Abstract
Polymorphisms have been identified to predispose to primary gouty arthritis (GA) and hyperuricemia (HUA). Here, we accessed the five polymorphisms of rs10754558, rs35829419, rs3738448, rs3806268, and rs7525979 in NLRP3 on GA and HUA susceptibility. We collected 1198 samples (314 GA, 377 HUA, and 507 controls) for this case-control study. Our data detected that the rs3806268 (GA vs. AA: OR = 0.65, p = 0.012) was significantly associated with the susceptibility to GA. The rs3738448 (TT vs. GG: OR = 2.05, p = 0.024) and rs7525979 (TT vs. CC: OR = 1.96, p = 0.037) were significantly associated with the susceptibility to HUA. The rs3806268 AG genotype presented decreased risk of GA among the hypertension (OR = 0.54, p = 0.0093), smoking (OR = 0.59, p = 0.018), and no obesity (OR = 0.60, p = 0.0097) subjects compared to the GG genotype group. The rs3738448 TT genotype demonstrated increased risk of HUA among the hypertension (OR = 4.10, p = 0.0056) and no drinking population (OR = 3.56, p = 0.016) compared to the GG genotype group. The rs7525979 TT genotype demonstrated increased risk of HUA among the hypertension (OR = 4.01, p = 0.0064) and no drinking population (OR = 3.24, p = 0.034) compared to the CC genotype group. Furthermore, a significant haplotype effect of rs10754558/C-rs35829419/C-rs3738448/G-rs3806268/A-rs7525979/C was found (OR = 1.60, p = 0.0046) compared with GCGAC haplotype. Bioinformatics analyses indicated that rs3738448, rs3806268, and rs7525979 might influence the gene regulation, while the T-allele of rs3738448 increased the stability of NLRP3-mRNA. Collectively, our case-control study confirms NLRP3 polymorphisms might participate in regulating immune and inflammation responses in GA and HUA.
1. Introduction
Gouty arthritis (GA) is a recurrent inflammatory disease caused by abnormal purine metabolism and/or excessive production of uric acid (uric acid (UA)) and/or decreased excretion of UA [1]. It is characterized by a continuous increase in the level of serum uric acid (serum uric acid (SUA)), resulting in the precipitation and deposition of urate (monosodium urate (MSU)) crystals in the synovium, cartilage, or other tissues of joints [2]. GA often affects joints and cartilage and can also endanger the kidney, cardiovascular system, and endocrine system [3]. At present, the prevalence rate of GA in China is between 1% and 3% and has a trend of increasing year by year [4]. The pathogenesis of GA is affected by both genetic and environmental factors, but the specific mechanism is not completely clear.
Hyperuricemia (HUA) is caused by the continuous increase of SUA level due to purine metabolism disorder and/or UA metabolism disorder in the human body [5]. Due to lack of obvious clinical symptoms, HUA patients are often ignored. In fact, HUA not only is the direct cause of GA but also is closely related to cardiovascular disease, chronic kidney disease, type 2 diabetes mellitus (T2DM), hypertension, obesity, hyperlipidemia, and so on [6]. At present, the prevalence of HUA in China is as high as 8.4% -13.3% [7]. Therefore, the prevention of HUA is imminent. However, the pathogenesis of HUA is complicated, and there are genetic, environmental, ethnic, and age differences.
NLR family pyrin domain containing 3 (NLRP3) encodes a pyrin-like protein containing a leucine-rich repeat domain and a nucleotide-binding domain [8]. This protein is located predominantly in peripheral blood leukocytes [9]. NLRP3 inflammasome mediates inflammatory process through proinflammatory cytokines in response to invading pathogens [10]. Recently, the polymorphisms of NLRP3 were reported to be involved in the genetic susceptibility to GA in a Chinese Han population [11]. However, another study showed that there is no association between NLRP3 polymorphisms and GA disease in Polynesia [12], suggesting that genetic risk factors for GA may differ between different populations. Therefore, independent population studies aimed at replicating these findings will help define the role of these SNPs in the development of GA. In addition, there is no research on the relationship between NLRP3 polymorphisms and HUA disease.
Therefore, in this study, we focus on the five polymorphisms of rs10754558, rs35829419, rs3738448, rs3806268, and rs7525979 in NLRP3 and assess the relationships between these gene polymorphisms and GA/HUA risks in the Chinese Xingjiang region population.
2. Materials and Methods
2.1. Ethics Approval of the Study Protocol
The local Ethics Committee of Xinjiang Medical University approved the protocol of this research (approval number: 20150225-127), and it was conducted according to the standards of the Declaration of Helsinki. Written informed consent was obtained from all subjects. Clinical data and blood DNA of all subjects were collected and for further analyses.
2.2. Study Population
Participants lived in the Xinjiang Uygur Autonomous Region of China. We recruited 691 cases (314 GA and 377 HUA subjects) from Affiliated Hospital of Xinjiang Medical University between January 2017 and January 2019, and the control group (507 controls) came from the same hospital in the same period. (1) The inclusion criteria of the GA group were as follows: diagnosed in accordance with the standards set of 2015 Gout Classification Criteria [13]. The exclusion criteria of the GA group were as follows: patients with secondary GA, such as GA patients secondary to hypertension, TDM, cardiovascular disease, nephropathy, and other diseases. (2) The inclusion criteria of the HUA group were as follows: on the same day, two fasting tests of SUA in men with normal purine diet were more than 420 μmol/L. The exclusion criteria of the HUA group were as follows: patients with GA, liver and kidney diseases, hyperthyroidism, inflammatory diseases, and recent use of drugs to reduce or promote UA metabolism. (3) The inclusion criteria of the control group were as follows: patients without GA, coronary heart disease, liver disease, HUA, and renal insufficiency and who have not recently taken drugs to reduce or promote UA metabolism.
2.3. Clinical Characteristics of the Study Participants
All subjects completed the standard test registration form and disclosed the following data: (1) general information—age and body mass index (BMI); (2) special test—serum uric acid (SUA), glucose (GLU), serum triglyceride (TG), total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), creatinine (Cre), and endogenous creatinine clearance rate (Ccr).
2.4. DNA Extraction and Genotyping
The HapMap Project is the most important functional genetic database from which Han-Chinese SNP information can be acquired [14]. First, genotypes for SNPs in NLRP3 representing Han-Chinese were downloaded from the HapMap database (http://www.hapmap.org). Second, we screened the gene loci whose MAF is greater than 0.1 and located in the exon region and promoter region. Then, it is found that the SNPs are related to the occurrence of gout, but there are few reports in China. Finally, based on the above screening criteria, five SNPs (rs10754558, rs35829419, rs3738448, rs3806268, and rs7525979) were selected in NLRP3 gene (supplement Table 1). Genomic DNA was isolated from peripheral blood samples by standard procedures (Promega). We used a custom-designed 2 × 48-Plex SNPscan™ Kit method (Cat#: G0104, Genesky Biotechnologies Inc., Shanghai, China) to genotype the five polymorphisms, which was based on double ligation and multiplex fluorescence PCR. Briefly, the ligation reaction was performed in an ABI2720 thermal cycler, and ABI3730XL sequencer was used to separate and detect PCR products by capillary electrophoresis. Raw data were further analyzed through the labeling dye color and fragment size of the allele-specific ligation-PCR product [15]. Then, 5% duplicate samples were tested to identify genotyping quality and were consistent with the original genotyping results.
2.5. Bioinformatics Analyses
We first predicted the effect of the SNPs on transcription factor binding using HaploReg (http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) [16]. Then, 3DSNP (http://cbportal.org/3dsnp/) [17], an informative tool for annotating human noncoding variants by discovering their functions in the distal interactions between genes and regulatory elements, was used to link the SNPs to their three-dimensional interacting genes. Furthermore, RNAsnp Web Server (https://rth.dk/resources/rnasnp/) [18] was used to predict SNP effects on local RNA secondary structures, and the significant structural change value was p value < 0.2. At last, the impacts of NLRP3 SNPs on gene expression in various tissues were assessed by the public GTEx (the Genotype-Tissue Expression) database (https://gtexportal.org/home/) [19], and the significant value was p value < 0.05 and m value > 0.9 [20, 21].
2.6. Statistical Analyses
Allelic frequencies, genotypic frequencies, Hardy–Weinberg equilibrium (HWE), Akaike information criterion (AIC) analysis, the pairwise linkage disequilibrium (LD), and haplotype analysis were performed using SNPStats software (http://bioinfo.iconcologia.net/SNPStats) [22]. All continuous variables (e.g., age, BMI, and TG) are presented as the means ± standard deviation (S.D.). The difference between the GA/HUA and control groups was analyzed using Student's t-test or the nonparametric Mann–Whitney U tests, as appropriate. The potential relationship of genotypic frequencies of the polymorphisms with the risk of GA/HUA was evaluated by the odds ratios (ORs) with their 95% confidence intervals (CIs) from logistic regression models. All statistical analyses were analyzed by the Statistical Package for Social Sciences software (SPSS, Windows version, release 22.0; SPSS Inc., Chicago, IL, USA). p values < 0.05 were defined as statistically significant level.
3. Results
3.1. Comparison of the Clinical Data between the Patient Group and the Control Group
A total of 1198 male samples were enrolled, consisting of 314 GA, 377 HUA, and 507 healthy controls in this case-control study. Table 1 shows the clinical characteristics of the GA, HUA, and control participants. For all subjects, there were no significant differences in age between GA/HUA and control subjects, indicating the study was an age-matched case-control study. Several risk factors for GA were significantly different between the GA and control groups: BMI, sUA, GLU, TG, Cre, and Ccr (p < 0.05). Moreover, significant differences were found between the HUA and control groups, including BMI, sUA, GLU, TG, HDL, LDL, Cre, and Ccr (p < 0.05).
Table 1.
Clinical characteristics of the patients and control subjects Characteristic.
| Characteristics | Control | GA | HUA | p1 | p2 |
|---|---|---|---|---|---|
| Number | 507 | 314 | 377 | ||
| Age (years) | 46.14 ± 13.24 | 47.63 ± 12.04 | 44.9 ± 13.88 | 0.103 | 0.184 |
| BMI (kg/m2) | 24.88 ± 2.88 | 26.69 ± 3.33 | 26.73 ± 3.52 | <0.001 | <0.001 |
| SUA (μmol/L) | 338.44 ± 49.8 | 508.57 ± 118.14 | 489.82 ± 61.46 | <0.001 | <0.001 |
| GLU (mmol/L) | 5.31 ± 1.15 | 5.8 ± 1.82 | 5.6 ± 1.85 | <0.001 | 0.004 |
| TG (mmol/L) | 1.66 ± 1.14 | 1.95 ± 1.32 | 2.75 ± 2.49 | 0.001 | <0.001 |
| TC (mmol/L) | 4.65 ± 0.89 | 4.59 ± 1.09 | 4.77 ± 1.16 | 0.431 | 0.064 |
| HDL (mmol/L) | 1.34 ± 0.26 | 1.42 ± 5.87 | 1.23 ± 0.3 | 0.751 | <0.001 |
| LDL (mmol/L) | 2.61 ± 0.71 | 2.7 ± 0.77 | 2.78 ± 0.89 | 0.092 | 0.002 |
| Cre (μmol/L) | 85.86 ± 12.34 | 92.57 ± 28.57 | 90.25 ± 21.76 | <0.001 | <0.001 |
| Ccr | 100.45 ± 22.8 | 106.43 ± 40.03 | 109.21 ± 35.47 | 0.007 | <0.001 |
BMI: body mass index; TG: serum triglyceride; TC: total cholesterol; HDL: high-density lipoprotein; LDL: low-density lipoprotein; SUA: serum uric acid; Cre: creatinine; Ccr: endogenous creatinine clearance rate; P1: GA vs. control; P2: HUA vs. control.
3.2. H-W Equilibrium Test and Association Analysis
All genotyped distributions of the control subjects were consistent with those expected from the Hardy-Weinberg equilibrium (p > 0.05), indicating that the samples were representative of the population, as shown in Table 2.
Table 2.
The association between the risk of GA and the genetic polymorphisms.
| SNP | WT Ho/ Ht/ VR Ho | Codominant model | Dominant model | Recessive model | VR allele vs. WT allele | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VR Ho vs. WT Ho | Ht vs. WT Ho | |||||||||||||||
| Control | GA | C_HWE | P | OR (95% CI) | P | OR (95% CI) | AIC | P | OR (95% CI) | AIC | P | OR (95% CI) | AIC | P | OR (95% CI) | |
| rs10754558(CC/CG/GG) | 132/266/109 | 100/158/56 | 0.29 | 0.064 | 0.68 (0.45-1.03) | 0.14 | 0.78 (0.57-1.09) | 1094.6 | 0.074 | 0.75 (0.55-1.03) | 1093.1 | 0.2 | 0.79 (0.55-1.13) | 1094.7 | 0.056 | 0.82 (0.67-1.01) |
| rs35829419(CC/CA/AA) | 503/4/0 | 313/1/0 | 1 | 0.38 | 0.40 (0.04-3.61) | 1095.6 | ||||||||||
| rs3738448(GG/GT/TT) | 325/165/17 | 204/98/11 | 0.57 | 0.94 | 1.03 (0.47-2.25) | 0.72 | 0.95 (0.70-1.28) | 1096.3 | 0.75 | 0.95 (0.71-1.28) | 1094.3 | 0.9 | 1.05 (0.49-2.27) | 1094.4 | 0.82 | 0.97 (0.75-1.25) |
| rs3806268(AA/AG/GG) | 124/265/117 | 98/136/75 | 0.33 | 0.3 | 0.81 (0.55-1.20) | 0.012 | 0.65 (0.46-0.91) | 1081.3 | 0.026 | 0.70 (0.51-0.96) | 1080.8 | 0.71 | 1.07 (0.76-1.49) | 1085.6 | 0.24 | 0.89 (0.73-1.08) |
| rs7525979(CC/CT/TT) | 325/165/17 | 211/92/11 | 0.57 | 0.99 | 1.00 (0.46-2.17) | 0.33 | 0.86 (0.63-1.17) | 1097.4 | 0.36 | 0.87 (0.65-1.17) | 1095.5 | 0.91 | 1.05 (0.48-2.26) | 1096.3 | 0.46 | 0.91 (0.70-1.17) |
OR: odds ratio; VR: variant; WT: wild-type; Ht: heterozygote; VR Ho: variant homozygote; WT Ho: wide-type homozygote.
For rs380628, the heterozygote vs. wide-type homozygote (AG vs. AA: OR = 0.65, 95%CI = 0.46-0.91, p = 0.012) and the dominant model (AG+GG vs. AA: OR = 0.70, 95%CI = 0.51-0.96, p = 0.026), showed a significant difference between GA and control participants. For rs3738448, the variant homozygote vs. wide-type homozygote (TT vs. GG: OR = 2.05, 95%CI = 1.09-3.87, p = 0.024) and the recessive model (TT vs. GG+GT: OR = 1.56, 95%CI = 1.02-2.38, p = 0.04), showed a significant difference between HUA and control subjects. For rs7525979, the variant homozygote vs. wide-type homozygote (TT vs. CC: OR = 1.96, 95%CI = 1.03-3.71, p = 0.037) and the recessive model (TT vs. CC+CT: OR = 2.05, 95%CI = 1.09-3.85, p = 0.024) showed a significant difference between HUA and control subjects. Then, a lower value in terms of AIC was used to find the most acceptable inheritance model. Among them, the dominant model is the best model for rs3806268 in GA, and the recessive model for rs3738448 and rs7525979 in HUA (Tables 2 and 3).
Table 3.
The association between the risk of HUA and the genetic polymorphisms.
| SNP | WT Ho/ Ht/ VR Ho | Codominant model | Dominant model | Recessive model | VR allele vs. WT allele | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VR Ho vs. WT Ho | Ht vs. WT Ho | |||||||||||||||
| Control | GA | C_HWE | P | OR (95% CI) | P | OR (95% CI) | AIC | P | OR (95% CI) | AIC | P | OR (95% CI) | AIC | P | OR (95% CI) | |
| rs10754558 | 132/266/109 | 113/189/75 | 0.29 | 0.27 | 0.80 (0.55-1.18) | 0.24 | 0.83 (0.61-1.13) | 1210.6 | 0.2 | 0.82 (0.61-1.11) | 1208.6 | 0.56 | 0.91 (0.65-1.26) | 1210 | 0.24 | 0.89 (0.73-1.08) |
| rs35829419 | 503/4/0 | 376/1/0 | 1 | 0.28 | 0.33 (0.04-3.00) | 1209.1 | ||||||||||
| rs3738448 | 325/165/17 | 242/109/26 | 0.57 | 0.024 | 2.05 (1.09-3.87) | 0.42 | 0.89 (0.66-1.19) | 1205.9 | 0.98 | 1.00 (0.75-1.32) | 1210.3 | 0.016 | 2.14 (1.14-3.99) | 1204.5 | 0.38 | 1.11 (0.88-1.39) |
| rs3806268 | 124/265/117 | 102/194/80 | 0.33 | 0.35 | 0.83 (0.56-1.22) | 0.48 | 0.89 (0.65-1.23) | 1208.5 | 0.38 | 0.87 (0.64-1.18) | 1206.7 | 0.51 | 0.90 (0.65-1.24) | 1207.1 | 0.34 | 0.91 (0.75-1.10) |
| rs7525979 | 325/165/17 | 357/199/158 | 0.57 | 0.037 | 1.96 (1.03-3.71) | 0.36 | 0.87 (0.65-1.17) | 1206.4 | 0.85 | 0.97 (0.74-1.29) | 1210.3 | 0.024 | 2.05 (1.09-3.85) | 1205.2 | 0.5 | 1.08 (0.86-1.36) |
OR: odds ratio; VR: variant; WT: wild-type; Ht: heterozygote; VR Ho: variant homozygote; WT Ho: wide-type homozygote.
3.3. Genotype of the Three Polymorphisms and the Clinical Characteristics of the Patients
The above association analysis showed that rs3806268, rs3738448, and rs7525979 were related to GA/HUA risk. Therefore, Table 4 further shows risk of GA/HUA based on these three polymorphisms taking into consideration obesity, smoking, hypertension, and drinking. Taking the A/A genotype group as reference, the rs3806268 A/G genotype group presented decreased risk of GA among the hypertension (OR = 0.54, 95%CI = 0.34-0.86, p = 0.0093), smoking (OR = 0.59, 95%CI = 0.38-0.92, p = 0.018), and no obesity (OR = 0.60, 95%CI = 0.41-0.88, p = 0.0097) group. The rs3738448 T/T genotype group demonstrated increased risk of HUA among the hypertension (OR = 4.10, 95%CI = 1.35-12.49, p = 0.0056) and no drinking population (OR = 3.56, 95%CI = 1.22-10.97, p = 0.016) compared to G/G genotype group. The rs7525979 T/T genotype group demonstrated increased risk of HUA among the hypertension (OR = 4.01, 95%CI = 1.32-12.23, p = 0.0064) and no drinking population (OR = 3.24, 95%CI = 1.06-9.92, p = 0.034) compared to C/C genotype group.
Table 4.
Association of NLRP3 polymorphisms with clinical characteristics and risk of GA/HUA.
| Category | rs3806268 | rs3738448 | rs7525979 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GA | Control | OR (95% CI) | p value | HUA | Control | OR (95% CI) | p value | HUA | Control | OR (95% CI) | p value | |
| Obesity | ||||||||||||
| WT Ho (no) | 72 | 109 | 1.00 | 162 | 277 | 1.00 | 163 | 277 | ||||
| Ht (no) | 91 | 229 | 0.60 (0.41-0.88) | 0.0097 | 85 | 143 | 1.02 (0.73-1.42) | 0.92 | 85 | 143 | 1.01 (0.73-1.41) | 0.95 |
| VR Ho (no) | 58 | 97 | 0.91 (0.58-1.41) | 0.66 | 17 | 13 | 1.94 (0.94-3.98) | 0.072 | 16 | 15 | 1.81 (0.87-3.76) | 0.11 |
| WT Ho (yes) | 15 | 26 | 1.00 | 80 | 48 | 1.00 | 81 | 48 | 1.00 | |||
| Ht (yes) | 36 | 45 | 0.72 (0.33-1.56) | 0.4 | 24 | 22 | 0.65 (0.33-1.29) | 0.22 | 23 | 22 | 0.62 (0.31-1.23) | 0.17 |
| VR Ho (yes) | 20 | 17 | 0.49 (0.20-1.21) | 0.12 | 9 | 2 | 2.70 (0.56-13.02) | 0.18 | 9 | 2 | 2.67 (0.55-12.86) | 0.18 |
| Smoking | ||||||||||||
| WT Ho (no) | 63 | 64 | 1.00 | 145 | 181 | 1.00 | 146 | 181 | 1.00 | |||
| Ht (no) | 90 | 154 | 0.59 (0.38-0.92) | 0.018 | 63 | 100 | 0.79 (0.54-1.15) | 0.22 | 62 | 100 | 0.77 (0.52-1.13) | 0.18 |
| VR Ho (no) | 44 | 71 | 0.63 (0.38-1.05) | 0.076 | 15 | 9 | 2.08 (0.88-4.89) | 0.087 | 15 | 9 | 2.07 (0.88-4.86) | 0.09 |
| WT Ho (yes) | 35 | 60 | 1.00 | 97 | 144 | 1.00 | 98 | 144 | 1.00 | |||
| Ht (yes) | 46 | 111 | 0.71 (0.41-1.22) | 0.22 | 46 | 65 | 1.05 (0.67-1.66) | 0.83 | 46 | 65 | 1.04 (0.66-1.64) | 0.87 |
| VR Ho (yes) | 31 | 46 | 1.16 (0.62-2.14) | 0.65 | 11 | 8 | 2.04 (0.79-5.26) | 0.14 | 10 | 8 | 1.84 (0.70-4.82) | 0.21 |
| Hypertension | ||||||||||||
| WT Ho (no) | 31 | 78 | 1.00 | 100 | 188 | 1.00 | 100 | 189 | 1.00 | |||
| Ht (no) | 40 | 144 | 0.70 (0.41-1.20) | 0.2 | 41 | 98 | 0.79 (0.51-1.22) | 0.28 | 42 | 97 | 0.82 (0.53-1.27) | 0.36 |
| VR Ho (no) | 21 | 77 | 0.69 (0.36-1.30) | 0.24 | 9 | 13 | 1.30 (0.54-3.15) | 0.56 | 8 | 13 | 1.16 (0.47-2.90) | 0.75 |
| WT Ho (yes) | 67 | 46 | 1.00 | 142 | 137 | 1.00 | 144 | 136 | 1.00 | |||
| Ht (yes) | 96 | 121 | 0.54 (0.34-0.86) | 0.0093 | 68 | 67 | 0.98 (0.65-1.48) | 0.92 | 66 | 68 | 0.92 (0.61-1.38) | 0.68 |
| VR Ho (yes) | 54 | 40 | 0.93 (0.53-1.61) | 0.79 | 17 | 4 | 4.10 (1.35-12.49) | 0.0056 | 17 | 4 | 4.01 (1.32-12.23) | 0.0064 |
| Drinking | ||||||||||||
| WT ho (no) | 60 | 69 | 1.00 | 104 | 190 | 1.00 | 105 | 189 | 1.00 | |||
| Ht (no) | 91 | 160 | 0.65 (0.42-1.01) | 0.054 | 51 | 95 | 0.98 (0.65-1.49) | 0.93 | 51 | 96 | 0.96 (0.63-1.45) | 0.83 |
| VR Ho (no) | 44 | 61 | 0.83 (0.49-1.39) | 0.48 | 10 | 5 | 3.65 (1.22-10.97) | 0.016 | 9 | 5 | 3.24 (1.06-9.92) | 0.034 |
| WT Ho (yes) | 38 | 55 | 1.00 | 138 | 135 | 1.00 | 139 | 136 | 1.00 | |||
| Ht (yes) | 45 | 105 | 0.75 (0.46-1.24) | 0.28 | 58 | 70 | 0.81 (0.53-1.24) | 0.33 | 57 | 69 | 0.81 (0.53-1.23) | 0.32 |
| VR Ho (yes) | 31 | 56 | 0.80 (0.44-1.46) | 0.47 | 16 | 12 | 1.30 (0.59-2.86) | 0.51 | 16 | 12 | 1.30 (0.60-2.86) | 0.5 |
OR (95% CI) and p values were obtained from logistic regression analysis.
3.4. Haplotype Analysis
To evaluate the correlations of the SNPs in NLRP3, we exerted haplotype analysis between GA/HUA and healthy controls. The linkage disequilibrium (LD) structures of five SNPs in NLRP3 are shown in Supplementary Table 2. There are six common haplotypes (>1%) among controls. The GCGAC showed the most frequently haplotype in GA, HUA, and healthy controls. Taking the most common haplotype as reference, the rs10754558/C-rs35829419/C-rs3738448/G-rs3806268/A-rs7525979/C haplotype presented increased risk of HUA (OR = 1.60, 95% CI: 1.16–2.22, p = 0.0046) (Tables 5 and 6).
Table 5.
Haplotypes of the NLRP3 gene with the risk of GA.
| Haplotypes | Control frequency | Case frequency | OR (95% CI) | p value |
|---|---|---|---|---|
| rs10754558/rs35829419/rs3738448/rs3806268/rs7525979 | ||||
| GCGAC | 0.3143 | 0.3005 | 1.00 | — |
| CCGGC | 0.2264 | 0.224 | 1.04 (0.78 - 1.38) | 0.81 |
| CCGAC | 0.1891 | 0.2302 | 1.26 (0.91 - 1.73) | 0.17 |
| CCTGT | 0.1062 | 0.1139 | 1.13 (0.78 - 1.66) | 0.52 |
| GCTGT | 0.0866 | 0.0661 | 0.79 (0.48 - 1.28) | 0.34 |
| GCGGC | 0.0695 | 0.0542 | 0.79 (0.46 - 1.37) | 0.41 |
OR (95% CI) and p values were obtained from logistic regression analysis.
Table 6.
Haplotypes of the NLRP3 gene with the risk of HUA.
| Haplotypes | Control frequency | Case frequency | OR (95% CI) | p value |
|---|---|---|---|---|
| rs10754558/rs35829419/rs3738448/rs3806268/rs7525979 | ||||
| GCGAC | 0.3143 | 0.2687 | 1.00 | — |
| CCGAC | 0.2264 | 0.2607 | 1.60 (1.16-2.22) | 0.0046 |
| CCGGC | 0.1891 | 0.1791 | 0.94 (0.70-1.25) | 0.66 |
| CCTGT | 0.1062 | 0.1066 | 1.21 (0.84-1.76) | 0.31 |
| GCTGT | 0.0866 | 0.1016 | 1.30 (0.86-1.97) | 0.22 |
| GCGGC | 0.0695 | 0.078 | 1.31 (0.79-2.17) | 0.29 |
OR (95% CI) and p values were obtained from logistic regression analysis.
3.5. Bioinformatics Analyses
Using HaploReg v4.1, rs3738448 was predicted to localize in promoter histone markers, enhancer histone markers, DNase hypersensitivity, and motifs changed (Nanog and STAT); it affected bound proteins such as TBP and POL2. Rs3806268 was predicted to localize in enhancer histone markers. Rs7525979 was predicted to localize in enhancer histone markers, and motifs changed (Gm397). 3D chromatin looping data showed that rs3806268, rs3738448, and rs7525979 may interact with GALNT2, GCSAML, GCSAML-AS1, OR2B11, OR2C3, and OR2W5 genes (Table 7). In the case of rs3806268, the minimum free energy of the G and A allele were −131.70 kcal/mol and −130.80 kcal/mol, respectively (p = 0.5327). The minimum free energy of the rs3738448 G and T allele, a significant structural change, were −148.40 kcal/mol and −151.50 kcal/mol, respectively (p = 0.0936). The minimum free energy of rs7525979 C and T allele were −129.90 kcal/mol and −132.30 kcal/mol, respectively (p = 0.4045) (Figure 1). Using the GTEx database, SNP rs3806268 was associated with NLRP3 expression and identified as expression quantitative trait locis (eQTLs) in artery-tibial, adipose-subcutaneous, whole blood, cell-cultured fibroblasts, muscle-skeletal, breast-mammary tissue, and esophagus-mucosa with significant p value and m value (Figure 2).
Table 7.
SNP functional annotation in HaploReg v4.1 and 3DSNP database.
| SNP | Ref | Alt | SNP functional annotation | 3D interacting gene |
|---|---|---|---|---|
| rs3738448 | G | T | Promoter histone marks, enhancer histone marks, DNAse, proteins bound, motifs changed | GALNT2, GCSAML, GCSAML-AS1, OR2B11, OR2C3, and OR2W5 |
| rs3806268 | G | C | Enhancer histone marks | GALNT2, GCSAML, GCSAML-AS1, OR2B11, OR2C3, and OR2W5 |
| rs7525979 | C | T | Enhancer histone marks, motifs changed | GALNT2, GCSAML, GCSAML-AS1, OR2B11, OR2C3, and OR2W5 |
SNP: single-nucleotide polymorphism; Ref: reference; Alt: alternation; eQTL: expression quantitative trait loci.
Figure 1.
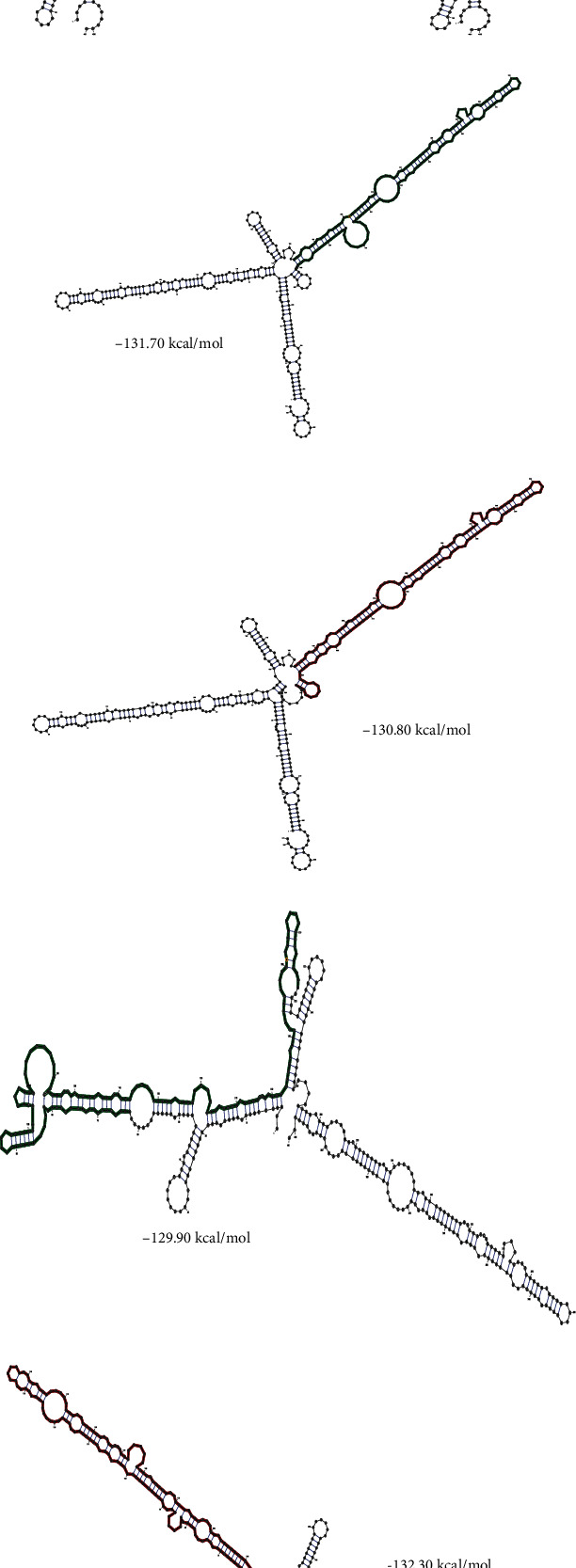
Analysis of rs3738448 G/T, rs3806268 G/A, and rs7525979 C/T, variant effects on local mRNA structure of NLRP3 using RNAfold server. (a, b) G and T allele of rs3738448, (c, d) G and A allele of rs3806268, (e, f) C and T allele of rs7525979. The most important structural change is related to rs2466294 C/G.
Figure 2.
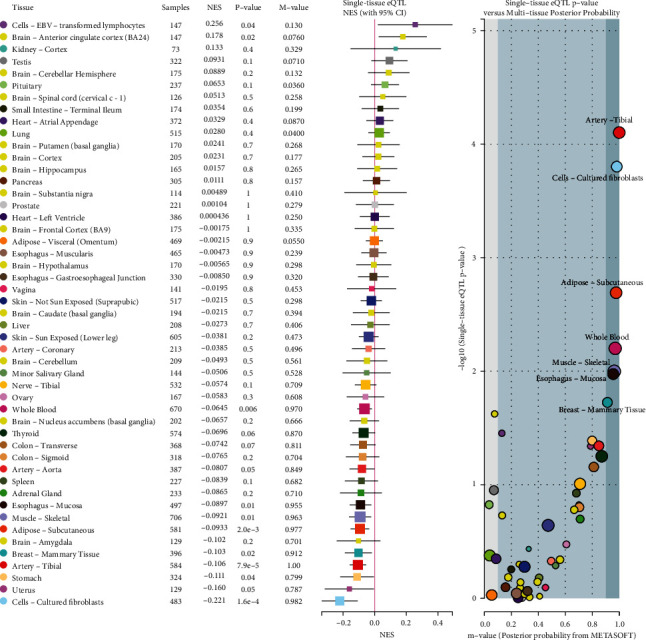
Multitissue expression quantitative trait loci (eQTL) comparison of rs3806268. NES: the slope of the linear regression of normalized expression data versus the three genotype categories using single-tissue eQTL analysis, representing eQTL effect size. m value: the posterior probability that an eQTL effect exists in each tissue tested in the cross-tissue meta-analysis; large m value (m value > 0.9): the tissue is predicted to have an eQTL effect.
4. Discussion
Our results showed that the NLRP3 rs3806268 polymorphism was significantly associated with GA; the NLRP3 rs3738448 and rs7525979 polymorphisms were significantly associated with HUA. Moreover, these three polymorphisms were significantly associated with clinical characteristics, such as hypertension, drinking, smoking and obesity.
Inflammasomes are composed of adaptors, receptors, and pro-caspase-1, and NLRP3 inflammasome is one of the most well reported [23, 24]. It is well-known that NLRP3 inflammasome is important inflammatory triggers during gout flare [25–28]. The synonymous SNP rs3806268 and rs7525979 are silent polymorphisms of the NLRP3 exon. Previous researches indicated that synonymous polymorphism can change the substrate specificity [29]. Rs7525979 can regulate NLRP3 translation and lead to the accumulation of an ubiquitinated, insoluble form of NLRP3 in Parkinson's disease [30]. AA genotype carriers of rs3806268 may directly increase IL-1β production in systemic lupus erythematosus disease [31]. Therefore, the NLRP3 rs3806268 and rs7525979 polymorphisms may alter the structure of substrate and inhibitor interaction sites, but it needs to be further elucidated in GA/HUA. However, inconsistent with the previous findings that NLRP3 rs10754558 was significantly associated with GA [11], we did not observe significant correlation between the polymorphism and GA/HUA risk, suggesting their potential interaction with environment, such as BMI, obesity, and age.
Additionally, the rs3806268 AG genotype presented decreased risk of GA among the hypertension, smoking, and no obesity subjects compared to GG genotype group. The rs3738448 TT genotype demonstrated increased risk of HUA among the hypertension and no drinking population compared to GG genotype group. The rs7525979 TT genotype demonstrated increased risk of HUA among the hypertension and no drinking population compared to the CC genotype group. Thus, understanding the mechanism of rs3806268 AG, rs3738448 TT, and rs7525979 TT genotypes with hypertension, smoking, drinking, and obesity interaction will require further studies.
Furthermore, bioinformatics analyses indicated that SNPs with statistical significance (rs3806268, rs3738448, and rs7525979) might be related to gene regulation, such as the promoter histone markers, enhancer histone markers, DNase hypersensitivity, and mRNA structure. Meanwhile, eQTL analysis underlined the correlation of rs3806268 with NLRP3 expression in different tissues. We thus speculated that these three genetic alterations could affect gene expression, which in turn affects GA/HUA susceptibility.
Nevertheless, our work has some limitations. First, the healthy and GA/HUA subjects were enrolled from hospitals which may have inherited biases. Second, the SNPs investigated in the present research may not be sufficiently comprehensive about genetic alteration in NLRP3 gene. And further fine-mapping researches in the NLRP3 susceptible region are needed. At last, further studies are needed to prove our findings, including the bioinformatics results and the potential effects of gene-gene and gene-environment interactions.
In summary, our case-control demonstrates that rs3806268 GA genotype is significantly decreased in GA cases compared with controls. The rs3738448 and rs7525979 TT genotypes were significantly increased in HUA cases compared to controls. Moreover, the rs10754558/C-rs35829419/C-rs3738448/G-rs3806268/A-rs7525979/C haplotype showed higher risk of HUA compared to the GCGAC control haplotype.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (81760169 and 81960169) and the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2019D01C219). We would like to thank all participants who agreed to participate in the study.
Data Availability
The datasets supporting the conclusions of this article are included within the article.
Additional Points
Key Points. Rs3806268 was significantly associated with the susceptibility to GA. Rs3738448 and rs7525979 were significantly associated with the susceptibility to HUA. Rs3738448, rs3806268, and rs7525979 might influence the gene regulation, while the T-allele of rs3738448 increased the stability of NLRP3-mRNA.
Ethical Approval
This study was approved by the Ethics Committee of Xinjiang Medical University, Urumqi, China. It was conducted according to the standards of the Declaration of Helsinki.
Consent
Written informed consent was obtained from all participants.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
YPS designed the research project. BZ, MTL, and WJC performed all the experiments. BZ, YZL, and KM helped with the result analysis. XBZ and TTT wrote the manuscript. All authors have reviewed and consented to publication of the paper.
Supplementary Materials
Table 1: primary information of genotyped SNPs. Supplementary table 2: the linkage disequilibrium coefficients among five SNPs of NLRP3.
References
- 1.Dehlin M., Jacobsson L., Roddy E. Global epidemiology of gout: prevalence, incidence, treatment patterns and risk factors. Nature Reviews Rheumatology . 2020;16(7):380–390. doi: 10.1038/s41584-020-0441-1. [DOI] [PubMed] [Google Scholar]
- 2.Ma Q., Honarpisheh M., Li C., et al. Soluble uric acid is an intrinsic negative regulator of monocyte activation in monosodium urate crystal-induced tissue inflammation. Journal of Immunology (Baltimore, Md: 1950) . 2020;205(3):789–800. doi: 10.4049/jimmunol.2000319. [DOI] [PubMed] [Google Scholar]
- 3.Marwah R. K. Comorbidities in gouty arthritis. Journal of Investigative Medicine . 2011;59(8):1211–1220. doi: 10.2310/JIM.0b013e318239f660. [DOI] [PubMed] [Google Scholar]
- 4.Ying Y., Chen Y., Li Z., Huang H., Gong Q. Investigation into the association between P2RX7 gene polymorphisms and susceptibility to primary gout and hyperuricemia in a Chinese Han male population. Rheumatology International . 2017;37(4):571–578. doi: 10.1007/s00296-017-3669-6. [DOI] [PubMed] [Google Scholar]
- 5.Battelli M., Bortolotti M., Polito L., Bolognesi A. The role of xanthine oxidoreductase and uric acid in metabolic syndrome. Biochimica et biophysica acta Molecular basis of disease . 2018;1864(8):2557–2565. doi: 10.1016/j.bbadis.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Mortada I. Hyperuricemia, type 2 diabetes mellitus, and hypertension: an emerging association. Current Hypertension Reports . 2017;19(9):p. 69. doi: 10.1007/s11906-017-0770-x. [DOI] [PubMed] [Google Scholar]
- 7.Liu R., Han C., Wu D., et al. Prevalence of hyperuricemia and gout in mainland China from 2000 to 2014: a systematic review and meta-analysis. BioMed Research International . 2015;2015:12. doi: 10.1155/2015/762820.762820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vong C., Tseng H., Yao P., et al. Specific NLRP3 inflammasome inhibitors: promising therapeutic agents for inflammatory diseases. Drug Discovery Today . 2021;26(6):1394–1408. doi: 10.1016/j.drudis.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Bauernfeind F., Rieger A., Schildberg F., Knolle P., Schmid-Burgk J., Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. Journal of Immunology (Baltimore, Md: 1950) . 2012;189(8):4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 10.Zhen Y., Zhang H. NLRP3 Inflammasome and inflammatory bowel disease. Frontiers in Immunology . 2019;10(276) doi: 10.3389/fimmu.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., Qing Y., He Y., Xie W., Zhou J. Association of NLRP3 polymorphisms with susceptibility to primary gouty arthritis in a Chinese Han population. Clinical Rheumatology . 2018;37(1):235–244. doi: 10.1007/s10067-017-3900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney C., Stamp L. K., Dalbeth N., et al. Multiplicative interaction of functional inflammasome genetic variants in determining the risk of gout. Arthritis Research & Therapy . 2015;17(1):288–288. doi: 10.1186/s13075-015-0802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neogi T., Jansen T. L., Dalbeth N., et al. 2015 gout classification criteria: an American College of Rheumatology/European League against rheumatism collaborative initiative. Annals of the Rheumatic Diseases . 2015;74(10):1789–1798. doi: 10.1136/annrheumdis-2015-208237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua F., Guo Y., Sun Q., Yang L., Gao F. HapMap-based study: CYP2A13 may be a potential key metabolic enzyme gene in the carcinogenesis of lung cancer in non-smokers. Thoracic Cancer . 2019;10(4):601–606. doi: 10.1111/1759-7714.12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu R., Song L., Jiang L., et al. Susceptible gene polymorphism in patients with three-vessel coronary artery disease. BMC Cardiovascular Disorders . 2020;20(1):p. 172. doi: 10.1186/s12872-020-01449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward L. D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research . 2012;40(D1):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y., Quan C., Chen H., Bo X., Zhang C. 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Research . 2017;45(D1):D643–D649. doi: 10.1093/nar/gkw1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabarinathan R., Tafer H., Seemann S. E., Hofacker I. L., Stadler P. F., Gorodkin J. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Research . 2013;41:W475–W479. doi: 10.1093/nar/gkt291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium GT. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science . 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B., Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genetics . 2012;8(3, article e1002555) doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han B., Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. American Journal of Human Genetics . 2011;88(5):586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics (Oxford, England) . 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 23.Bakele M., Joos M., Burdi S., et al. Localization and functionality of the inflammasome in neutrophils. The Journal of Biological Chemistry . 2014;289(8):5320–5329. doi: 10.1074/jbc.M113.505636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M. F., Shu C. C., Wang J. Y., et al. NLRP3 inflammasome is attenuated in patients with _Mycobacterium avium_ complex lung disease and correlated with decreased interleukin-1 β response and host susceptibility. Scientific Reports . 2019;9(1):p. 12534. doi: 10.1038/s41598-019-47609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J., Lin W., Chen Y., et al. rs3806268 of NLRP3 gene polymorphism is associated with the development of primary gout. International Journal of Clinical and Experimental Pathology . 2015;8(10):13747–13752. [PMC free article] [PubMed] [Google Scholar]
- 26.Meng D. M., Zhou Y. J., Wang L., et al. Polymorphisms in the NLRP3 gene and risk of primary gouty arthritis. Molecular Medicine Reports . 2013;7(6):1761–1766. doi: 10.3892/mmr.2013.1429. [DOI] [PubMed] [Google Scholar]
- 27.Wang L. F., Ding Y. J., Zhao Q., Zhang X. L. Investigation on the association between NLRP3 gene polymorphisms and susceptibility to primary gout. Genetics and molecular research: GMR . 2015;14(4):16410–16414. doi: 10.4238/2015.December.9.10. [DOI] [PubMed] [Google Scholar]
- 28.Yang G., Lee H. E., Moon S. J., et al. Direct binding to NLRP3 pyrin domain as a novel strategy to prevent NLRP3-driven inflammation and gouty arthritis. Arthritis & rheumatology . 2020;72(7):1192–1202. doi: 10.1002/art.41245. [DOI] [PubMed] [Google Scholar]
- 29.Kimchi-Sarfaty C., Oh J. M., Kim I. W., et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science . 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 30.von Herrmann K. M., Salas L. A., Martinez E. M., et al. _NLRP3_ expression in mesencephalic neurons and characterization of a rare _NLRP3_ polymorphism associated with decreased risk of Parkinson 's disease. NPJ Parkinson's disease . 2018;4(1):p. 24. doi: 10.1038/s41531-018-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Z., Niu Q., Huang Z., Yang B., Zhang J. Association of nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3 polymorphisms with systemic lupus erythematosus disease activity and biomarker levels: a case-control study in Chinese population. Medicine . 2020;99(35, article e21888) doi: 10.1097/MD.0000000000021888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: primary information of genotyped SNPs. Supplementary table 2: the linkage disequilibrium coefficients among five SNPs of NLRP3.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


