Abstract
Objective
To study the role of hepatitis B virus with A1762T/G1764A double mutation in liver cirrhosis and hepatocellular carcinoma, and create a sensitive, fast, accurate assay for detection of A1762T/G1764A double mutation.
Methods
We developed an accurate and fast real-time amplification refractory mutation system to detect A1762T/G1764A double mutation. Cloned hepatitis B virus genome was used as a control. Assay sensitivity was determined by serial dilution and mixed template experiments. Specificity was determined by cross experiments with wild and mutant hepatitis B virus. Fifty clinical samples were tested by the real-time amplification refractory mutation system and the results were compared with sequencing.
Results
The real-time amplification refractory mutation system had a sensitivity of 100 copies of virus with these mutations, and 0.1% weak population virus with double mutation could be found in mixtures. A total of 50 randomly collected clinical samples were detected by real-time amplification refractory mutation system, and the results were consistent with those by DNA sequencing. Hepatitis B virus genotype C was more prevalent in 39 of 50 samples than genotype B (11 samples), and about 75% of genotype C carried a double mutation compared to 45% of genotype B. However, the percentage of A1762T/G1764A double mutation in hepatitis B e antigen-negative (58.3%) samples was almost the same as in hepatitis B e antigen-positive (61%) samples.
Conclusion
The real-time amplification refractory mutation system is sensitive and specific for detection of hepatitis B virus double mutation.
Keywords: Hepatitis B virus, Basal core promoter mutations, Amplification refractory mutation system, PCR
Introduction
Chronic infection with hepatitis B virus (HBV) is one of the major serious risk factors for liver cirrhosis and hepatocellular carcinoma (HCC). HBV infection is associated with up to 80–90% of HCC patients in the Asia–Pacific region.1 Eight HBV genotypes (A–H) have been identified based on the divergence over the entire HBV genomic sequence.2, 3 In China, the most prevalent genotypes of HBV are B and C. Most of the HBV infection cases in China are chronic.3, 4, 5 In addition, HBV genotype C can carry a higher risk for liver cirrhosis and HCC.6, 7, 8, 9
Among the many factors that may contribute to pathogenesis and carcinogenesis,10 scientists have found that HBV A1762T/G1764A double mutation is a predictive biomarker for HCC development.8, 9, 11, 12 This mutation is involved in the mechanism of infection with hepatitis B e antigen (HBeAg)-negative virus.13, 14 A1762T/G1764A double mutation is more prevalent in HBV genotype C, and correlates with increased replication capacity.6 Furthermore, HBV genotype C with 1762T/1764A mutations is associated with a higher risk of HCC, and this association is independent of serum HBV DNA level.6, 15 A recent study has indicated that quantification of the A1762T/G1764A mutant virus helps predicting the risk of HCC.16, 17, 18 Currently, A1762T/G1764A mutation is routinely detected by nucleotide sequencing of PCR products. This method can only detect the mutant viruses when it comprises at least 25% of the total virus population.19 Although a sensitive, fast, and accurate assay for detecting A1762T/G1764A double mutation has been reported, it was a laboratory-specific assay and could not be widely used in other laboratories.19
In this study, we report an amplification refractory mutation system (ARMS) PCR for detecting A1762T/G1764A double mutation.
Materials and methods
Patient samples and DNA purification
A total of 50 patient serum samples were randomly retrieved from the Third People's Hospital of Changzhou. HBV viral DNA was isolated from 200 μL patient serum samples using the QIAamp DNA blood mini kit (Qiagen) and eluted into 50 μL buffer according to manufacturer's instructions.
Plasmid standards and other reagents
Full-length HBV genome was cloned according to the method of Parekh et al.18, 20 Briefly, clinical samples with chronic HBV infection were used. HBV DNA isolated by commercial kits was amplified by PCR in 50 μL buffer containing 5 μL 10× LAmp buffer, 20 μM dNTP, 0.3 μM each primer (p1 and p2), 2.5 mM Mg2+ and 5 U LAmp DNA Polymerase (Beijing Cowin Bioscience Co. Ltd., Beijing, China). PCR was performed using a BioRad Mycyle PCR (Bio-Rad, Hercules, CA, USA) under the following conditions: 94 °C for 2 min, followed by 94 °C for 30 s, 60 °C for 1 min, and 72 °C for 3 min for 35 cycles, with a final extension of 5 min. The PCR products were purified (Axygen Scientific, Shanghai, China) and cloned into the pGEM-T Easy Vector System (Promega, Madison, WI, USA). The plasmid was identified by sequencing. The clones that carried the A1762T/G1764A double mutation, A1762T and G1764A single mutations, and wild template were used as the assay standards and quantified by using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
Direct PCR sequencing
For identification of the double mutation in the basal core promoter (BCP) region, HBV DNA was amplified and sequencing using the p3 and p4 primers and LAmp DNA Polymerase. The thermal program was 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, for a total of 40 cycles. The PCR products were separated by electrophoresis, purified using a Qiagen gel extraction kit, and sequencing was done with the BigDye terminator cycle-sequencing reaction kit and Prism 3730 DNA analyzer (Life Technology, Shanghai, China).
Real-time amplification refractory mutation system (RT-ARMS)
RT-ARMS was developed for detection of the A1762T/G1764A double mutation. The RT-ARMS was performed in a 50-μL reaction mixture containing: 0.15 μM ARMS primer (p6), 0.15 μM forward primer (p5), 0.15 μM TaqMan probe (p8), optimized concentration of Mg2+ ions, dNTP and polymerase mix (Takara Premix Ex Taq, TaKaRa Bioech, Dalian, China). The cycle conditions were: 95 °C for 10 min, 10 cycles of 95 °C for 20 s, 52 °C for 40 s, 72 °C for 30 s, followed by: 35 cycles of 95 °C for 20 s, 54 °C for 40 s, and 72 °C for 30 s. The PCR was carried on using 7500 real-time PCR (Applied Biosystems).
Results
Primer and probe design
To avoid confusion of base differences among genotypes A–E, a total of 839 full-length HBV genome sequences were downloaded from GenBank sequence database and analyzed by MEGA 4.0 (http://www.ncbi.nlm.nih.gov). According to the ARMS technique, the reverse primer specifically designed for detecting A1762T/G1764A double mutation, an additional mismatch at the n−1 position next to the 3′-end base of the ARMS primer decreased the amplification efficiency of the single mutation and wild target sequence to a greater extent than that of the double mutation target sequence. For the signal system, TaqMan probe was used instead of molecular beacon (Table 1). Primers and probes were designed according to the primer and TaqMan probe design requirements. After 48 experiments screening the ARMS primer, the reverse primer (p6 in Table 1) with G:G mismatch at the n − 1 position was better than others for detection of A176T/G1764A double mutation (data not shown). The reaction conditions were selected after optimization of the concentrations of Mg2+ ions, primers and polymerase. The optimal primer annealing temperature for the matched template was 54 °C.
Table 1.
Primers and probes for detecting A1762/G1764A double mutation.
| Primer | 5′–3′ |
|---|---|
| Primer for full-length HBV clone | |
| p1 | CCGGAAAGCTTGAGCTCTTCTTTTTCACCTCTGCCTAATCA |
| p2 | CCGGAAAGCTTGAGCTCTTCAAAAAGTTGCATGGTGCTGG |
| Primer for A1762T/G1764A sequencing | |
| p3 | GGACTCTTGGACTCTCAGC |
| p4 | ATTTATGCCTACAGCCTCCTA |
| Primer and probe for RT-ARMS | |
| p5 | acCCGACCTTGAGGMRTACTT |
| p6 | ACAGCCTCCTAGTACAAAGATgA |
| p7 | TACAGCCTCCTAGTACAAAGATCA |
| p8 | Fam-TGGGAGGAGTTGGGGGAGGA-TAMRA |
Evaluation of sensitivity and specificity
To test the stability and sensitivity of RT-ARMS, a full-length sequence of HBV genome was cloned and used. Gradient-diluted double mutation template, single mutation template, wild and mixed template were performed separately to determine the sensitivity and specificity of the RT-ARMS. Five repeats with six serially diluted samples revealed that RT-ARMS showed a sensitivity of 100 copies/mL for detection of the double mutation and the Ct value was 31.06 ± 0.182; about one cycle ahead of detection of 107 copies/mL of non-double mutation template. When RT-ARMS was used to amplify 107 copies/mL A1762T single mutation template, the Ct value was 32.33 ± 0.24 (Fig. 1). For detection of the same copies of G1764A single mutation template, the Ct value was 32.1 ± 0.27 and 33.23 ± 0.36 cycles for wild template (Fig. 1). There was a delay of >18 cycles in comparison with amplification of double mutant template at the same concentration (Table 2). For higher concentrations (such as 108 and 109 copies/mL) of single mutation and wild templates, the Ct value was the same as for detection of 107 copies/mL, with <0.5 cycles fluctuation. When we detected a low concentration of non-double-mutation templates, such as 106, 105, and 104 copies/mL, the Ct value was >34 cycles or underdetermined (data not shown).
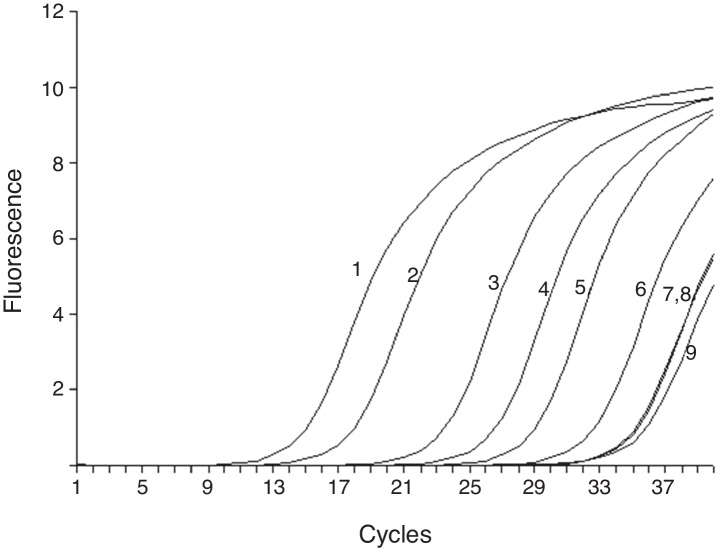
Fig. 1.
Sensitivity of RT-ARMS detection of A1762T/G1764A double mutation template. The assay could detect 100 copies/mL A1762T/G1764A double mutation (6). 1–6 were 107, 106, 105, 104, 103 and 100 copies/mL of A1762T/G1764A double mutation template; and 7–9 were 107 copies/mL A1762T and G1764A single mutation and wild template.
Table 2.
Ct value of amplification of different concentrations of A1762T/G1764A double mutation template, single mutation template and wild template by RT-ARMS.
| A1762T/G1764A | A1762T | G1764A | Wild | |||
|---|---|---|---|---|---|---|
| Copies/mL | 1.00E+07 | 1.00E+03 | 1.00E+02 | 1.00E+07 | 1.00E+07 | 1.00E+07 |
| Ct value | 13.57 ± 0.60 | 28.28 ± 0.18 | 31.06 ± 0.17 | 32.33 ± 0.24 | 32.01 ± 0.26 | 33.29 ± 0.36 |
To confirm the threshold Ct value, a large number of repeats of 107 copies/mL single mutation template, 107 copies/mL wild template, 103 copies/mL double mutation template, and 107 copies/mL mixed plasmid containing 103–106 copies/mL double mutation were tested by RT-ARMS. We obtained a threshold Ct value of 28 cycles, therefore confirming development of a sensitive and accurate RT-ARMS.
To evaluate the distinguishing ability of the assay, 3 × 107 copies/mL mixed population, which contained 107 copies/mL of each single mutation template, 107 copies/mL of wild template and different concentrations of double mutation template were tested. We found 0.1% (∼3 × 104 copies/mL) of double mutant templates in the mixtures, and the Ct value was 28 ± 0.24, which was five cycles ahead of detection of 107 copies/mL wild template (Table 3), and the results matched the standard Ct value.
Table 3.
RT-ARMS for detecting weak populations with A1762T/G1764A double mutation in the mixture.
| 100% | 10% | 1% | 0.10% | 0.01% | Wild | |
|---|---|---|---|---|---|---|
| Copies/mL | 1.00E+07 | 1.00E+06 | 1.00E+05 | 1.00E+04 | 1.00E+03 | 1.00E+07 |
| Ct value | 13.57 ± 0.60 | 28.28 ± 0.18 | 31.06 ± 0.17 | 32.33 ± 0.24 | 32.01 ± 0.26 | 33.29 ± 0.36 |
Detection of clinical samples
A total of 50 clinical samples were collected from HBV-infected patients and analyzed by the RT-ARMS and sequencing in a double-blind manner in two different laboratories. The results are summarized in Table 4. Twenty-eight samples (56%) were A1762/G1764A dual mutation by RT-ARMS and sequencing. Three samples were identified with double mutations by sequencing an additional TA clone, but the concentration was outside the limit of the RT-ARMS. Another three samples had double mutations by RT-ARMS, but could not be detected by sequencing due to low mutant copy numbers contained in the HBV population. Of all 50 patients, 40 were diagnosed clinically; there were 27 patients with chronic hepatitis B and 13 with severe liver disease (seven patients with cirrhosis, one primary hepatic carcinoma, one hepatic encephalopathy, and four with chronic severe hepatitis). Eleven out of 13 patients (84%) with severe liver disease carried A1762T/G1764A double mutation. The genotypes of the samples were tested by sequencing, and 11 (22%) were genotype B and 39 (78%) were genotype C. Among those with A1762T/G1764A double mutation, there were 45% of genotype B and 74% of genotype C.
Table 4.
Comparison of the results of ARMS and sequencing for detecting hepatitis B virus A1762T/G1764A mutant.
| Results of ARMS | Results of sequencing |
|
|---|---|---|
| Double mutation | Single mutation or wild type | |
| Double mutation (n = 31) | 28 | 3 |
| Single mutation or wild type (n = 19) | 3 | 16 |
Discussion
Accumulated evidence shows that HBV carriers with A1762T/G1764A double mutation in the BCP region are significantly associated with HCC. Patients with these mutations die at an earlier age from HCC than those without the mutations.6, 7, 8, 9 Beyond that, there is ∼70% increased risk of HCC in individuals with these mutations and total HBV plasma level of >10 000 copies/mL.16 The A1762T/G1764A double mutation is not only a predictive biomarker for HCC development, but also a prognostic indicator of life expectancy of the patient with HCC.8 Thus, it is very important to detect these mutations.
Detection of HBV BCP mutations by using a TaqMan real-time PCR and LNA-blocker Simple-Probe-based real-time PCR has been previously reported.16, 19, 21 Since there are more single nucleotide polymorphisms (SNPs) next to A1762T and G1764A double mutations TaqMan real-time PCR is not a good choice of method to detect these mutations. Although it is easy to avoid this by using the LNA-clamp PCR assay, it is a two-step PCR assay and only available for research use.19 The INNO LiPA assay is simple and accurate, but the entire assay from DNA extraction to development of color reaction is completed within 12 h, and the hybridization is complex. Detection of HBV mutations has been greatly dependent on direct DNA sequencing, which is considered to be the gold standard. However, DNA sequencing only distinguishes mutations >25% in mixed virus populations.
RT-ARMS is an accurate and stable assay and widely used to detect SNPs and mutations.22, 23, 24, 25, 26, 27, 28 We developed an RT-ARMS to detect A1762T/G1764A double mutation, even at concentrations below the currently used thresholds. The RT-ARMS enabled the monitoring of double mutations in the BCP region. The limitation of the assay is that novel polymorphisms not yet registered in the GenBank sequence database are not incorporated within the primer design. The RT-ARMS had high sensitivity, specificity and stability for detecting these mutations, and ∼103 copies/mL double mutation templates and 0.1% percent double mutation template contained in 3 × 107 copies/mL mixture could be detected. Although it is not as sensitive as PNA-LNA-Clamp PCR (PBPPO), the RT-ARMS does not need nested PCR, and the assay effectively eliminates single mutation interference, which other assays do not do. As a result, the RT-ARMS is a functional, simple and accurate assay that can be used in the clinical laboratory for detecting HBV A1762T/G1764A double mutation.
Patients have a high risk of severe liver disease in Changzhou Jiangsu province, China, because the A1762T/G1764 mutations and HBV genotype C are highly prevalent in this area. It is a major concern that patients with HBV genotype C infection carrying these mutations have a high risk of HCC.6, 7, 8, 9, 13, 29 Our results matched those in a recent report.30 Among 13 patients with severe liver disease, 11 of them tested positive for A1762T/G1764A double mutation, and seven of them had these two markers. Although the patient sample size is small, these mutations in the BCP region are found in patients with serious liver disease such as cirrhosis and post-hepatitis B cirrhosis. These mutations could be an indicator of serious liver disease development.29 However, among HBeAg-positive and negative patients, the percentage with the double mutation was almost the same, which differs from previously reported data.31 This suggests some divergence in different cities or provinces in China and other areas.
Although it is difficult to design an RT-PCR to detect A1762T/G1764A double mutation,19 we developed an RT-ARMS that has the potential for early and accurate detection of minor viral populations, before they emerge as major viral populations that may influence the natural history of HBV infection. In conclusion, the RT-ARMS represents a simple but efficient method for the identification of minor variants in mixed viral populations and may provide a helpful tool in monitoring BCP double mutation.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Zhao Y., Zhang X.Y., Guo J.J., et al. Simultaneous genotyping and quantification of hepatitis B virus for genotypes B and C by real-time PCR assay. J Clin Microbiol. 2010;48:3690–3697. doi: 10.1128/JCM.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidd-Ljunggren K., Miyakawa Y., Kidd A.H. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 3.Liaw Y.F., Lau G.K., Kao J.H., Gane E. Hepatitis B e antigen seroconversion: a critical event in chronic hepatitis B virus infection. Dig Dis Sci. 2010;55:2727–2734. doi: 10.1007/s10620-010-1179-4. [DOI] [PubMed] [Google Scholar]
- 4.Liaw Y.F., Chu C.M. Hepatitis B virus infection. Lancet. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 5.Yang H.I., Sherman M., Su J., et al. Nomograms for risk of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. J Clin Oncol. 2010;28:2437–2444. doi: 10.1200/JCO.2009.27.4456. [DOI] [PubMed] [Google Scholar]
- 6.Qin Y., Tang X., Garcia T., et al. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol. 2011;85:10167–10177. doi: 10.1128/JVI.00819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacke F., Gehrke C., Luedde T., Heim A., Manns M.P., Trautwein C. Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of lamivudine-resistant mutants. J Virol. 2004;78:8524–8535. doi: 10.1128/JVI.78.16.8524-8535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuang S.Y., Jackson P.E., Wang J.B., et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci U S A. 2004;101:3575–3580. doi: 10.1073/pnas.0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuang S.Y., Lekawanvijit S., Maneekarn N., et al. Hepatitis B 1762T/1764A mutations, hepatitis C infection, and codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol Biomarkers Prevent. 2005;14:380–384. doi: 10.1158/1055-9965.EPI-04-0380. [DOI] [PubMed] [Google Scholar]
- 10.Block T.M., Mehta A.S., Fimmel C.J., Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 11.Fang Z.L., Sabin C.A., Dong B.Q., et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Tanaka Y., Huang Y., et al. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laskus T., Rakela J., Nowicki M.J., Persing D.H. Hepatitis B virus core promoter sequence analysis in fulminant and chronic hepatitis B. Gastroenterology. 1995;109:1618–1623. doi: 10.1016/0016-5085(95)90651-7. [DOI] [PubMed] [Google Scholar]
- 14.Lindh M., Horal P., Dhillon A.P., Furuta Y., Norkrans G. Hepatitis B virus carriers without precore mutations in hepatitis B e antigen-negative stage show more severe liver damage. Hepatology. 1996;24:494–501. doi: 10.1002/hep.510240305. [DOI] [PubMed] [Google Scholar]
- 15.Yang H.I., Yeh S.H., Chen P.J., et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan J.M., Ambinder A., Fan Y., Gao Y.T., Yu M.C., Groopman J.D. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol Biomarkers Prevent. 2009;18:590–594. doi: 10.1158/1055-9965.EPI-08-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Zhang H., Gu C., et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parekh S., Zoulim F., Ahn S.H., et al. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie H., Evans A.A., London W.T., Block T.M., Ren X.D. Ultrasensitive quantification of hepatitis B virus A1762T/G1764A mutant by a simpleprobe PCR using a wild-type-selective PCR blocker and a primer-blocker-probe partial-overlap approach. J Clin Microbiol. 2011;49:2440–2448. doi: 10.1128/JCM.02472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther S., Li B.C., Miska S., Kruger D.H., Meisel H., Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang A., Yuen M.F., Yuan H.J., Lai C.L., Kwong Y.L. Real-time quantification of hepatitis B virus core-promoter and pre-core mutants during hepatitis E antigen seroconversion. J Hepatol. 2004;40:1008–1017. doi: 10.1016/j.jhep.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 22.Ntziora F., Paraskevis D., Haida C., et al. Quantitative detection of the M204V hepatitis B virus minor variants by amplification refractory mutation system real-time PCR combined with molecular beacon technology. J Clin Microbiol. 2009;47:2544–2550. doi: 10.1128/JCM.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacerra G., Musollino G., Di Noce F., Prezioso R., Carestia C. Genotyping for known Mediterranean alpha-thalassemia point mutations using a multiplex amplification refractory mutation system. Haematologica. 2007;92:254–255. doi: 10.3324/haematol.10736. [DOI] [PubMed] [Google Scholar]
- 24.Newton C.R., Graham A., Heptinstall L.E., et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura H., Kasahara K., Kawaishi M., et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 26.Bates J.A., Taylor E.J. Scorpion ARMS primers for SNP real-time PCR detection and quantification of Pyrenophora teres. Mol Plant Pathol. 2001;2:275–280. doi: 10.1046/j.1464-6722.2001.00074.x. [DOI] [PubMed] [Google Scholar]
- 27.Whitcombe D., Theaker J., Guy S.P., Brown T., Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 28.Kotoula V., Charalambous E., Biesmans B., et al. Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics. PLoS ONE. 2009;4:e7746. doi: 10.1371/journal.pone.0007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu C.M., Lin C.C., Chen Y.C., Jeng W.J., Lin S.M., Liaw Y.F. Basal core promoter mutation is associated with progression to cirrhosis rather than hepatocellular carcinoma in chronic hepatitis B virus infection. Br J Cancer. 2012;107:2010–2015. doi: 10.1038/bjc.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan C.H., Zhao C.Y., Ding H., et al. Hepatitis B virus basal core promoter mutations A1762T/G1764A are associated with genotype C and a low serum HBsAg level in chronically-infected HBeAg-positive Chinese patients. Antiviral Res. 2012;96:108–114. doi: 10.1016/j.antiviral.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Nie H., Evans A.A., London W.T., Block T.M., Ren X.D. Quantitative dynamics of hepatitis B basal core promoter and precore mutants before and after HBeAg seroconversion. J Hepatol. 2012;56:795–802. doi: 10.1016/j.jhep.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]