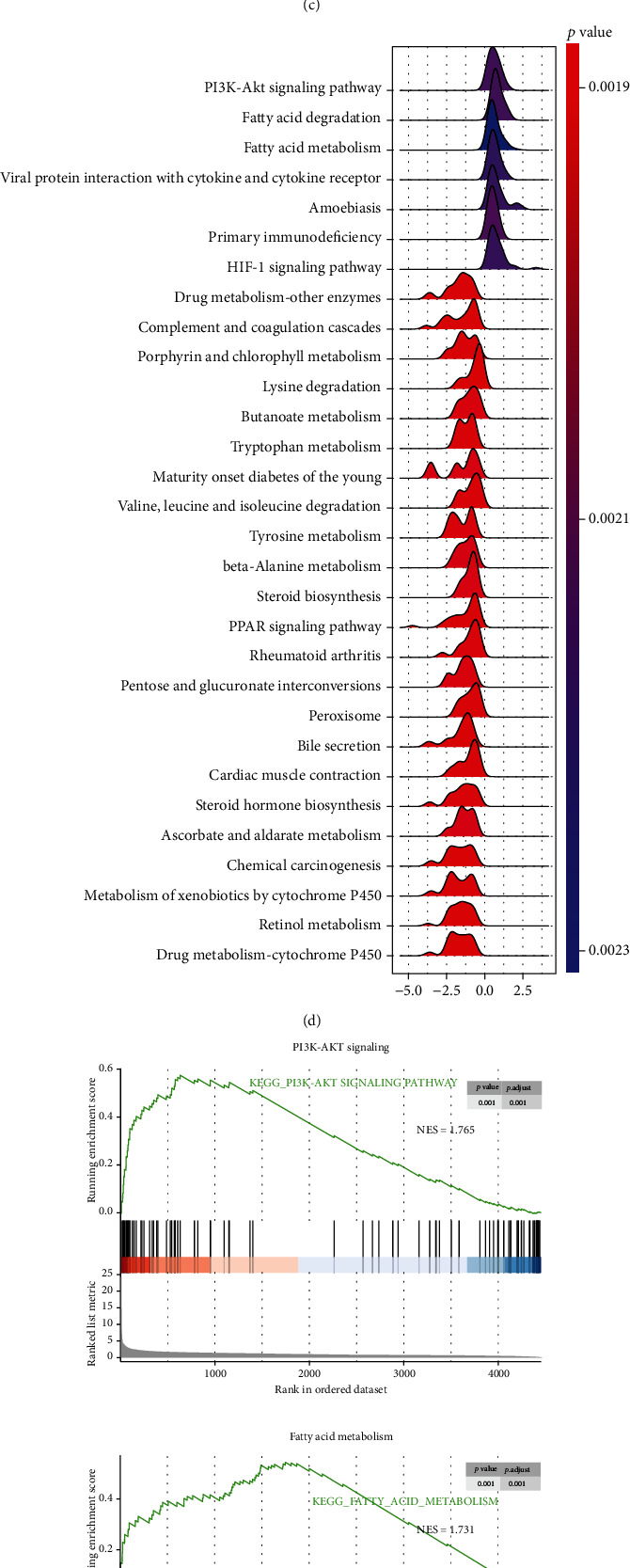
PDZK1 has oncogenic roles by enhancing PI3K/AKT and fatty acid metabolism. (a) Heat map of all DEGs obtained by comparing PDZK1 top 20 high and top 20 low samples. (b) GO enrichment for biological process (BP) based on the DEGs. (c) KEGG enrichment based on the DEGs. (d) GSEA analysis for gene expression fold change between PDZK1 top 20 high and top 20 low samples. (e) GSEA plot for PI3K-AKT, fatty acid metabolism, and fatty acid degradation pathways. (f) Heat map for key genes representing PI3K-AKT, fatty acid metabolism, and fatty acid degradation signaling.
Additionally, we used our clinical samples (PDZK1High, n = 25, and PDZK1Low, n = 25) to verify the relationship between PDZK1 expression and multiple signaling pathways. First, six genes, MET, IL6R, FGFR3, BCL2L1, RAF1, and ITGA6, were selected to be associated with the PI3K-Akt signaling pathway. We found that these six genes were significantly overexpressed in HCC samples with high PDZK1 expression compared with the HCC samples with low PDZK1 expression (Figure 4(a)). Simultaneously, the protein expressions of ITGA6, RAF1, and FGFR3 were detected using immunohistochemical staining, and we found that these three proteins were also overexpressed in PDZK1 high-HCC samples (Figure 4(b)). Next, real-time PCR technology was used to analyze some components of genes belonging to “fatty acid degradation” and “fatty acid metabolism” signal pathways. Fatty acid degradation-related genes included ALDH3A2, ADH5, ADH1B, and ALDH1B1 and fatty acid metabolism-related genes HACD2, FASN, HSD17B8, and HACD3. These genes were relatively high in HCC samples with high PDZK1 expression (Figures 4(c) and 4(d)). Finally, we used immunohistochemistry to detect the expression of ADH5 and FASN proteins in HCC tissues, suggesting that the expression of these two key enzymes in the metabolic process was high in HCC samples with high PDZK1 expression (Figure 4(e)).
Figure 4.
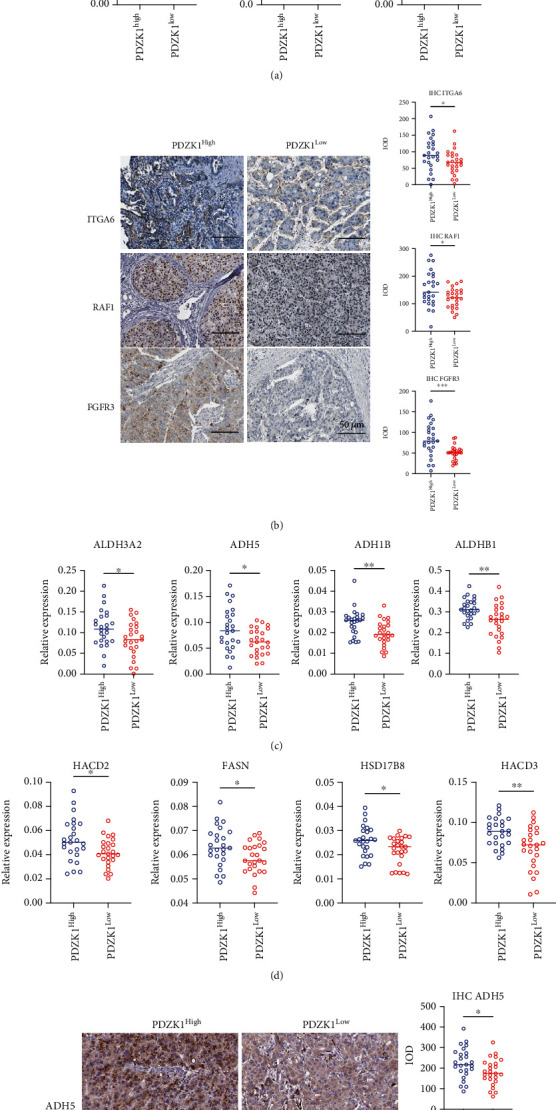
PDZK1 was correlated to increased PI3K/AKT and fatty acid metabolism. (a) Real-time PCR detection PI3K-AKT signaling-related genes indicated in the figure in PDZK1 high and low expression samples. (b) IHC staining of ITGA6, RAF1, and FGFR3 in PDZK1 high and low expression samples. (c, d) Real-time PCR detection fatty acid metabolism and degradation-related genes indicated in the figure in PDZK1 high and low expression samples. (e) IHC staining of ADH5 and FASN in PDZK1 high and low expression samples. Data are presented as mean ± SEM and were analyzed using Student's t-test (∗P < 0.05, ∗∗P < 0.01; ns: not significant, P > 0.05).
3.4. Expression of PDZK1 in HCC Tissues Was Associated with Treg and TAM-Induced Immunosuppression
To study the relationship between the PDZK1 gene and the tumor microenvironment, we used single-sample GSEA to analyze the possible immune cell infiltration in the samples from the TCGA-LIHC database. We found that tissues with high PDZK1expression had more abundant infiltration of macrophages and Treg cells (Figure 5(a)). Then, this result was confirmed using GSEA. The results suggested that the DEGs between PDZK1 high/low comparison were enriched in the genes of macrophages and regulatory T cells (Figure 5(b)). Then, we validated the bioinformatics analysis by staining macrophages and TAMs with IHC staining for CD68 and CD163, and we found a high infiltration of macrophages, especially TAMs, in PDZK1-high expression samples (Figure 5(c)). Simultaneously, we found that the proportion of Treg cells in HCC tissues with high PDZK1 expression was also higher (Figure 5(c)).
Figure 5.
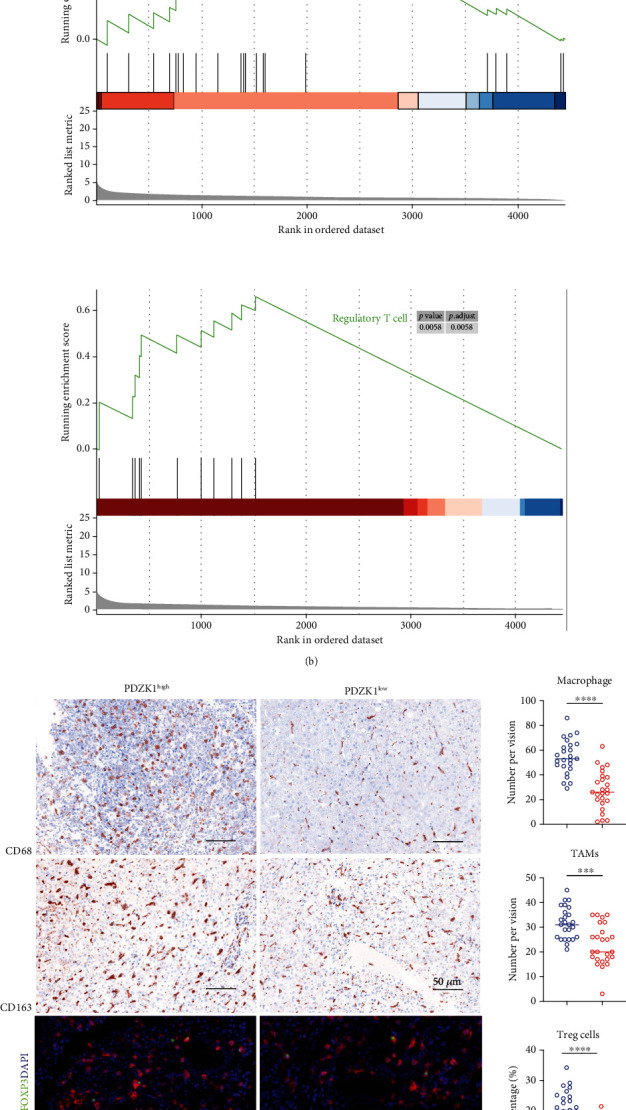
Expression of PDZK1 in HCC tissues was associated with Treg, and tumor-associated macrophages (TAMs) induced immunosuppression. (a) ssGSEA analysis for PDZK1 top 20 high and low expression TCGA samples by using previously reported gene sets. (b) GSEA analysis for PDZK1 top 20 high and low expression TCGA samples. (c) IHC and multiple color IHC staining for CD68, CD163, and CD4/FOXP3 in PDZK1 high and low expression samples. Data are presented as mean ± SEM and were analyzed using Student's t-test (∗P < 0.05, ∗∗P < 0.01; ns: not significant, P > 0.05).
4. Discussion
Our study found that PDZK1 is a gene highly linearly correlated with HBV infection and may promote the occurrence and development of liver cancer by promoting the PI3K-Akt pathway and fatty acid metabolism. PDZK1 is an adaptor protein containing four PDZ domains, which binds to the carboxy-terminal cytoplasmic tail of SR-B1 through PDZ domain 1 (PDZ1) and PDZ3, whereas PDZ4 mediates interaction with membrane lipids [13, 14]. It tissue specifically protects SR-B1 from degradation. In the absence of PDZK1, SR-B1 protein levels were significantly reduced in the liver (95%) and moderately decreased in the intestinal tissue (50%) but were unaffected in steroids, endothelial cells, or macrophages [14, 15]. SR-B1 and PDZK1 also play an important role in HDL-mediated signal transduction in vascular cells [16, 17]. Mice deficient in PDZK1 showed an increase in HDL-C, consistent with the loss of SR-B1 protein in the liver [18, 19]. PDZK1/ApoE DKO mice developed greater diet-induced aortic atherosclerosis compared with control ApoE knockout mice when fed a high-fat, Western-induced atherosclerotic diet [20, 21]. From the reports above, we can conclude that PDZK1is related positively to fatty acid metabolism in different types of normal cells. However, we first reported that PDZK1is related significantly to fatty acid metabolism in human liver cancer.
In this study, we found that increased PDZK1 in human liver cancer, especially HBV-positive cancers, was related to enhanced P3IK-Akt signaling. However, we found that the reports concerning the relationship between PDZK1 and PI3K-Akt signaling in different tissues are controversial. In studies of intestinal inflammation, soluble uric acid increased the expression of PDZK1 and ABCG2 [22]. Stimulation of soluble uric acid also promoted the transport of ABCG2 from the intracellular compartment to the plasma membrane and increased its transport activity. TLR4-NLRP3 inflammasome inhibitors or PI3K-Akt signaling inhibitors can partially reduce the upregulation of PDZK1 and ABCG2 by soluble uric acid. Additionally, PDZK1downregulation significantly inhibited ABCG2 expression and transport activity. However, it was unaffected by the activation of soluble uric acid, suggesting that PDZK1 plays a key role in regulatingABCG2. These results suggest that uric acid promotes excretion by activating the TLR4-NLRP3 inflammasome and PI3K-Akt signaling pathway to upregulate the expression of PDZK1 and ABCG2 in intestinal cells [22]. However, other studies have shown that PDZK1 can inhibit PI3K-Akt activation. In this study, PTEN was identified as a new PDZK1 binding protein by binding to the PDZ domain of PDZK1 through its carboxyl terminal. Through direct interaction with PTEN, PDZK1 inhibited PTEN phosphorylation at S380/T382/T383, further enhancing the ability of PTEN to inhibit PI3K-Akt activation. In vivo and in vitro studies have shown that PDZK1 inhibits PI3K-Akt activation by inhibiting PTEN phosphorylation, thereby inhibiting the proliferation of gastric cancer cells. Frequent downregulation of PDZK1 expression in gastric cancer specimens is associated with the disease progression and poor prognosis of study subjects with gastric cancer [23]. In clinical specimens, PDZK1downregulation is associated with PTEN inactivation, Akt signaling pathway, and cell proliferation activation. Our studies in liver cancer have shown that high expression of PDZK1 in liver cancer correlates highly with PI3K-Akt pathway activation. This difference in the role of PDZK1 in different tumors may be related to tissue differences.
Thus far, there are no reports concerning the effect of PDZK1 on the tumor microenvironment, and we reported for the first time that high expression of PDZK1 was associated with immunosuppression caused by increased TAMs and Treg cells.
In summary, we found that the PDZK1 gene correlated highly with HBV infection, and PDZK1 might play oncogenic roles by relating positively with PI3K-Akt activation, fatty acid metabolism, and immunosuppression in human HCC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant Number 82171737 to W.Z.), State Key Program of the National Natural Science Foundation of China (Grant Number 81930086 to B.S.), Nanjing Medical Science and Technology Development Project (Grant Number YKK19071 to W.Z.), and Clinical Trials from the affiliated Nanjing Drum Tower Hospital, Medical School of Nanjing University (Grant Number 2021-LCYJ-PY-28 to W.Z.). B.S. is a Yangtze River Scholar of Distinguished Professor.
Contributor Information
Wenjie Zhang, Email: drzhangwj@163.com.
Beicheng Sun, Email: sunbc@nju.edu.cn.
Data Availability
The data were obtained from the TCGA database.
Conflicts of Interest
The authors declared no conflict of interest.
Authors' Contributions
Xin Chen and Xiaodong Wang contributed equally to this work.
References
- 1.Duffell E., Cortez-Pinto H., Simonova M., et al. Estimating the attributable fraction of cirrhosis and hepatocellular carcinoma due to hepatitis B and C. Journal of Viral Hepatitis . 2021;28(8):1177–1189. doi: 10.1111/jvh.13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lapinski T. W., Tarasik A., Januszkiewicz M., Flisiak R. Clinical aspects and treatment of hepatocellular carcinoma in North-Eastern Poland. Clinical and Experimental Hepatology . 2021;7(1):79–84. doi: 10.5114/ceh.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J., Althoff K. N., Jing Y., et al. Trends in hepatocellular carcinoma incidence and risk among persons with HIV in the US and Canada, 1996-2015. JAMA Network Open . 2021;4(2) doi: 10.1001/jamanetworkopen.2020.37512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Huang C. J., Huang Y. H., et al. HLA zygosity increases risk of hepatitis B virus-associated hepatocellular carcinoma. The Journal of Infectious Diseases . 2021;224(10):1796–1805. doi: 10.1093/infdis/jiab207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manrai M., George E., Kapoor R. Profile of hepatobiliary dysfunction in hematopoietic stem cell transplant recipients - an Indian perspective. Journal of Clinical and Experimental Hepatology . 2021;11(1):14–20. doi: 10.1016/j.jceh.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langton M., Pandelia M. E. Hepatitis B virus Oncoprotein HBx IsNotan ATPase. ACS Omega . 2020;5(27):16772–16778. doi: 10.1021/acsomega.0c01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albas S., Koc E. M., Nemli S. A., et al. Vitamin D levels and vitamin D receptor (VDR) gene polymorphisms in inactive hepatitis B virus carriers. Journal of College of Physicians and Surgeons Pakistan . 2021;30(4):393–398. doi: 10.29271/jcpsp.2021.04.393. [DOI] [PubMed] [Google Scholar]
- 8.Arfianti A., Sumpena A. S., Djojosugito F. A., Sari D. K., Paulina A. J. Genotype distribution of vitamin D receptor polymorphisms among Indonesian patients with chronic hepatitis B. Reports of Biochemistry & Molecular Biology . 2021;9(4):463–469. doi: 10.52547/rbmb.9.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y., Xia T., Xia X., Zhang A. M. Genetic polymorphisms of the HLA-DP and HLA-DQ genes could influence hepatitis B virus infection in Yunnan population. Immunological Investigations . 2021;50(1):47–57. doi: 10.1080/08820139.2020.1733010. [DOI] [PubMed] [Google Scholar]
- 10.Wei Y., Zhao Z., Wang Z., Zhang K., Tang Z., Tao C. Relationships between IL-1 β, TNF-α genetic polymorphisms and HBV infection: a meta-analytical study. Gene . 2021;791, article 145617 doi: 10.1016/j.gene.2021.145617. [DOI] [PubMed] [Google Scholar]
- 11.Tang K. W., Alaei-Mahabadi B., Samuelsson T., Lindh M., Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nature Communications . 2013;4(1):p. 2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research . 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira C., Meyer R., Meyer ZuSchwabedissen H. E. The nuclear receptors PXR and LXR are regulators of the scaffold protein PDZK1. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms . 2019;1862(4):447–456. doi: 10.1016/j.bbagrm.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Trigatti B. L. SR-B1 and PDZK1: partners in HDL regulation. Current Opinion in Lipidology . 2017;28(2):201–208. doi: 10.1097/MOL.0000000000000396. [DOI] [PubMed] [Google Scholar]
- 15.Trigattiand B. L., Fuller M. HDL signaling and protection against coronary artery atherosclerosis in mice. Journal of Biomedical Research . 2016;30(2):94–100. doi: 10.7555/JBR.30.20150079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung K. Y., Wang C., Nyegaard S., Heit B., Fairn G. D., Lee W. L. SR-BI mediated transcytosis of HDL in brain microvascular endothelial cells is independent of caveolin, clathrin, and PDZK1. Frontiers in Physiology . 2017;8:p. 841. doi: 10.3389/fphys.2017.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mineo C., Shaul P. W. Regulation of signal transduction by HDL1. Journal of Lipid Research . 2013;54(9):2315–2324. doi: 10.1194/jlr.R039479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pal R., Ke Q., Pihan G. A., et al. Carboxy-terminal deletion of the HDL receptor reduces receptor levels in liver and steroidogenic tissues, induces hypercholesterolemia, and causes fatal heart disease. American Journal of Physiology. Heart and Circulatory Physiology . 2016;311(6):H1392–H1408. doi: 10.1152/ajpheart.00463.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukamoto K., Wales T. E., Daniels K., et al. Noncanonical role of the PDZ4 domain of the adaptor protein PDZK1 in the regulation of the hepatic high density lipoprotein receptor scavenger receptor class B, type I (SR-BI) The Journal of Biological Chemistry . 2013;288(27):19845–19860. doi: 10.1074/jbc.M113.460170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yesilaltay A., Daniels K., Pal R., Krieger M., Kocher O. Loss of PDZK1 causes coronary artery occlusion and myocardial infarction in Paigen diet-fed apolipoprotein E deficient mice. PLoSOne . 2009;4(12):p. e8103. doi: 10.1371/journal.pone.0008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocher O., Yesilaltay A., Shen C. H., et al. Influence of PDZK1 on lipoprotein metabolism and atherosclerosis. Biochimica et Biophysica Acta . 2008;1782(5):310–316. doi: 10.1016/j.bbadis.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M., Lu X., Lu C., et al. Soluble uric acid increases PDZK1 and ABCG2 expression in human intestinal cell lines via the TLR4-NLRP3 inflammasome and PI3K/Akt signaling pathway. Arthritis Research & Therapy . 2018;20(1) doi: 10.1186/s13075-018-1512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao C., Tao T., Yang L., et al. Loss of PDZK1 expression activates PI3K/AKT signaling via PTEN phosphorylation in gastric cancer. Cancer Letters . 2019;453:107–121. doi: 10.1016/j.canlet.2019.03.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data were obtained from the TCGA database.


