Abstract
Activation of the final sporulation-specific transcription factor, ςK, is regulated by a signal emanating from the forespore which interacts with the pro-ςK processing complex, comprising SpoIVFA, BofA, and the pro-ςK processing protease, SpoIVFB. Mature ςK then directs late gene expression in the parental compartment of the developing sporangial cell. The nature of this complex and how it is activated to process pro-ςK are not understood. All three proteins are predicted to be integral membrane proteins. Here, we have analyzed the membrane topology of SpoIVFA and SpoIVFB by constructing chimeric forms of spoIVFA and spoIVFB with the complementary reporters phoA and lacZ and analyzing activity in Escherichia coli. SpoIVFA was found to have a single transmembrane-spanning domain, while SpoIVFB was shown to have six transmembrane-spanning domains (6-transmembrane configuration). Further, SpoIVFA is required to stabilize SpoIVFB in the membrane. SpoIVFB was shown to have a 4-transmembrane configuration when expressed on its own but was found to have a 6-transmembrane configuration when coexpressed with SpoIVFA, while BofA had a positive effect on the assembly of both SpoIVFA and SpoIVFB. The single transmembrane domain of SpoIVFA (approximately residues 73 to 90) was shown to be the principle determinant in stabilizing the 6-transmembrane configuration of SpoIVFB. Although the bofB8 allele, which uncouples the ςK checkpoint, did not appear to promote a conformational change from a 6- to 4-transmembrane configuration of SpoIVFB (apparently ruling out a profound conformational change as the mechanism of activating SpoIVFB proteolytic activity), instability of SpoIVFB may be an important factor in SpoIVFB-mediated processing of pro-ςK.
In Bacillus subtilis, checkpoints regulate differential gene expression during spore formation, ensuring developmental fidelity and enforcing a dependence to a sequence of events (4, 8, 9, 11, 24). In the ςK checkpoint, activation of the transcription factor, ςK, in the mother cell chamber of the sporangial cell is coupled to events under the control of ςG-dependent gene expression in the opposed (forespore) compartment (4). The inactive form of ςK, termed pro-ςK, is synthesized in the mother cell and must be proteolytically cleaved at its N terminus to be rendered active (13). Pro-ςK has itself been shown to target and associate with the outer forespore membrane (OFM) of the developing forespore (28), and this event presumably allows pro-ςK to interact with the processing complex embedded in the OFM. Genetic experiments have identified three components of this complex, SpoIVFB, SpoIVFA, and BofA. SpoIVFB has been proposed to be the putative protease (5) which cleaves pro-ςK; this protein contains some homology with Zn2+ metalloproteases (10), although activity in vitro has not been shown to date (12, 21). SpoIVFA and BofA, however, have been shown to regulate the proposed proteolytic activity of SpoIVFB both positively, by preserving the stability of SpoIVFB and, negatively, by rendering the SpoIVFB molecule inactive until an appropriate signal is received (4, 5, 19, 21, 22). SpoIVFA and SpoIVFB have also been localized to the OFM by immunofluorescence microscopy (20), while BofA has been shown to assemble into a phospholipid membrane in Escherichia coli and its topology has been determined by using the analysis of complementary bofA-phoA or bofA-lacZ gene fusions (26). So far, genetic experimentation has shown that SpoIVFA and SpoIVFB are likely to interact, and BofA may also interact directly to form a hetero-oligomeric complex (4, 5).
Pro-ςK proteolysis is coupled to a signal emanating from the opposed forespore chamber. This signal is comprised of one ςG-transcribed gene, spoIVB, whose 46-kDa product is proposed to be secreted through the inner forespore membrane (IFM), where it can subsequently interact with the pro-ςK processing complex (3). This interaction ultimately relieves SpoIVFA- and BofA-mediated inhibition of SpoIVFB, leading to pro-ςK proteolysis, activation of ςK-directed gene expression, and completion of the terminal stages of spore formation.
In this work, we have investigated the topology of the SpoIVFA and SpoIVFB polypeptides in a phospholipid bilayer. Using the analysis of alkaline phosphatase (AP) in different spoIVFA-phoA and spoIVFB-phoA gene fusions in E. coli, together with complementary analysis of lacZ gene fusions, we show that SpoIVFB is an intrinsically unstable molecule which can assume two membrane configurations, dependent on the presence or absence of SpoIVFA and, to a lesser degree, BofA.
MATERIALS AND METHODS
Bacterial strains.
E. coli XL1-Blue recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] (Stratagene) was used for all routine plasmid constructions. E. coli CC118 araD139 Δ(ara,leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE argE(Am) recA1 (16) was used for analysis of AP. E. coli TG1 (23) was used for analysis of β-galactosidase activity. SC1256 (spoIVFA181), SC1257 (spoIVFB587), VO138 (spoIVFAΔ91), SC1358 (spoIVFAΔ91 spoIVFBVI235), SC1363 (spoIVFAΔ91 spoIVFBVM207), SC1365 (spoIVFAΔ91 spoIVFBLV221), and SC1366 (spoIVFAΔ91 spoIVFBAV273) have been described previously (5). DG675 (spoIVFAΔ91 bofA::erm) was constructed by transforming cells of VO138 with chromosomal DNA from ER76 (bofA::erm spoIIIGΔ1) with selection for erythromycin resistance (erm). All strains used in this study are congenic with the prototrophic wild-type (Spo+) strain PY79 (27).
Plasmids.
pMV100, containing the intact phoA gene, has been described elsewhere (6). pJBZ280 contains a full-length but promoterless lacZ gene (gift of D. Alley). pK184 (ATCC 37766) carries the p15A replicon (kanr) and can be used in conjunction with ColE1 replicons. pMUTIN2 (25) is a vector containing the bla gene (selectable in E. coli), the erm gene (erythromycin resistance in B. subtilis), and the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspac. pSL13 (Pspac spoIVFB), pSL14 (Pspac spoIVFABbofB8), and pSL31 (Pspac spoIVFAB) have been described previously (12) and contain segments of the wild-type spoIVF or mutant spoIVF(bofB8) operon cloned downstream of the IPTG-inducible Pspac promoter of the B. subtilis vector pDG148, modified to remove the origin for autonomous replication in B. subtilis. pDG5981 contains the bofA cistron under the control of the IPTG-inducible Pspac promoter. 5′ (GCCGGATCCATTAAGAAGAGAGTTTG) and 3′ (TATGCTCGAGCCTATTTTTTATTATTG) oligonucleotide primers were used to PCR amplify the bofA operon, which was then cloned into pMUTIN2 by using restriction endonuclease sites contained within the PCR primers. Using the resulting clone, a second PCR was performed to amplify upstream of Pspac the termination codon of bofA. This PCR product was cloned in pK184 (see above), and the construct (pDG5981) was verified by nucleotide sequencing.
General methods.
General Bacillus methods (transformations, induction of sporulation, etc.) are described elsewhere (7).
Plasmid constructions. (i) spoIVF-phoA and spoIVF-lacZ chimeric plasmids.
Plasmids pSL13, pSL31, and pSL14 were used as the basis for the construction of the chimeric spoIVF-phoA or -lacZ genes in a two-step procedure. Each contains the spoIVFAB(bofB8) (pSL14), spoIVFB (pSL13), or spoIVFAB (pSL31) cistrons fused downstream of the IPTG-inducible Pspac promoter (12). First, a sense primer, specific for a region upstream of Pspac, and specific antisense primers to various regions of the spoIVF coding region were used to generate PCR products of appropriate segments of spoIVFA, spoIVFB, or spoIVFA and spoIVFB. Next, each PCR product was digested with XbaI and BamHI or XbaI and EcoRI for phoA or lacZ constructs, respectively (restriction sites were contained within the sense and antisense primers), and ligated with pMV100 (phoA) or pJBZ280 (lacZ) to create an in-frame fusion between the spoIVF coding sequence and phoA or lacZ. We found that it was not possible to construct spoIVFA and spoIVFB fusions to lacZ at every position. Attempting this produced extremely unstable clones which underwent a frameshift mutation, preventing expression of LacZ in E. coli. We attribute this to the inappropriate expression and translocation of a chimeric SpoIVFA- or SpoIVFB-LacZ chimera, which is toxic to cell growth. Fusion junctions were confirmed by DNA sequencing.
(ii) spoIVFAΔ164-spoIVFB-phoA chimeric plasmids.
pMV100-derived plasmids, created as described above with phoA fused to the entire spoIVF operon at positions immediately following codons 32 (A32), 112 (Q112), and 180 (L180) within the spoIVFB cistron, were used as templates for an inverse PCR to delete codons 100 to 264 of spoIVFA. The in-frame deletion was verified by DNA sequencing.
(iii) spoIVFAΔ91-spoIVFB-phoA chimeric plasmids.
The same procedure for spoIVFAΔ164-spoIVFB-phoA as that described above was used, and an inverse PCR was used to delete codons 2 to 92 of spoIVFA.
(iv) spoIVFAΔ67-spoIVFB-phoA chimeric plasmids.
The same procedure for spoIVFAΔ164-spoIVFB-phoA as that described above was used, and an inverse PCR was used to delete codons 3 to 70 of spoIVFA while retaining the putative transmembrane domain.
Analysis of AP activity.
Activity was monitored on solid medium by using the AP substrate BCIP (5-bromo-4-chloro-3-indolylphosphate sodium salt; Sigma) at 40 μg/ml. For liquid assays, CC118 derivative strains containing chimeric phoA genes were grown overnight at 30 or 37°C in Luria-Bertani (LB) broth (containing 50 μg of ampicillin per ml and 0.2% glucose), diluted 1:100 in 20 ml of fresh LB broth, and incubated at either 30 or 37°C with orbital shaking (200 rpm) until an approximate density of 0.3 A600 was reached. IPTG was then added to a final concentration of 1 mM, and incubation was continued for a further 2 h (37°C) or 3 h (30°C), at which point 1-ml volumes were removed and the A595 and AP activity determined. AP activity was assayed from 1 ml of culture by resuspending the cell pellet in 800 μl of distilled water. Cells were permeabilized by adding 20 μl of 0.1% sodium dodecyl sulfate and 20 μl of chloroform and vortexed for 1 min. A 100-μl volume of 10 mg of para-nitrophenolphosphate (in 1 M Tris-HCl, pH 9.0) per ml was added, and incubation was commenced at 37°C until either a straw-yellow color had developed or 1 h had elapsed, and the reaction was stopped by adding 100 μl of 10 M NaOH. Specific activity was calculated according to the following formula: 1,000 × {A420/[reaction time (min) × volume of cells (ml) × A595]} (2).
Coexpression experiments.
To coexpress BofA with SpoIVF-AP fusions, E. coli XL1-Blue carrying pDG5981 (Pspac-bofA; see above) was used as a host. pMV100 derivatives carrying Pspac-spoIVF-phoA chimeras were introduced into competent cells by selection for Apr (carried by the pMV100 vector) on LB plates containing ampicillin and kanamycin. Kanr was encoded by pDG5981. AP activity was determined by growth of these XL1-Blue strains in the presence of ampicillin and kanamycin and was found to be identical to that obtained in CC118. Background levels of AP activity were provided by an XL1-Blue strain containing both pK184 and pMV100.
To coexpress SpoIVFA and/or SpoIVFAB with BofA-AP fusions, we first constructed plasmids that expressed SpoIVFA or both SpoIVFA and SpoIVFB. The spoIVFA or complete spoIVF operon, including the upstream Pspac promoter from plasmid pSL13 (Pspac-spoIVFB), was cloned into pK184, which contains the p15A replicon. These plasmids were then transformed into XL1-Blue cells containing the four bofA-AP fusions described by Varcamonti et al. (26) (these are pMV100 derivatives and contain the ColE1 replicon), and AP activity was measured following expression in the presence of IPTG.
Western analysis.
The entire open reading frame (ORF)-encoding pro-ςK was cloned in a pET28b expression vector (Novagen) such that the encoded pro-ςK protein was fused to a polyhistidine tag at its C terminus. Pro-ςK was expressed in E. coli [strain BL21-(DE3)], and polyclonal antisera was raised in rabbits. For Western blotting, (NH4)2SO4-precipitated sera were preadsorbed with E. coli lysates prepared from BL21 (DE3) pLysS and then used at a 1:5,000 dilution. Anti-rabbit horseradish peroxidase antibody conjugate was used at a dilution of 1:2,000. Reactive antibodies were visualized by using the ECL Western blotting system (Amersham).
RESULTS
Topology of the SpoIVFA and SpoIVFB polypeptides.
To define the topology of both SpoIVFA and SpoIVFB, we used the analysis of chimeric SpoIVF proteins fused to the E. coli AP protein (encoded by phoA) or β-galactosidase (encoded by lacZ). This strategy for determining protein topology in membranes has been validated for a large number of membrane proteins and is based on the observation that AP is rendered active when exported across a phospholipid bilayer, while it remains inactive if retained in the cytoplasm. Conversely, LacZ is active when retained in the cytoplasm and is inactivated if transported across the membrane (15, 17).
To construct chimeric genes that express SpoIVFA or SpoIVFB fused to AP or LacZ, we first cloned defined segments of spoIVFA or spoIVFB in an E. coli plasmid. Our constructions provided an IPTG-inducible promoter (Pspac) to drive expression of the B. subtilis gene in E. coli. Next, we subcloned these constructs into the E. coli plasmid pMV100 (6), which contains the phoA gene, or pJBZ280, which contains lacZ, such that an in-frame fusion between spoIVFA-spoIVFB and phoA or lacZ was created.
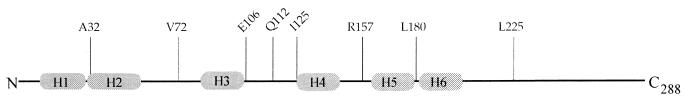
Expression of chimeric gene-AP was confirmed by Western blotting of E. coli cells with an anti-PhoA monoclonal serum. In total, we constructed four SpoIVFA-AP fusions and eight SpoIVFB-AP fusions; their positions are shown in Fig. 1 and 2, respectively.
FIG. 1.
Positions of four SpoIVFA-AP fusions.
FIG. 2.
Positions of eight SpoIVFB-AP fusions.
Since activity of the SpoIVFA-SpoIVFB processing complex has been proposed to be temperature sensitive (5), we analyzed AP activity at both 30 and 37°C and in solid and liquid media. For SpoIVFA at either 30 or 37°C, we found that the three AP fusions lying downstream of the single, putative transmembrane sequence of SpoIVFA (residues 73 to 90) were blue on agar plates containing the AP substrate BCIP, indicating that AP was active (Table 1). This was confirmed by growth in LB medium, where the level of AP-specific activity was 100 times greater than that in cells containing no fusion. In contrast, the one SpoIVFA-AP fusion (S72) which was white on a BCIP agar plate produced essentially no activity in liquid. The spoIVFA-lacZ at S72 fusion was positive for β-galactosidase (see Table 3), complementing the negative spoIVFA-AP S72 fusion. This demonstrated that the N terminus of SpoIVFA is in the cytoplasm and the C terminus of SpoIVFA transverses a phospholipid bilayer with at least 164 residues (residues 100 to 264) of SpoIVFA being exported to the outer face. Presumably, the hydrophobic sequence of amino acids (residues 73 to 90) anchors SpoIVFA in the phospholipid bilayer.
TABLE 1.
Analysis of SpoIVFA-AP fusions in E. coli at 30 and 37°Ca
| Temperature (°C) | Expressed proteins | Activity of AP insertion at the following position
|
|||
|---|---|---|---|---|---|
| S72 | Q100 | V217 | E264 | ||
| 30 | SpoIVFA-AP | 4 | 268 | 211 | 331 |
| SpoIVFA-AP + BofA | 6 | 365 | 161 | 463 | |
| 37 | SpoIVFA-AP | 1.3 | 137 | 138 | 99 |
| SpoIVFA-AP + BofA | 2 | 225 | 214 | 285 | |
AP was fused in frame after the amino acids indicated in SpoIVFA. Enzyme activity was measured in E. coli cultures expressing the chimeric protein alone or in the presence of the indicated coexpressed proteins. Mean specific AP activities (milliunits) from a minimum of four experiments are given. Background levels of activity found in E. coli CC118 cells (approximately 4 to 5 milliunits) containing the promoterless phoA plasmid, pMV100, have been subtracted.
TABLE 3.
Qualitative analysis of SpoIVFA and SpoIVFB LacZ fusionsa
| Protein expressed | Fusion position | Activity on agarb |
|---|---|---|
| SpoIVFA-LacZ | S72 | +++ |
| SpoIVFB-LacZ | A32 | − |
| V72 | +++ | |
| R157 | ++ | |
| L180 | − | |
| L225 | +++ | |
| A273 | +++ |
Activity is expressed as weakly (++) or strongly (+++) positive or negative (−), compared to a control strain which contained the promoterless lacZ plasmid pJBZ280 only. Activity was assessed following the introduction of DNA sequence-confirmed fusions into E. coli TG1 and grown on LB agar plates containing ampicillin (50 μg/ml), IPTG (32 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml).
We were unable to determine β-galactosidase activity in liquid culture for the A32 and L180 fusions due to premature lysis of E. coli cultures. We attribute this phenomenon to the disruption of the E. coli cell membrane caused by projection of LacZ through the phospholipid bilayer, and this may be a peculiarity of the SpoIVFB protein, although this has been noted for other LacZ protein fusions (1).
For SpoIVFB, our analysis showed that at either 30 or 37°C (Table 2) only two SpoIVFB-AP fusions were active, those with AP fused at A32 and L180. The available SpoIVFB-LacZ data (Table 3) complemented the SpoIVFB-AP data. The additional fusion of SpoIVFB-LacZ A273 was also positive, clearly positioning the C terminus of SpoIVFB in the cytoplasm. The membrane configuration predicted from this data is that of four transmembrane-spanning domains (4-transmembrane configuration), consisting of four transmembrane anchors, as shown in Fig. 3. In B. subtilis, this four-transmembrane conformation would dictate that both the N and C termini of SpoIVFB are positioned in the mother cell chamber of the sporulating cell, as shown in Fig. 3.
TABLE 2.
Analysis of SpoIVFB-AP fusions in E. coli at 30 and 37°Ca
| Expressed proteins | Temperature (°C) | Activity of AP insertion at the following position
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A32 | V72 | E106 | Q112 | I125 | R157 | L180 | L225 | ||
| IVFB-AP | 30 | 40 | 2 | 1 | 4 | 2 | 0 | 44 | 0 |
| 37 | 54 | 1.3 | 2.3 | 3.3 | 5.3 | 0 | 36 | 0.3 | |
| IVFB-AP + IVFA | 30 | 68 | 1 | 0.5 | 29 | 0.4 | 2 | 38 | 1 |
| 37 | 86 | 0.3 | 2.3 | 27 | 2.3 | 2.3 | 41.3 | 0 | |
| IVFB-AP + BofA | 30 | 173 | 5 | 2 | 19 | 8 | 0 | 150 | 0 |
| 37 | 236 | 0.4 | 0.5 | 6.5 | 5.5 | 0 | 85 | 1.5 | |
| IVFB-AP + IVFA + BofA | 30 | 219 | 3 | 1 | 44 | 1 | 3 | 80 | 0 |
| 37 | 198 | 1.5 | 1.5 | 28.5 | 2 | 3 | 79 | 1 | |
| IVFB-AP + IVFAΔ164 | 30 | 49 | 15 | 14 | |||||
| 37 | 64.3 | 27.3 | 24.3 | ||||||
| IVFB-AP + IVFAΔ164 + BofA | 30 | 194 | 35 | 52 | |||||
| 37 | 224 | 26 | 66 | ||||||
| IVFB-AP + IVFA(BofB8) + BofA | 30 | 4 | 48 | 2 | |||||
| 37 | 2 | 29 | 10 | ||||||
| IVFB-AP + IVFAΔ91 | 30 | 38 | 1 | 49 | |||||
| 37 | 45 | 0 | 63 | ||||||
| IVFB-AP + IVFAΔ67 | 30 | 83 | 24 | 39 | |||||
| 37 | 102 | 23 | 58 | ||||||
AP was fused in frame after the amino acids indicated in SpoIVFB. Enzyme activity was measured in E. coli cultures expressing the chimeric protein alone or in the presence of the indicated coexpressed proteins. Mean specific AP activities (milliunits) from a minimum of four experiments are given. Background levels of activity found in E. coli CC118 cells (approximately 4 to 5 milliunits) containing the promoterless phoA plasmid, pMV100, have been subtracted.
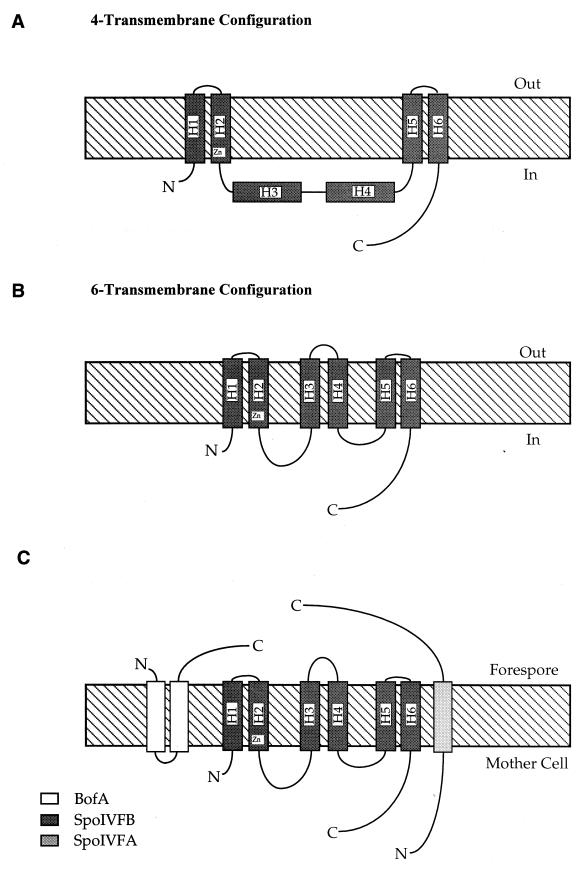
FIG. 3.
Membrane topology of processing complex. (A) Shows the 6-transmembrane configuration of SpoIVFB found in the absence of SpoIVFA. (B) Shows the 6-transmembrane configuration assumed by SpoIVFB in the presence of SpoIVFA. (C) Shows the conformation of the pro-ςK processing complex comprising SpoIVFA, BofA, and the putative protease, SpoIVFB, deduced from this work and that of Varcamonti et al. for BofA topology (26). Hydrophobic membrane-spanning segments (H1 to H6) are numbered where appropriate, and the putative Zn2+-binding domain of the SpoIVFB Zn2+ metalloprotease is shown.
Membrane topology of the putative ςK protease is stabilized by SpoIVFA and BofA.
Since the putative pro-ςK processing enzyme, SpoIVFB, is thought to form a hetero-oligomeric complex together with SpoIVFA and BofA proteins (5), we coexpressed SpoIVFB-AP fusions with SpoIVFA and BofA.
To accomplish this, we constructed a further series of E. coli expression vectors where the complete spoIVF operon was cloned adjacent to an E. coli-recognized and IPTG-inducible promoter, Pspac, and with phoA fused at eight positions to the spoIVFB ORF. Apart from the C-terminal phoA fusion and the Pspac promoter, the spoIVFA and spoIVFB cistrons were identical in structure to that of the native operon. This is important since the spoIVFA and spoIVFB ORFs overlap and it has been proposed that these cistrons are translationally coupled to ensure the correct stoichiometric balance of the SpoIVFA and SpoIVFB polypeptides during sporulation (5). Analysis of SpoIVFB-AP activity in the presence of SpoIVFA revealed that at either 30 or 37°C the Q112 SpoIVFB-AP fusion produced active AP in both liquid and solid media (Table 2). Since the Q112 SpoIVFB-AP was inactive in the absence of SpoIVFA, we suggest that SpoIVFB, in the presence of SpoIVFA, can assume a 6-transmembrane configuration (Fig. 3), requiring six transmembrane anchors in the phospholipid bilayer. Interestingly, the E106 and I125 AP insertions, constructed to map the boundaries of the transmembrane-spanning regions surrounding Q112, did not show AP activity. This suggests that the transmembrane domains H3 and H4 are clustered closer together than previously predicted (5), and hence, the AP moiety was not being efficiently translocated. Our data show that SpoIVFB assumes a 6-transmembrane configuration, which agrees with the computer-generated topological model of SpoIVFB proposed previously (5). Importantly, our results show that this conformation is dependent upon the presence of SpoIVFA.
Next, we coexpressed these eight SpoIVFA-SpoIVFB-AP fusions together with the third member of the pro-ςK processing complex, BofA. To achieve simultaneous expression of BofA, we used a second autonomously replicating plasmid (with a p15A replicon) containing the bofA gene under the control of the Pspac promoter (see Materials and Methods). As shown in Table 2, coexpression of BofA at 30 or 37°C resulted in the 6-transmembrane configuration of SpoIVFB. In each case, we observed that the relative levels of AP activity were greater (>2-fold) in the presence of BofA than in its absence. Since different plasmid replicons were used to express bofA and the spoIVFA-spoIVFB-phoA genes, we can not state that the stoichiometric levels of BofA, SpoIVFA, and SpoIVFB were balanced or equivalent to that which might be expected in B. subtilis. However, our results do suggest that BofA promotes the 6-transmembrane configuration of SpoIVFB.
We also coexpressed BofA in E. coli together with the eight SpoIVFB-AP fusions but in the absence of SpoIVFA. Although, as mentioned above, we cannot assume equivalent stoichiometric levels of BofA and SpoIVFA as in B. subtilis, coexpression of BofA did appear to promote the 6-transmembrane configuration of SpoIVFB at 30 but not at 37°C (Table 2).
Coexpression of BofA with SpoIVFA-AP fusions resulted in enhanced levels of AP activity of the AP-positive fusions. This result, as with the coexpression of BofA with SpoIVFB, shows that BofA has a positive effect on the assembly of SpoIVFA and SpoIVFB. Anti-PhoA Western blotting demonstrated that coexpression of BofA with SpoIVFA-PhoA did not show an enhanced accumulation of the SpoIVFA-PhoA chimeras, compared to the expression of SpoIVFA-PhoA chimeras alone (data not shown). This may suggest that BofA enhances the PhoA activity by aiding the translocation and assembly of SpoIVFA into the membrane, not the accumulation of total chimeric protein. Resnekov (19) has shown in vegetative B. subtilis cells that BofA enhances the accumulation of SpoIVFA by preventing its degradation. The disparity between these two results may be because the PhoA moiety artificially protects SpoIVFA from degradation when BofA is not present.
The membrane topology of BofA has been established in E. coli and predicts that in B. subtilis two transmembrane segments project the hydrophobic C terminus of BofA into the space between the IFM and OFM (Fig. 3). Although no temperature-sensitive alleles have been identified in bofA, it is possible that BofA is also unstable. Such instability might, for example, prevent the C terminus of BofA from transversing the membrane. We asked whether the SpoIVFA and/or SpoIVFB polypeptides affect the topology of BofA. By coexpressing either SpoIVFB or SpoIVFA and SpoIVFB with each of four BofA-AP fusions used in the study of Varcamonti et al. (26), we were unable to detect any change in the topology of BofA in E. coli membranes (data not shown). We conclude that BofA topology is intrinsically stable and is independent of the products of the spoIVF operon.
Functional domains of SpoIVFA.
Previous genetic experimentation has suggested that SpoIVFA has two functions (5): positively, to stabilize SpoIVFB in the membrane and, negatively, to inhibit the proteolytic activity of SpoIVFB until the appropriate signal (SpoIVB) is received from the forespore chamber. Data, as presented in this paper, show that SpoIVFA is required to stabilize the 6-transmembrane configuration of SpoIVFB.
To examine the positive function of SpoIVFA, we constructed plasmid vectors where a modified spoIVF operon was fused to AP in three positions. AP was fused to spoIVFB at positions A32, Q112, and L180. In each case, the spoIVFA cistron contained an in-frame deletion of codons 100 to 264 (spoIVFAΔ164). This construct was expressed by using the Pspac promoter and would encode a polypeptide of 100 residues, including the hydrophobic transmembrane sequence (residues 73 to 90). Analysis of these SpoIVFAΔ164-SpoIVFB-AP fusions in the presence of BofA in E. coli (Table 2) revealed that SpoIVFB assumed a 6-transmembrane configuration. This shows that to stabilize SpoIVFB in a 6-transmembrane configuration, only the first 100 amino acids of SpoIVFA are required. Since these residues are exposed to the inner face of the membrane, this stabilization may require interaction between the N-terminal SpoIVFA domain in the cytoplasm and the hydrophobic transmembrane region.
To further limit the region of SpoIVFA required for stabilization of SpoIVFB, we created two more deletions of spoIVFA, spoIVFAΔ91 and spoIVFAΔ67. To achieve this, we used the three spoIVFA-spoIVFB-AP fusions at positions A32, Q112, and L180 and constructed an in-frame deletion of spoIVFA between residues 1 and 92 (spoIVFAΔ91), deleting the transmembrane region, and an in-frame deletion of spoIVFA between residues 3 and 70 (spoIVFAΔ67), which retains the transmembrane region and the C terminus of spoIVFA. The truncated SpoIVFAΔ91 product produced was unable to transverse or translocate the membrane. AP activity in the three strains (Table 2) showed that SpoIVFB assumed a 4-transmembrane configuration at either 30 or 37°C. Conversely, the SpoIVFAΔ67 product was able to translocate the membrane, and all three of the SpoIVFB-AP fusions were positive at 30 and 37°C (Table 2). This indicates that SpoIVFB assumes a 6-transmembrane configuration. These results show, first, that the insertion of SpoIVFA through the membrane is required for the 6-transmembrane configuration of SpoIVFB and, second, that the transmembrane domain of SpoIVFA (residues 72 to 90) is primarily responsible for the stabilization of the 6-transmembrane configuration of SpoIVFB.
The proposed negative function of SpoIVFA stems from the observation that bofB alleles in the extreme C terminus of SpoIVFA allow constitutive activity of SpoIVFB (5). This disruption of the SpoIVFA C terminus may allow premature access of the signalling molecule to the SpoIVFB protease, allowing SpoIVFB to be responsive to the forespore-derived signal or simply promote a conformational change in SpoIVFB, rendering it active. Since our work has shown that SpoIVFB can assume at least two quite distinct conformations in the membrane, we asked what topological conformation SpoIVFB would assume in the presence of SpoIVFA carrying a bofB8 allele (a nonpolar, nonsense mutation at codon 259 of spoIVFA). In other words, is the change of SpoIVFB from a 6-transmembrane configuration to a 4-transmembrane configuration the mechanism responsible for activating pro-ςK processing? We used three pMV100-derived plasmids containing the spoIVF operon carrying the bofB8 allele fused to Pspac and with phoA fused after codons 106, 112, and 125 of SpoIVFB. Analysis of these fusions in E. coli revealed that SpoIVFB assumes a 6-transmembrane configuration at both 30 and 37°C, since the Q112-AP fusion was active and must therefore be exported to the outer face of the phospholipid bilayer (Table 2). These results imply, but do not prove, that disruption of the C terminus of SpoIVFA while producing a Bof phenotype (constitutive processing of pro-ςK) does not lead to a detectable conformational change in SpoIVFB.
Temperature sensitivity.
Two spoIVFA mutations exist which are temperature sensitive (4): spoIVFA181, which is a nonsense allele at codon 100 and truncates the C terminus of SpoIVFA, and spoIVFAΔ91, which encodes SpoIVFA with an in-frame deletion of residues 2 to 92. Both alleles are Spo+ at 30°C but render cells Spo− at 37°C. SpoIVFA181 would contain the hydrophobic transmembrane sequence at positions 73 to 90 and so, like the Q100 SpoIVFA-AP fusion polypeptide (see above), should target and insert into a membrane. In contrast, SpoIVFAΔ91 lacks the membrane-targeting sequence, and the C-terminal fragment of SpoIVFA encoded by this truncated gene and which would normally be translocated across the membrane should remain in the cytoplasm. We have described above that SpoIVFB assumes a 6-transmembrane configuration in the presence of the spoIVFAΔ164 deletion (Table 2), this truncation being similar to the spoIVFA181 nonsense allele. In contrast, the spoIVFAΔ91 deletion mutant, by encoding a form of SpoIVFA unable to associate with the membrane, prevents the formation of the 6-transmembrane configuration in SpoIVFB (Table 2). Since both the spoIVFA181 and spoIVFAΔ91 mutants support sporulation at a permissive temperature, it must follow that both membrane configurations (four and six) are able to facilitate pro-ςK processing.
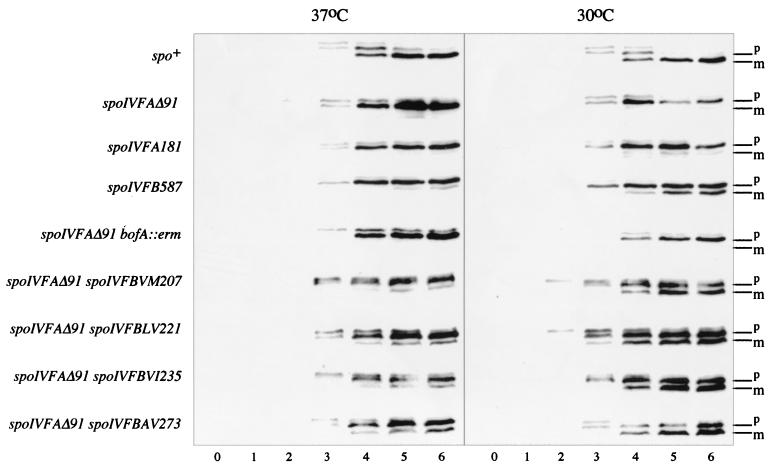
To address this, we examined processing of the transcription factor pro-ςK in sporulating cells of spo+, spoIVFA181, and spoIVFAΔ91 strains at 30 and 37°C. We found that at both the permissive and restrictive temperatures, very little levels of pro-ςK processing occurred as determined from Western blotting in the spoIVFA mutants (Fig. 4). In contrast, spore formation was reduced only 10-fold at the permissive temperature (Table 4). Since spores are produced in significant numbers at 30°C, we inferred that some ςK must be produced by proteolysis of pro-ςK, but this is not or is just detectable by immunoblotting (Fig. 4). We also examined pro-ςK processing in a double mutant, spoIVFAΔ91 bofA::erm (Fig. 4). Here again, pro-ςK processing cannot be detected, but at 30°C the strain is Spo+.
FIG. 4.
Pro-ςK processing in temperature-sensitive spoIVF strains. Sporulation was induced by resuspension at 30 and 37°C in strains PY79 (spo+), VO138 (spoIVFAΔ91), SC1256 (spoIVFA181), SC1257 (spoIVFB587), DG675 (spoIVFAΔ91 bofA::erm), SC1363 (spoIVFAΔ91 spoIVFBVM207), SC1365 (spoIVFAΔ91 spoIVFBLV221), SC1358 (spoIVFAΔ91 spoIVFBVI235), and SC1366 (spoIVFAΔ91 spoIVFBAV273). Samples were collected at hourly intervals (0 to 6) after the initiation of sporulation. Whole-cell extracts were prepared and analyzed with a polyclonal anti-pro-ςK serum. The unprocessed (p) and processed (m) forms of ςK are indicated by the bars. Occasionally, three bands were seen, the upper band being a nonspecific species caused by inadequate blocking.
TABLE 4.
Temperature-sensitive spoIVF allelesa
| Strain | Genotype | % Sporesb at
|
|
|---|---|---|---|
| 30°C | 37°C | ||
| PY79 | spo+ | 50 | 93 |
| SC1256 | spoIVFA181 | 12 | 0.41 |
| VO138 | spoIVFAΔ91 | 11 | 0.5 |
| SC1257 | spoIVFB587 | 58 | 8 |
| DG675 | spoIVFAΔ91 bofA::erm | 23 | 0.62 |
| SC1363 | spoIVFAΔ91 spoIVFBVM207 | 91 | 88 |
| SC1365 | spoIVFAΔ91 spoIVFBLV221 | 81 | 78 |
| SC1358 | spoIVFAΔ91 spoIVFBVI235 | 66 | 60 |
| SC1366 | spoIVFAΔ91 spoIVFBAV273 | 80 | 71 |
Strains were induced to sporulate at 30 or 37°C by the resuspension method (18).
At 9 h after the initiation of sporulation at 37°C and at 12 h after the initiation of sporulation at 30°C, samples were heated at 65°C for 45 min and the CFU per milliliter were measured. Spore counts are shown as a percentage of total, unheated CFU per milliliter.
It has been reported that some ςK-controlled genes are susceptible to very low threshold levels of ςK (14). In turn, it must also follow that the pro-ςK processing complex is active in the absence of the C terminus of SpoIVFA (in spoIVFA181 cells) as well as in the complete absence of the SpoIVFA protein in the membrane (in spoIVFAΔ91 cells) and SpoIVFA and BofA (in spoIVFAΔ91 bofA::erm cells). From our E. coli topological analysis, we predict that in spoIVFA181 cells SpoIVFB assumes a 6-transmembrane configuration, while in spoIVFAΔ91 and spoIVFAΔ91 bofA::erm cells, it assumes only a 4-transmembrane configuration. Interestingly, the complete absence of SpoIVFA in the membrane (i.e., in a spoIVFAΔ91 mutant) can be relieved by compensatory mutations in SpoIVFB. Four intragenic suppressor alleles have been identified which mutate residues 207, 221, 235, and 273, bypass the temperature sensitivity of a spoIVFAΔ91 mutant, and have been reported briefly in an earlier work (5). Here we show that pro-ςK processing of these mutants at 37°C is clearly restored, albeit at perhaps a slower rate than the wild type (Fig. 4). These missense alleles all lie within the C-terminal segment of SpoIVFB, which would be exposed to the mother cell chamber of the sporangial cell and may define a domain (between residues 207 and 273) that is required to interact with SpoIVFA. Supporting this, one temperature-sensitive allele, spoIVFB587, has been identified in this region (at position 226, producing a Pro to Leu change [5]). We found here that sporulation in spoIVFB587 cells was reduced 10-fold, and pro-ςK processing was reduced at the nonpermissive temperature (Table 4 and Fig. 4).
DISCUSSION
We have investigated the topology of the SpoIVFA and SpoIVFB polypeptides in a phospholipid membrane with the aim of defining the location and functional domains of the pro-ςK processing complex in B. subtilis. Our analysis has used an established method for topological analysis by using expression of gene-phoA and, to a lesser extent, gene-lacZ fusions in E. coli and analysis of AP and β-galactosidase activity encoded by the chimeric gene. At the outset, we must stress that any conclusions we draw are subject to the possibility, however remote, that the protein topology may be different in the outer forespore membranes of B. subtilis.
The most important finding from this work is that SpoIVFA is required to stabilize SpoIVFB, as is BofA, but to a lesser degree. This work shows that SpoIVFB, the putative zinc metalloprotease which cleaves pro-ςK, can assume two distinct conformations in a phospholipid membrane which we have referred to as the 4- or 6-transmembrane configurations. This may imply that SpoIVFB is intrinsically unstable, and in turn, this may be important for the signalling process. In the presence of SpoIVFA, SpoIVFB can integrate with six transmembrane segments, while in the absence of SpoIVFA, SpoIVFB is anchored by four transmembrane segments (Fig. 3). As a 6-transmembrane configuration, the topology of SpoIVFB and SpoIVFA is consistent with the topology predicted from a computer-based analysis of SpoIVFA-SpoIVFB (4).
A previous work, using a similar approach as that described here, has also defined the topology of the third member of the pro-ςK processing complex, BofA (26). This topology (shown in Fig. 3) shows that the small BofA protein anchors to the membrane with two transmembrane anchors, with its C and N termini exposed to the outer face of the membrane. With the obvious caveat mentioned above, we can propose a topological model (shown in Fig. 3) for the pro-ςK processing complex in the OFM of the developing spore (or forespore) comprising the three protein components of the complex (note that in this model the space between the IFM and OFM is equivalent to the periplasmic space of E. coli).
Characterization of the spoIVF-AP fusions has allowed us to further refine the domain functions of SpoIVFA and SpoIVFB. Analysis of SpoIVFB-AP gene products in the presence of three different truncated forms of SpoIVFA (spoIVFAΔ164, spoIVFAΔ91, and spoIVFAΔ67) has revealed that the transmembrane region of SpoIVFA (residues 73 to 90) is the primary determinant for stabilizing SpoIVFB in the 6-transmembrane configuration. A deletion of either the C-terminal (SpoIVFAΔ164) or N-terminal (SpoIVFAΔ67) domain of SpoIVFA did not disrupt the 6-transmembrane configuration of SpoIVFB. A deletion of the N-terminal and transmembrane-spanning region did result in a loss of stability of SpoIVFB, producing a 4-transmembrane configuration. Because SpoIVFBQ112-AP was the only fusion profoundly affected by the presence or absence of SpoIVFA, it is possible that the transmembrane domain of SpoIVFA interacts with the third transmembrane domain (H3) of SpoIVFB.
The observation that SpoIVFB had two distinct membrane conformations lead us to believe initially that a conformational change from a 6- to 4-transmembrane configuration might be involved in the activation of SpoIVFB proteolytic activity. To this end, we asked whether the bofB8 allele, which uncouples the ςK checkpoint by allowing constitutive processing of pro-ςK, might cause a change to the membrane topology of SpoIVFB. Analysis of the three AP fusions positioned between H3 and H4 showed no change in activity when the C-terminal tip of SpoIVFA was deleted (BofB8). This indicated, but does not rule out the possibility, that in vivo activation of SpoIVFB does not involve a change in its membrane topology from six- to four-transmembrane domains.
What then is the significance of this apparent instability of SpoIVFB and its reliance on the presence of SpoIVFA to stabilize it? Does this simply imply that in vivo SpoIVFB requires an additional protein (SpoIVFA) to stabilize it, or does the instability of SpoIVFB implicate that this fluid or flexible region is important in transducing the forespore signal?
In terms of the biological significance of the two SpoIVFB topologies, spores can be produced when SpoIVFB is in its 4-transmembrane configuration. We have found that at 30°C in spoIVFA181 (6-transmembrane configuration), spoIVFAΔ91, and spoIVFAΔ91 bofA::erm (4-transmembrane configuration) temperature-sensitive mutants, spores are produced, but very low levels of pro-ςK processing were observed. If our topological model is correct, then we predict that in both the configurations SpoIVFB is proteolytically active, albeit at very low levels. We cannot exclude the possibility, however, that in spoIVFAΔ91 and spoIVFAΔ91 bofA::erm cells at the nonpermissive temperature a low number of SpoIVFB molecules are able to assume the 6-transmembrane configuration sufficient to process pro-ςK.
In B. subtilis development, pro-ςK is synthesized in the outer (mother cell) chamber of the sporulating cell. The proteolytic processing complex is activated by receipt of the forespore signalling (the SpoIVB protein), which is synthesized in the opposed forespore chamber and is thought to be secreted through the IFM, where it then initiates processing of pro-ςK. Substantial evidence now exists that SpoIVFB is the processing enzyme, and a zinc-binding site has been identified (12) along with the putative third active residue, aspartate-137 (D137), by ourselves and others (L. Kroos, personal communication; D. Rudner and R. Losick, personal communication). Site-directed mutagenesis has also shown that E44 and D137 are both critical to processing of pro-ςK in vivo (L. Kroos, personal communication; D. Rudner and R. Losick, personal communication). Our topological analysis identifies both active residues to be located in transmembrane domains H2 and H4, respectively. The significance of the positioning of these residues, and the location of the catalytic site, may make sense since pro-ςK localizes to the membrane (28), where the cleavage site could be positioned in close proximity to the putative catalytic site of SpoIVFB. SpoIVFB has recently been assigned to a new and novel group of zinc metalloproteases that characteristically cleave their substrate at the cytosolic membrane face (10). From these studies, H2 and H4 would form a membrane pocket which could initiate cleavage of pro-ςK.
How is SpoIVFB rendered active? The intrinsic instability of this molecule and its apparent membrane fluidity suggest that it could alter its conformation in the presence of pro-ςK and also in response to the forespore signalling molecule, SpoIVB. Perhaps we can not measure such a minute change experimentally, but we have evidence that this is at least a possibility.
ACKNOWLEDGMENTS
We thank Lee Kroos for the gift of plasmids pSL13, pSL14, and pSL31 and helpful comments prior to publication, Margerita Sacco for the gift of plasmids containing the bofA-phoA constructs, Dickon Alley for the lacZ-containing plasmid pJBZ280, and Phil Wakeley for making the ςK antibody.
This work was supported by grants from the MRC and BBSRC to S.M.C.
REFERENCES
- 1.Alexeyev M F, Winkler H H. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J Mol Biol. 1999;285:1503–1513. doi: 10.1006/jmbi.1998.2412. [DOI] [PubMed] [Google Scholar]
- 2.Brockman R W, Heppel L A. On the localisation of alkaline phosphatase and cyclic phosphodiesterase in Escherichia coli. Biochemistry. 1968;7:2554–2562. doi: 10.1021/bi00847a016. [DOI] [PubMed] [Google Scholar]
- 3.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 4.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 5.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. [Google Scholar]
- 8.Ireton K, Grossman A D. A developmental checkpoint couples the initiation of sporulation to DNA replication in Bacillus subtilis. EMBO J. 1994;13:1566–1573. doi: 10.1002/j.1460-2075.1994.tb06419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroos L, Zhang B, Ichikawa H, Nicco Yu Y-T. Control of ς factor activity during Bacillus subtilis. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 10.Lewis A P, Thomas P J. A novel clan of zinc-metalloproteases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 12.Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-ςK in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu S, Halberg R, Kroos L. Processing of the mother-cell ς factor, ςK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu S, Kroos L. Overproducing the Bacillus subtilis mother cell sigma factor precursor, pro-ςK, uncouples ςK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 19.Resnekov O. Role of the sporulation protein BofA in regulating activation of the Bacillus subtilis developmental transcription factor ςK. J Bacteriol. 1999;181:5384–5388. doi: 10.1128/jb.181.17.5384-5388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 21.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Stragier P, Bonamy C, Karmazyn-Campbelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 25.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 26.Varcamonti M, Marasco R, De Felice M, Sacco M. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-ςK processing. Microbiology. 1997;143:1053–1058. doi: 10.1099/00221287-143-4-1053. [DOI] [PubMed] [Google Scholar]
- 27.Youngman P, Perkins J, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]