Abstract
Phenylacetate-coenzyme A ligase (PA-CoA ligase; AMP forming, EC 6.2.1.30), the enzyme catalyzing the first step in the aerobic degradation of phenylacetate (PA) in Azoarcus evansii, has been purified and characterized. The gene (paaK) coding for this enzyme was cloned and sequenced. The enzyme catalyzes the reaction of PA with CoA and MgATP to yield phenylacetyl-CoA (PACoA) plus AMP plus PPi. The enzyme was specifically induced after aerobic growth in a chemically defined medium containing PA or phenylalanine (Phe) as the sole carbon source. Growth with 4-hydroxyphenylacetate, benzoate, adipate, or acetate did not induce the synthesis of this enzyme. This enzymatic activity was detected very early in the exponential phase of growth, and a maximal specific activity of 76 nmol min−1 mg of cell protein−1 was measured. After 117-fold purification to homogeneity, a specific activity of 48 μmol min−1 mg of protein−1 was achieved with a turnover number (catalytic constant) of 40 s−1. The protein is a monomer of 52 kDa and shows high specificity towards PA; other aromatic or aliphatic acids were not used as substrates. The apparent Km values for PA, ATP, and CoA were 14, 60, and 45 μM, respectively. The PA-CoA ligase has an optimum pH of 8 to 8.5 and a pI of 6.3. The enzyme is labile and requires the presence of glycerol for stabilization. The N-terminal amino acid sequence of the purified protein showed no homology with other reported PA-CoA ligases. The gene encoding this enzyme is 1,320 bp long and codes for a protein of 48.75 kDa (440 amino acids) which shows high similarity with other reported PA-CoA ligases. An amino acid consensus for an AMP binding motif (VX2SSGTTGXP) was identified. The biochemical and molecular characteristics of this enzyme are quite different from those of the isoenzyme catalyzing the same reaction under anaerobic conditions in the same bacterium.
Phenylacetic acid and its mono- or dihydroxylated derivatives are formed in nature from a wide variety of natural as well as synthetic compounds. Their microbial biodegradation and biotransformation have received the attention of many research groups for a long time. A number of bacteria and fungi have been isolated and studied to reveal the metabolic pathways of these acids under both aerobic and anaerobic growth conditions.
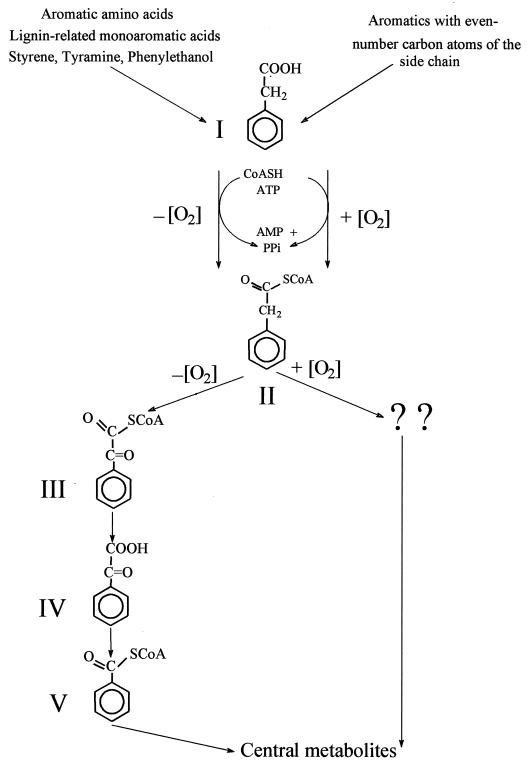
Recently, the complete anaerobic metabolism of phenylacetic acid in the bacterium Thauera aromatica has been shown to follow a novel α-oxidation of the side chain of the coenzyme A (CoA)-activated acid, leading to the formation of the central intermediate benzoyl-CoA (Fig. 1). Further metabolism of benzoyl-CoA subsequently leads to the formation of three acetyl-CoA molecules and one CO2 molecule (21, 22, 24, 31, 35, 38, 39).
FIG. 1.
Aerobic and anaerobic catabolic pathways of phenylacetic acid in A. evansii and T. aromatica. I, phenylacetic acid; II, PACoA; III, phenylglyoxyl-CoA; IV, phenylglyoxylate; V, benzoyl-CoA; CoASH, coenzyme A.
It has been established that the aerobic metabolism of most aromatic compounds starts by ring hydroxylation reactions carried out by mono- and dioxygenases. These oxic reactions, which are widely distributed in microorganisms, bring the aromatic rings of many aromatic compounds to the redox state present in catechol, protochatechuic and homoprotochatechuic acids, and gentisic and homogentisic acids. These compounds are considered common intermediates in the aerobic metabolism of most aromatic compounds (14, 17).
The aerobic metabolism of the hydroxylated derivatives of phenylacetic acid in many bacterial species follows this general pattern of oxic attack of the aromatic ring, leading to the formation of homogentisic acid (2,5-dihydroxyphenylactate) or homoprotocatechuic acid (3,4-dihydroxyphenylacetate) (4, 5, 12, 13, 28, 34, 41). However, none of the possible routes which might be expected for phenylacetic acid catabolism by analogy to those of its hydroxy derivatives appears to operate in phenylacetate (PA)-degrading bacteria. The inability to demonstrate the hydroxylation of PA by cell extracts of different bacteria in the presence of different cofactors has been reported by several research groups (7, 8, 10, 15, 25, 41, 43). Therefore, there is still uncertainty about the aerobic pathway followed by bacteria for phenylacetic acid metabolism, which has to be considered unknown.
Recently, it has been shown for Pseudomonas putida that the initial reaction in aerobic metabolism of PA is CoA thioesterification of the aromatic acid (29, 33), a reaction which is common in the anaerobic catabolism of aromatic compounds (20). CoA ligase activities against PA have also been reported for other bacterial species (45). Moreover, there is evidence for a role of aromatic-CoA thioesters in the aerobic metabolism of 2-aminobenzoate (1), benzoate (B) (2), and 4-chlorobenzoate (26, 37). The gene coding for phenylacetate-CoA ligase (PA-CoA ligase) has been recently sequenced from P. putida (33), Pseudomonas sp. strain Y2 (44), and Escherichia coli W (16); similar genes are present in Bacillus halodurans (42) and E. coli K-12 (9), suggesting that these bacteria are also able to oxidize PA via phenylacetyl-CoA (PACoA). However, PA-CoA ligase of P. putida is the only characterized gene product (29).
The β-subclass proteobacterium Azoarcus evansii has shown a versatile ability to utilize phenylacetic acid as a carbon and energy source under both aerobic and anaerobic growth conditions. A PA-CoA ligase involved in the anaerobic metabolism of this acid has been purified and characterized from this bacterium (30). During aerobic growth of A. evansii with PA, a PA-CoA ligase activity which was consistent with the rate of substrate utilization by whole cells was detected. There was some doubt as to whether this activity was due to the enzyme catalyzing the same reaction under anaerobic conditions in the same bacterium or to another isoenzyme which is specifically induced under aerobic conditions.
In this report, I describe the purification, characterization, and gene sequence of the isoenzyme catalyzing the formation of PACoA under aerobic conditions in A. evansii.
MATERIALS AND METHODS
Growth of bacteria.
A. evansii KB740 (DSM 6898 [3]) was grown under aerobic conditions in chemically defined medium on 5 mM PA, 4-hydroxyphenylacetate (4-OHPA), B, Phe, acetate, or adipate. The growth medium contained 40 mM potassium phosphate buffer (pH 7.4), 10 mM ammonium chloride, 0.1 mM calcium chloride, and 0.8 mM magnesium sulfate and was supplemented with SL-10 trace element solution and VL-7 vitamin solution as previously described (30). The medium was dispensed in Erlenmeyer flasks (300 ml/1-liter flask), inoculated, and incubated at 37°C with shaking (180 rpm). Anaerobic growth with PA plus nitrate, growth measurement, and cell harvest were as described previously (30). Growth on a large scale with phenylacetic acid was carried out in a 200-liter fermentor with stirring at 200 rpm and a flow of sterile air at 90 liters min−1. Cells were frozen in liquid nitrogen until used.
Preparation of cell extracts.
Frozen or fresh cells were suspended in Tris-HCl buffer, pH 7.8 (1 g of cells/2 ml of buffer), in the presence of DNase I, 2 mM MgCl2, 2 mM dithioerythritol (DTE), and 20% (wt/vol) glycerol. The cell slurry was passed twice through a French pressure cell at 137 MPa, and the lysate was centrifuged at 4°C for 2 h at 100,000 × g.
CoA ligase activity.
The activation of the aromatic acids to their corresponding CoA thioesters by CoA ligases was monitored in a coupled enzyme assay as described before (30). This assay couples the formation of AMP or ADP, which results from CoA ester formation, to an ATP-regenerating system containing myokinase, phosphoenol pyruvate, and pyruvate kinase, leading to the formation of pyruvate. The reduction of pyruvate by equimolar amounts of NADH to lactate in the presence of lactate dehydrogenase was monitored spectrophotometrically at 365 nm. A stoichiometry of 1 mol of NADH oxidized per mol of CoA thioester formed was taken as evidence for ADP formation, and a stoichiometry of 2 mol of NADH oxidized was taken as evidence for AMP formation. Cell extract precipitated at a 60% saturation of ammonium sulfate was used, since some interfering substances were present in the cell extract. Extracts of cells grown aerobically with PA, 4-OHPA, B, Phe, adipate, and acetate and anaerobically with PA plus nitrate were screened for these activities. The specific enzyme activity refers to micromoles of PACoA formed per minute per milligram of protein in the protein fraction precipitated at 60% ammonium sulfate. For identification of the products of the CoA ligase reaction, the assay mixture was acidified to pH 3.5 with 10% formic acid and centrifuged at 14,000 rpm (Eppendorf centrifuge) and the supernatant was analyzed by high-performance liquid chromatography (HPLC). The products of the reaction were also subjected to alkaline hydrolysis in KOH (pH 10 at 70°C for 30 min), and the liberated free acids were determined by HPLC. In other assays [C1-14C]PA was used and the formed labeled CoA thioester was identified by autoradiography after separation on thin-layer chromatography (TLC) plates.
In vivo formation of PACoA.
To demonstrate the formation of PACoA in whole A. evansii cells as a result of the activity of PA-CoA ligase, cell suspensions (2 ml, with an optical density at 578 nm of about 25) of PA- and Phe-grown cells were suspended in growth medium lacking a carbon source. These cells were fed with 0.5 mM PA plus 37 kBq of [14C]PA or 0.5 mM Phe plus 37 kBq of [14C]Phe and incubated at 37°C under aeration. At different time intervals (0 and 30 s and 1, 2, and 5 min) samples of 300 μl were withdrawn and rapidly centrifuged at 4°C for 5 min at 14,000 rpm. The cell pellet was washed once with ice-cold buffer lacking an aromatic substrate and extracted with hot acidic ethanol (pH 4, 80°C). The ethanolic extract was evaporated, and the resulting residue was dissolved in 50 μl of H2O and analyzed by HPLC and TLC for the presence of PACoA.
Purification of PA-CoA ligase.
PA-CoA ligase was purified from 20 g of cells. In the following chromatographic steps, the equilibration buffer used contained 10 mM Tris-HCl (pH 7.8), 2 mM MgCl2, 2 mM DTE, and 10% (wt/vol) glycerol. The different protein fractions collected during the purification steps were tested for PA-CoA ligase activity by the coupled enzyme assay.
Anion-exchange chromatography on DEAE Bio-Gel A (Bio-Rad) (negative chromatography).
A column with an 85-cm3 matrix was equilibrated with 500 ml of equilibration buffer supplemented with 70 mM KCl at a flow rate of 3 ml min−1. Cell extract (52 ml) was applied to the column, followed by washing with 200 ml of equilibration buffer. Fractions of 15 ml were collected. The PA-CoA ligase activity was detected in the fractions collected during this washing step. The rest of the contaminating proteins remained bound to the matrix and were washed off with 300 ml of equilibration buffer containing 500 mM KCl. The active fractions were pooled (118 ml) and diluted with the equilibration buffer (1 volume plus 2 volumes of buffer) to adjust the concentration of KCl to around 20 mM.
Anion-exchange chromatography on DEAE Bio-Gel A (Bio-Rad) (positive chromatography).
The diluted protein sample was applied to another DEAE column (32-cm3 matrix) which had been equilibrated with 200 ml of equilibration buffer containing 20 mM KCl. The loaded column was washed with 100 ml of the same buffer at a flow rate of 2 ml min−1, and fractions of 10 ml were collected. The elution of PA-CoA ligase activity was achieved with a linear KCl gradient from 20 to 70 mM in 150 ml of equilibration buffer and then with 50 ml of buffer containing 70 mM KCl. The CoA ligase activity was eluted at the end of this gradient step between 60 and 70 mM KCl.
Anion-exchange chromatography on Q-Sepharose.
The active protein sample from the above-described step (59 ml) was applied directly at a flow rate of 2 ml min−1 to a fast protein liquid chromatography Q-Sepharose column (25 cm3; Pharmacia) which had been equilibrated with 150 ml of equilibration buffer containing 70 mM KCl. The loaded column was washed with 75 ml of the same buffer followed by 150 ml of a linear 70 to 150 mM KCl gradient and 50 ml of 150 mM KCl in the equilibration buffer. Fractions of 5 ml were collected. The fractions that eluted at around 90 mM KCl contained the main PA-CoA ligase activity. The active fractions were pooled (42 ml), and the concentration of KCl was adjusted to be about 50 mM by adding equilibration buffer containing no KCl.
Affinity chromatography.
A 10-cm3 column of Reactive-Green 19-agarose (Sigma, Munich, Germany) was washed with equilibration buffer containing 50 mM KCl at a flow rate of 1 ml min−1. The column was loaded with 15 ml of the active protein sample obtained from the preceding step and then washed with 50 ml of the equilibration buffer and 50 ml of buffer lacking KCl. Fractions of 3 ml were collected. The specific elution of the enzyme was carried out with 5 mM PA in 50 ml of buffer without KCl.
Chromatography on hydroxyapatite (Bio-Rad).
A Macro-Prep ceramic hydroxyapatite column (10 cm3) was activated with 50 ml of 400 mM potassium phosphate buffer and then with 100 ml of equilibration buffer at a flow rate of 1 ml min−1. The pooled active protein collected after the Reactive-Green 19 column (28 ml) was applied and the column was washed with 40 ml of equilibration buffer. Fractions of 5 ml were collected. A linear 0 to 700 mM KCl gradient in 70 ml was passed through the column, followed by 40 ml of buffer lacking KCl. The column was washed with 60 ml of a linear 10 to 60 mM KH2PO4 gradient in the presence of 2 mM MgCl2, 2 mM DTE, and 10% glycerol. The fractions collected at a concentration of KH2PO4 around 20 mM (15 ml) contained the enzyme activity.
Gel permeation chromatography.
The molecular mass of the native protein was determined by gel permeation chromatography on a calibrated Superdex 200 HR 10/30 column (Pharmacia). Half of the purified protein sample obtained from the preceding chromatographic step was precipitated with ammonium sulfate at a 60% saturation, and the resulting protein pellet was dissolved in 300 μl of equilibration buffer. The protein sample was applied to the column, which had been equilibrated with 5 bed volumes of the equilibration buffer at a flow rate of 0.8 ml min−1. Protein elution was monitored at an A280, and fractions of 0.5 ml were collected and tested for activity. The molecular mass of the purified enzyme was estimated as described before (30), using protein molecular weight standards which contained catalase, aldolase, bovine serum albumin, ovalbumin, and chymotrypsinogen (molecular masses, 240, 158, 67, 43, and 25 kDa, respectively).
Cloning and DNA manipulations.
Standard protocols were used for DNA cloning, transformation, amplification, and purification (6, 36). A λ-ZAP Express gene library containing chromosomal DNA of A. evansii after Sau3AI digestion was constructed as described in the ZAP Express cloning kit instruction manual (Stratagene). A 17-mer degenerated oligonucleotide (P1) was designed on the basis of the determined N-terminal amino acid sequence. Another four reverse 20-mer degenerate oligonucleotides (PR1 to PR4) of conserved DNA regions of putative E. coli and P. putida PA-CoA ligase genes were synthesized. Different PCR assays were performed with various combinations of P1 and PR1 to PR4 primers. A 450-bp PCR product was obtained with a combination of P1 and PR2. This PCR product was labeled with [γ-32P]dATP and was used as a probe to screen the constructed gene library. Five positive clones were obtained, and the recombinant plasmids were maintained in E. coli XL1-Blue MRF′ strain.
DNA sequencing and computer analysis.
Purification of plasmid DNA was performed according to a spin miniprep kit protocol (Qiagen). Sequencing of the DNA insert was carried out by J. Alt-Mörbe (Labor für DNA-Analytik, Freiburg, Germany). DNA and amino acid sequences were analyzed with the BLAST network service at the National Center for Biotechnology Information (Bethesda, Md.).
Chemicals and biochemicals.
Chemicals and biochemicals were from Gerbu (Gaiberg, Germany), Boehringer (Mannheim, Germany), Sigma, and Fluka (Neu-Ulm, Germany). Radiolabeled [C1-14C]PA and [U-14C]Phe were from American Radiolabeled Chemicals/Biotrend Chemikalien (Cologne, Germany). PACoA was chemically synthesized according to established methods (18, 24).
Electrophoretical methods.
For one-dimensional gel electrophoresis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (11% polyacrylamide) was carried out with a discontinuous buffer system (23). Proteins were visualized by silver staining (32), and the molecular mass of the purified protein was determined with molecular mass protein standards (phosphorylase, 97 kDa; bovin serum albumin, 67 kDa; ovalbumin, 45 kDa; lactate dehydrogenase, 34 kDa; and carbonic anhydrase, 29 kDa). One-hundred-microliter samples of the active proteins obtained from the affinity chromatography and hydroxyapatite chromatography were precipitated by adding 30 μl of 24% trichloroacetic acid. The resulting protein pellet was dissolved in 18 μl of 0.1 M NaOH and loaded on the gel.
Isoelectric focusing and determination of the pI of the purified enzyme.
Determination of the pI of the purified enzyme obtained after the hydroxyapatite step was carried out by two-dimensional gel electrophoresis with the Immobiline Dry-Strip system according to the instruction manual (Pharmacia). The first dimension (isoelectric focusing [IEF]) was performed with IEF Dry-Strips and 0.2% ampholyte (pH 3 to 10; 40%, wt/vol; Bio-Rad). The second dimension was SDS-PAGE as described above.
Electrophoretic transfer of protein and determination of N-terminal amino acid sequence.
Proteins separated by SDS-PAGE were transferred to nitrocellulose filters (pore size, 0.45 μm), and the transblotted proteins were visualized by staining with Ponceau S (11). The protein bands were cut off, and the amino acid sequence was determined by gas phase sequencing by H. Schägger (University of Frankfurt).
Protein determination.
Protein was determined by using the modified Lowry method (27) with bovine serum albumin as the standard.
TLC.
For separation and preliminary identification of the PA-CoA ligase reaction product on silica gel aluminum TLC plates (thickness, 0.2 mm; 20 by 20 cm; Kieselgel type 60 F 254; Merck, Darmstadt, Germany), the following solvent system was used: n-butanol-acetic acid-water (12:3:5, vol/vol/vol) with the following Rf values: 0.84, 0.26, and 0.50 for PA, PACoA, and Phe, respectively. These compounds were visualized under UV light at 254 nm, and radioactive spots were localized either by autoradiography on X-ray films or with a phosphorimaging plate (Fuji Photo Film Co., Ltd., Kanagawa, Japan).
HPLC.
For separation, identification, and quantification of PA and PACoA in acid-stopped or alkali-treated samples, an HPLC equipped with a variable-wavelength UV- or visible-light monitor and a flowthrough radioactivity detector with a solid scintillator cell was used. The separation was carried out at room temperature at a flow rate of 1 ml min−1 and monitored at a wavelength of 260 nm. Two separation systems were used. (i) For separation of the free acids PA and Phe, a reversed-phase C8 column (Ultrasphere, octyl, 5-μm particle size, 4.6 mm by 25 cm; Beckman) was used. The mobile phase (2% methanol in 40 mM formic acid) was applied isocratically for 13 min, followed by a linear gradient of from 2 to 80% methanol in 40 mM formic acid within 27 min. The retention times for PA and Phe were 33 and 15 min, respectively. (ii) For separation of PA and PACoA a reversed-phase C18 column (Grom-Sil octadecyl silane-4HE, 5-μm particle size, 120 by 4 mm; Grom) was used. The mobil phase was a gradient of acetonitrile (2 to 40%) in 50 mM potassium phosphate buffer (pH 4.5) within 45 min. The retention times of PA and PACoA were 14 and 17 min, respectively.
Radioactivity determination.
Radioactive spots separated by TLC were scraped from the plates, and their radioactivities were determined by liquid scintillation counting. Radioactive peaks separated by HPLC were monitored by a flowthrough radioactivity detector with a solid scintillator cell. The radioactivities of these peaks were quantified by determination of peak area integration in comparison to that of a standard labeled compound.
Nucleotide sequence accession number.
The sequence data reported in this article were submitted to the EMBL database (accession no. AF176259).
RESULTS
Growth characteristics and time course of PA-CoA ligase activity in aerobically growing cells.
The aerobic growth of A. evansii on PA in chemically defined medium containing a 5 mM concentration of the aromatic substrate as the sole carbon source was studied. A. evansii consumed PA rapidly, with a maximal growth rate of 0.23 h−1 at 37°C. The molar growth yield was 56 g of dry cell mass formed per mol of PA consumed. The calculated specific substrate consumption rate of the culture was 114 nmol min−1 mg of protein−1, assuming that 60% of the cell dry matter was protein (31).
The level of PA-CoA ligase activity in extracts (the protein fraction precipitated at 60% ammonium sulfate) of cells obtained during aerobic growth with PA was determined. The catalytic ability to activate PA was present early in the exponential growth phase and increased rapidly, reaching a maximal specific activity of 76 nmol min−1 mg of protein−1 within 6 h of incubation. This level of activity was maintained as long as PA was present in the medium; thereafter, about a 60% drop in activity was observed when PA was completely exhausted.
Induction pattern of PA-CoA ligase activity.
To study the induction of PA-CoA ligase in cells grown with different aromatic or aliphatic substrates as the sole carbon source, extracts of cells grown aerobically with PA, 4-OHPA, Phe, B, acetate, and adipate and anaerobically with PA were screened for this activity (Table 1). The protein fraction (precipitated at 60% ammonium sulfate) of the cell extract centrifuged at 100,000 × g was used, since the cell extracts of A. evansii have a high level of activity of endogenous NADH oxidation which interferes with the detection of CoA ligase activity coupled to NADH oxidation. In addition, it turned out that the assay was inhibited by unknown compounds present in the soluble protein fraction.
TABLE 1.
CoA-ligase activities in a protein fraction of A. evansii cell extracts precipitated at 60% ammonium sulfatea
| Substance with which cells were grown aerobically | CoA ligase activity (nmol min−1 mg of protein−1) with substrate:
|
||
|---|---|---|---|
| PA | 4-OHPA | B | |
| PA | 153 | 18 | 37 |
| 4-OHPA | 17 | 23 | 7 |
| B | 6 | 4 | 196 |
| Phe | 194 | 20 | 34 |
| PA (anaerobic) | 139 | 21 | 106 |
| Adipate | 5 | 4 | NDb |
| Acetate | 4 | 3 | ND |
The cell extracts were the supernatant after centrifugation at 100,000 × g. Enzyme activities were determined by a coupled enzyme-spectrophotometric assay.
ND, not determined.
When PA was the aromatic substrate to be tested, it was activated to its CoA thioester by extracts of PA- or Phe-grown cells at specific activities of 153 and 194 nmol min−1 mg of protein−1, respectively. Much lower activities (about 11% or less) were measured in other cell extracts. In contrast, when 4-OHPA was the test substrate, much lower activities were measured (≤15% of those with PA) and they were nearly at the same rate in PA, Phe, and 4-OHPA cell extracts. High CoA ligase activities against B were detected in cell extracts aerobically grown with B and anaerobically grown with PA. These results clearly indicate that aerobic growth of A. evansii on PA or Phe and anaerobic growth on PA induces the synthesis of specific PA-CoA ligases. This activity was nearly or totally lost when cells were grown with other aromatic or aliphatic substrates. The trace activities toward 4-OHPA or B in extracts of PA- or Phe-grown cells may be due to the presence of other CoA ligases.
In vivo formation of PACoA.
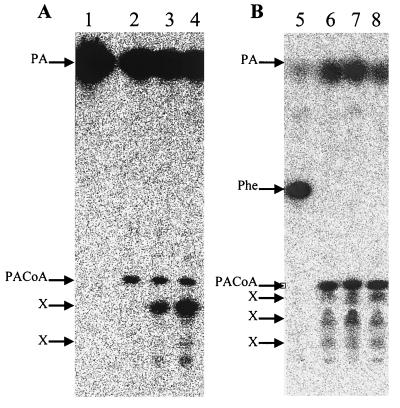
Cells incubated with labeled PA or Phe rapidly synthesized a labeled product which was detected very early after 30 s (Fig. 2). The retention time and the Rf value of this product matched exactly those of the authentic PACoA sample. The amount of this early product increased with time, and later on the product was consumed and other labeled products were formed which have not yet been identified. Alkaline hydrolysis of this product generated labeled PA. This result indicated that PACoA is the first true metabolite formed in cells growing with PA as the sole carbon source.
FIG. 2.
In vivo formation of [14C]PACoA in A. evansii cells grown aerobically in the presence of [14C]PA (A) or [14C]phenylalanine (B). The reactions were stopped with 10% formic acid and separated by TLC followed by autoradiography. Lanes 1 to 4, samples taken at 0 and 30 s and 1 and 2 min in the presence of PA; lanes 5 to 8, samples taken at 30 s and 1, 2, and 5 min in the presence of phenylalanine. X indicates an unknown product.
Purification of PA-CoA ligase.
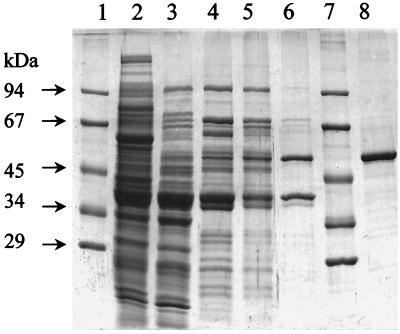
The purification of the PA-CoA ligase was achieved by a protocol which involved five chromatographic steps (Table 2). The recovered activity obtained after the first step (negative chromatography on DEAE-Sepharose) was considered 100% because of the difficulty of estimating accurately the activity in the soluble protein fraction. Although, the specific elution of the enzyme by its substrate from an affinity matrix (Reactive-Green 19) resulted in loss of two-thirds of the activity, a 15-fold purification was gained, with only one major contaminating protein in addition to some minor proteins, which were removed by hydroxyapatite chromatography (Fig. 3). There was no change in the specific activity of the enzyme after gel permeation chromatography. Therefore, this step was not considered in the purification protocol described in Table 2.
TABLE 2.
Purification protocol for PA-CoA ligase from A. evansii cells aerobically grown with PAa
| Purification step | Total protein (mg) | Total enzyme activity (μmol min−1) | Sp act (μmol min−1 mg of protein−1) | Yield (%) | Level of purification (fold) |
|---|---|---|---|---|---|
| Soluble protein fraction (100,000 × g) | 1,714 | 130.3 | 0.076 | —b | —b |
| DEAE (negative chromatography) | 463 | 191.2 | 0.413 | 100 | 1 |
| DEAE (positive chromatography) | 191 | 167.1 | 0.875 | 87.0 | 2 |
| Q-Sepharose | 67 | 131.3 | 1.960 | 69.0 | 5 |
| Reactive-Green | 1.6 | 46.9 | 29.31 | 24.5 | 71 |
| Hydroxyapatite | 0.8 | 38.7 | 48.40 | 20.0 | 117 |
Twenty grams of cells and 40 ml of buffer was used. The enzyme activities in the collected protein fractions were measured by the coupled enzyme assay.
Enzyme activity is inhibited in crude cell extract and increases after the first chromatographic step. Therefore, values for yields and levels of purification (fold) were calculated in relation to the activity obtained after the first chromatographic step.
FIG. 3.
SDS-PAGE of PA-CoA ligase protein fractions collected during the purification scheme. Lanes 1 and 7, molecular mass protein standards containing phosphorylase (97 kDa), bovine serum albumin (67 kDa), ovalbumin (45 kDa), lactate dehydrogenase (34 kDa), and carbonic anhydrase (29 kDa); lane 2, crude extract (supernatant after centrifugation at 100,000 × g, 130 μg of protein); lane 3, DEAE fraction (negative chromatography, 85 μg of protein); lane 4, DEAE fraction (positive chromatography, 60 μg of protein); lane 5, Q-Sepharose fraction (45 μg of protein); lane 6, reactive-green fraction (6 μg of protein); lane 8, hydroxyapatite fraction (5 μg of protein).
Molecular properties of the PA-CoA ligase.
The molecular mass of the purified enzyme was determined by gel permeation chromatography to be about 52 kDa. By SDS-PAGE, the protein migrated as a single band corresponding to a molecular mass of 50 kDa (Fig. 3). Hence, native PA-CoA ligase is a monomeric enzyme of only one polypeptide. The enzyme migrated in two-dimensional gel electrophoresis as a band at a pI between 6.1 and 6.5.
N-terminal amino acid sequence.
The N-terminal amino acid sequence of the aerobically induced PA-CoA ligase was determined to be MPVKTPSPG. This N-terminal amino acid sequence differed totally from that determined for the anaerobically induced PA-CoA ligase from the same bacterium (Table 3).
TABLE 3.
Biochemical and molecular characteristics of two PA-CoA ligase isoenzymes activating PA in A. evansii under aerobic and anaerobic growth conditionsa
| Characteristic | Result for isoenzyme induced:
|
|
|---|---|---|
| Aerobically | Anaerobicallyb | |
| Molecular mass (kDa) | 48.752 (monomer) | 49.376 (monomer) |
| N-terminal amino acid sequence | MPVKTPSPG | SARDGFAVP |
| pI | 6.3 | ND |
| Maximal sp act in growing cultures (nmol min−1 mg of protein−1) | 76 | 48 |
| Km (μM) with: | ||
| PA | 14 | 60 |
| ATP | 60 | 290 |
| CoA | 45 | 150 |
| Turnover no. (kcatc [s−1]) | 40 | 22 |
| Substrate specificity | Highly specific, PA only | Highly specific, PA only |
| Optimal pH | 8 to 8.5 | 8.5 |
| Induction growth conditions | Aerobic growth with PA or Phe | Anaerobic growth with PA or Phe plus nitrate |
| Enzyme stability | Requires glycerol | Requires glycerol |
Products and stoichiometry.
The PA-CoA ligase reaction was dependent on PA, ATP, Mg2+, and CoA. The formation of PACoA was monitored spectrophotometrically at a wavelength of 365 nm in a reaction which allowed the rate of AMP or ADP formation to be determined by coupling the reaction via myokinase, pyruvate kinase, and lactate dehydrogenase to the oxidation of NADH.
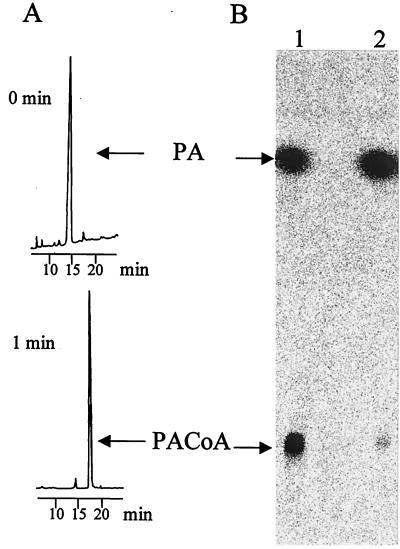
A stoichiometry of 2.0 mol of NADH oxidized per mol of PA added was observed. This ratio indicates that the products are PACoA and AMP plus pyrophosphate, rather than ADP plus phosphate. The formed PACoA was identified by HPLC (Fig. 4A) before and after alkaline hydrolysis to liberate free PA. With [14C]PA, the formation of labeled PACoA was also analyzed by TLC and identified by its Rf value by autoradiography (Fig. 4B). TLC and HPLC analyses of acid-stopped samples showed two labeled compounds which cochromatographed with authentic samples of PA and PACoA. After alkaline hydrolysis of the same samples, mainly one labeled compound which matched PA was detected by TLC (Fig. 4B) and HPLC. The radioactivity content of this compound was nearly equal to the total radioactivity present in the acid-stopped sample.
FIG. 4.
(A) HPLC separation of PA-CoA ligase reaction product. (B) Results of TLC and autoradiography of PA-CoA ligase in the presence of [14C]PA. Lane 1, acid-stopped sample after 1 min; lane 2, the same sample after alkaline hydrolysis.
Substrate specificity.
The substrate specificity of PA-CoA ligase was tested with the active fractions obtained with the hydroxyapatite column and also with the protein fractions showing the highest activities during the different chromatographic steps. The followings groups of molecules were tested: (i) aromatic acids and their ring-substituted (hydroxy-, carboxy-, amino-, and chloro-) derivatives, such as phenylbutyric, phenylpropionic, cinnamic, phenylacetic, mandelic, phenylglyoxylic, homophthalic, phthalic, and benzoic acids and phenylglycine and (ii) different aliphatic acids such as acetic, propionic, pyruvic, maleic, fumaric, succinic, butyric, acetoacetic, and oxaloacetic acids. All active protein fractions obtained after the first purification step and the purified enzyme activated only phenylacetic acid, whereas all the other tested compounds were not used as substrates by PA-CoA ligase (≤2%).
Catalytic properties.
The catalytic properties of the protein purified after the hydroxyapatite step were measured by the coupled enzyme assay described above (see Materials and Methods). The dependence of the activity on the pH was tested in Na-citrate (pHs 5 to 6), K-phosphate (pHs 6 to 8), and Tris-HCl (pHs 7 to 9) buffers, (100 mM each). The enzyme showed maximal activity at pHs 8 to 8.5, with a dramatic drop of activity (55%) at pH 9. Less than 10% activity was observed at pH 6, and half the maximal activity was measured at pH 7. ATP was the only nucleotide triphosphate accepted (100%), while the other tested nucleotides (CTP, GTP, and UTP) were not used (<2%). Also, N-acetylcysteamine (2 mM) could not substitute for CoA.
The apparent Km values of the purified enzyme were determined for PA (10 μM to 5 mM), ATP (10 μM to 2 mM), and CoA (10 μM to 1 mM) at 37°C in assays containing 15 μg of protein. The reaction was started by adding various concentrations of one substrate to an assay containing nonlimiting concentrations of the other cosubstrates. The Km values were determined from linear Lineweaver-Burk plots for each substrate. The enzyme showed high affinities towards its substrate and cosubstrates (Km values of 14, 60, and 45 μM for PA, ATP, and CoA, respectively). A turnover number (catalytic constant) of 40 s−1 at 37°C was calculated from the molecular mass of the purified enzyme (50 kDa) and the maximal specific activity of 48 μmol min−1 mg of protein−1.
In presence of thiol group-modifying reagents such as N-ethylmaleimide and iodoacetamide (1 mM), no activity was detected. Severe inhibition of activity was observed in the presence of a 1 mM concentration of the divalent cations Zn2+, Cu2+, and Ni2+ (≥80% inhibition). Also, a dramatic loss of activity (84%) was observed in the presence of NaF (2 mM), a reagent that inhibits some Mg2+-requiring enzymes. The activity was absolutely dependent on the cocatalyst Mg2+ (5 mM); Mn2+ (5 mM) could partially replace Mg2+, with 36% of the activity being recovered.
The enzyme was extremely labile, and the activity was totally lost within 48 h at 4°C. Glycerol was necessary for stabilizing the enzyme activity. At 10% glycerol (wt/vol), about 65% of the activity was recovered after 72 h at 4°C. The enzyme activity was more stable in 20% glycerol (80% recovery after 72 h at 4°C), but for practical reasons, the purification protocol was carried out in buffers containing only 10% glycerol. At −20°C in the presence of 10% glycerol, the enzyme activity was retained for 2 months without remarkable loss.
Cloning and sequencing of the gene coding for PA-CoA ligase.
A 17-mer degenerated oligonucleotide primer was derived from the determined N-terminal amino acid sequence and was used in PCR assays against different reverse primers, which were deduced from conserved regions of the putative E. coli and P. putida PA-CoA ligases (see Materials and Methods). A 450-bp DNA fragment which showed similarity with other PA-CoA ligase genes was obtained. This PCR product was used to screen a λ-ZAP Express gene library (Stratagene) containing chromosomal DNA of A. evansii. Five positive clones were obtained and subsequently analyzed for the presence of the correct insert. Three of the clones contained 600 bp of the gene for PA-CoA ligase, while the other two clones carried the complete gene. The gene codes for 440 amino acids (1,320 bp) which correspond to a polypeptide of 48.75 kDa. This mass agrees well with the determined mass of the purified PA-CoA ligase (50 kDa by SDS-PAGE). The N-terminal amino acid sequence determined for the purified enzyme was identical with that deduced from the nucleotide sequence of PA-CoA ligase gene, thus confirming it as the gene product and showing that no processing of its N terminus occurs. A theoretical pI value of 6.26 was calculated from the deduced amino acid sequence of PA-CoA ligase, which agrees with the experimentally determined values (6.1 to 6.5) for the purified protein.
No potential open reading frames were found within 100 bp upstream of the 5′ end or within 200 bp downstream of the 3′ end of the gene.
DISCUSSION
CoA thioesterification of aromatic compounds is often necessary under anaerobic conditions to activate these compounds as a prerequisite to destabilize the inert aromatic ring prior to further transformations (20). The alternative widely distributed strategy followed under the aerobic conditions to destabilize the aromatic ring involves ring hydroxylation reactions which render the aromatic ring suitable for oxygenolytic ring fission.
Recently, an unusual new aerobic strategy that resembles the anaerobic degradation mechanism in CoA activation of the aromatic compounds prior to ring attack has been reported for some bacterial species (see the introduction).
The β-subclass proteobacterium A. evansii efficiently utilizes PA under both aerobic and anaerobic growth conditions. A specific PA-CoA ligase, which has been purified (30), was found to be induced when cells were grown anaerobically with PA. The same activity was detected also in cells grown aerobically with PA. Hence, it was not clear whether this activity in cells grown aerobically and anaerobically with PA is due to the same enzyme or to another isoenzyme whose induction is restricted to the presence of PA under aerobic conditions. The induction of this activity under aerobic conditions was dependent on the presence of PA or Phe. The very low CoA-ligase activity detected with PA in 4-OHPA cell extract may be due to the presence of another nonspecific CoA ligase with a broad substrate spectrum or to the weak induction of PA-CoA ligase by the substrate analogue 4-OHPA. The substrate specificity of the purified enzyme indicated that the CoA ligase activities towards 4-OHPA and B detected in extracts of cells aerobically grown with PA are due to other CoA ligases. The following data indicate that the potential inducer molecule for the synthesis of the aerobic PA-CoA ligase in A. evansii may be PA or PACoA but not other aromatic substrates: (i) the detection of this activity very early in the exponential growth phase; (ii) the decrease in activity when PA was exhausted in the medium; (iii) the detectability of this activity also in Phe-grown cells, which most likely metabolize Phe via PA and PACoA; and (iv) the high substrate affinity and specificity of this enzyme. All these data in addition to the absence of hydroxylase activity with the free acid show that PACoA is the first true metabolite derived from PA during the aerobic metabolism of this acid.
The N-terminal amino acid sequences of the induced PA-CoA ligases under aerobic and anaerobic conditions in A. evansii were different (Table 3); hence, it was clear that PA is activated by two isoenzymes whose expression in this organism depends on the prevailing growth conditions. The two isoenzymes could also be differentiated by their kinetic properties (Table 3). The aerobically induced isoenzyme has a greater affinity for the aromatic substrate and the cosubstrates. This finding is consistent with an enzyme having a role in the PA aerobic degradation pathway. This enzyme initiates the activation of PA, facilitating the subsequent ring hydroxylation reactions, which, so far, are not easily demonstrated. The enzyme purified in this work is the second characterized CoA ligase activating PA under aerobic conditions. The substrate affinity of the A. evansii isoenzyme (14 μM for PA) is about a thousandfold higher than that reported for the first purified enzyme from P. putida (Km values for PA, ATP, and CoA were 16.5, 9.7, and 1.0 mM, respectively [29]). Thus, the sequences 239DIYGLSE245 and 305YRTRD309 (Fig. 5), which are conserved in all putative or proven PA-CoA ligases and match motifs II and III in acyl-adenylate-forming enzymes (16), probably do not contribute to the substrate-binding sites in PA-CoA ligases.
FIG. 5.
Amino acid alignment of PA-CoA ligases from different microorganisms. Ae, A. evansii; Ec, E. coli (9); Pp, P. putida (33); Psp, Pseudomonas sp. strain Y2 (44); Bh, B. halodurans (42); MtF390, coenzyme F390 M. thermoautotrophicum (40). The Arabic numbers refer to the amino acid sequence deduced from the nucleotide sequence. The Roman numerals above the sequence denote postulated binding motifs. I, AMP-binding motif; II and III, postulated substrate-binding site motifs. Note that only the A. evansii and P. putida genes have been proven to code for this enzyme.
Although the reported PA-CoA ligases have different N-terminal amino acid sequences, they share many biochemical and catalytic features, such as (i) highly conserved amino acid motifs (Fig. 5), (ii) similar molecular masses of about 50 kDa, (iii) high substrate specificities, (iv) the requirement of Mg2+ for activity, (v) maximal catalytic activities in alkaline pHs (pHs 8 to 8.5), (vi) extreme lability and the requirement for glycerol for their stabilization, and (vii) inhibition of activity by divalent cations (Cu2+, Ni2+, and Zn2+). These data support the opinion that the specific induction of these enzymes in PA-degrading bacteria occurs when PA or other aromatic substrates metabolized via PA or PACoA (such as aromatic amino acids, lignin-related monoaromatic acids, aromatics with even-number carbon atoms of the side chain, styrene, phenylethanol, 2-phenylethylamine, and tyramine) serve as the sole carbon and energy sources.
A typical ribosome-binding site (AGGAG) is found 8 bases upstream of the potential ATG start codon of this gene. The gene product of A. evansii showed that it is closely related to other sequenced PA-CoA ligases. The derived amino acid sequence showed high identity to the corresponding putative PA-CoA ligase gene products of E. coli (PaaK, 64.6%) (9), Pseudomonas sp. strain Y2 (PaaK, 64.7%) (44), and B. halodurans (open reading frame 12, 50%) (42); the PA-CoA ligase gene product of P. putida (PhaE, 66.5%) (33); and the coenzyme F390 gene product of Methanobacterium thermoautotrophicum (MTH 161, 46.8%) (40). As shown by protein sequence alignments, a typical amino acid consensus sequence for an AMP binding motif (VX2SSGTTGKPTV) which is shared in other PA-CoA ligases (Fig. 5) was identified.
However, these sequence alignments have not allowed the identification of the consensus sequence essential for enzyme catalysis. Also, no sequence information data are available to explain the lability of these enzymes and their stabilization in the presence of glycerol.
The PA-CoA ligase gene of A. evansii was designated paaK according to the abbreviations for the putative genes probably involved in PA catabolism in E. coli K-12.
ACKNOWLEDGMENTS
I am very grateful to G. Fuchs for his support, valuable discussions, and critical reading of the manuscript. I am indebted to H. Heider for his help and fruitful discussions. Thanks also go to H. Schägger for determination of the N-terminal amino acid sequence and to J. Alt-Mörbe for DNA sequencing.
I gratefully acknowledge the financial support of this work by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Altenschmidt U, Fuchs G. Novel aerobic 2-aminobenzoate metabolism. Purification and characterization of 2-aminobenzoate–CoA ligase, localization of the gene on a 8-kbp plasmid, and cloning and sequencing of the gene from a denitrifying Pseudomonas sp. Eur J Biochem. 1992;205:721–727. doi: 10.1111/j.1432-1033.1992.tb16835.x. [DOI] [PubMed] [Google Scholar]
- 2.Altenschmidt U, Oswald B, Steiner E, Herrmann H, Fuchs G. New aerobic benzoate oxidation pathway via benzoyl-coenzyme A and 3-hydroxybenzoyl–coenzyme A in a denitrifying Pseudomonas sp. J Bacteriol. 1993;175:4851–4858. doi: 10.1128/jb.175.15.4851-4858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders H-J, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int J Syst Bacteriol. 1995;45:327–333. doi: 10.1099/00207713-45-2-327. [DOI] [PubMed] [Google Scholar]
- 4.Anderson J J, Dagley S. Catabolism of aromatic acids in Trichosporon cutaneum. J Bacteriol. 1980;141:534–543. doi: 10.1128/jb.141.2.534-543.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arunachalam U, Massey V, Vaidyanathan C S. p-Hydroxyphenylacetate-3-hydroxylase. A two-protein component enzyme. J Biol Chem. 1992;267:25848–25855. [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7.Baggi G, Boga M M, Catelani D, Galli E, Treccani V. Styrene catabolism by a strain of Pseudomonas fluorescens. Syst Appl Microbiol. 1983;4:141–147. doi: 10.1016/S0723-2020(83)80042-3. [DOI] [PubMed] [Google Scholar]
- 8.Blakley E R, Kurz W, Halvorson H, Simpson F J. The metabolism of phenylacetic acid by Pseudomonas. Can J Microbiol. 1967;13:147–157. doi: 10.1139/m67-021. [DOI] [PubMed] [Google Scholar]
- 9.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Chapman P J, Dagley S. Oxidation of homogentisic acid by cell-free extracts of a Vibrio. J Gen Microbiol. 1961;28:251–256. doi: 10.1099/00221287-28-2-251. [DOI] [PubMed] [Google Scholar]
- 11.Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T. Current protocols in protein science. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 12.Cooper R A, Skinner M A. Catabolism of 3- and 4-hydroxyphenylacetate by the 3,4-dihydroxyphenylacetate pathway in Escherichia coli. J Bacteriol. 1980;143:302–306. doi: 10.1128/jb.143.1.302-306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford R L. Degradation of homogentisate by strains of Bacillus and Moraxella. Can J Microbiol. 1976;22:276–280. doi: 10.1139/m76-037. [DOI] [PubMed] [Google Scholar]
- 14.Dagley S. Catabolism of aromatic compounds by micro-organisms. Adv Microb Physiol. 1971;6:1–46. doi: 10.1016/s0065-2911(08)60066-1. [DOI] [PubMed] [Google Scholar]
- 15.Dagley S, Wood J M. Oxidation of phenylacetic acid by a Pseudomonas. Biochim Biophys Acta. 1965;99:381–383. doi: 10.1016/s0926-6593(65)80140-0. [DOI] [PubMed] [Google Scholar]
- 16.Ferrandez A, Minambres B, Garcia B, Olivera E R, Luengo J M, Garcia J L, Diaz E. Catabolism of phenylacetic acid in Escherichia coli. J Biol Chem. 1998;273:25974–25986. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 17.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker; 1984. pp. 181–252. [Google Scholar]
- 18.Gross G G, Zenk M H. Darstellung und Eigenschaften von Coenzyme A-Thioestern substituierter Zimtsäuren. Z Naturforsch Teil B. 1966;21:683–690. [Google Scholar]
- 19.Hass S, Burchhardt G, Fuchs G, Herrmann H. Cloning, sequencing and overexpression of phenylacetate-CoA ligase from A. evansii (KB740). Biospektrum Sonderausg.; 1996. p. 106. [Google Scholar]
- 20.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch W, Schägger H, Fuchs G. Phenylglyoxylate: NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism in the denitrifying bacterium Azoarcus evansii. Eur J Biochem. 1998;251:907–915. doi: 10.1046/j.1432-1327.1998.2510907.x. [DOI] [PubMed] [Google Scholar]
- 22.Koch J, Eisenreich W, Bacher A, Fuchs G. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur J Biochem. 1993;211:649–661. doi: 10.1111/j.1432-1033.1993.tb17593.x. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Laempe D, Jahn M, Fuchs G. 6-Hydroxycyclohex-1-ene-1-carbonyl–CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl–CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1999;263:1–12. doi: 10.1046/j.1432-1327.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee C-W, Desmazeaud M J. Evaluation of the contribution of the tyrosine pathway to the catabolism of phenylalanine in Brevibacterium linens 47. FEMS Microbiol Lett. 1986;33:95–98. [Google Scholar]
- 26.Löffler F, Müller R, Lingens F. Purification and properties of 4-halobenzoate-coenzyme A ligase from Pseudomonas sp. CBS3. Biol Chem Hoppe-Seyler. 1992;373:1001–1007. doi: 10.1515/bchm3.1992.373.2.1001. [DOI] [PubMed] [Google Scholar]
- 27.Markwell M A K, Haas S M, Bieber L I, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 28.Martin M, Gibello A, Fernandez J, Ferrer E, Garrido-Pertierra A. Catabolism of 3- and 4-hydroxyphenylacetic acid by Klebsiella pneumoniae. J Gen Microbiol. 1991;132:621–628. doi: 10.1099/00221287-137-3-621. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Blanco H, Reglero A, Rodriguez-Aparicio L B, Luengo J M. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J Biol Chem. 1990;265:7084–7090. [PubMed] [Google Scholar]
- 30.Mohamed M E, Fuchs G. Purification and characterization of phenylacetate-coenzyme A ligase from a denitrifying Pseudomonas sp., an enzyme involved in the anaerobic degradation of phenylacetate. Arch Microbiol. 1993;159:554–562. doi: 10.1007/BF00249035. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed M E, Seyfried B, Tschech A, Fuchs G. Anaerobic oxidation of phenylacetate and 4-hydroxyphenylacetate to benzoyl-CoA and CO2 in denitrifying Pseudomonas sp. Evidence for an α-oxidation mechanism. Arch Microbiol. 1993;159:563–573. doi: 10.1007/BF00249036. [DOI] [PubMed] [Google Scholar]
- 32.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 33.Olivera E R, Minambers B, Garcia B, Muniz C, Moreno M A, Ferrandez A, Diaz E, Garcia J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prieto M A, Garcia L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 35.Rhee S-K, Fuchs G. Phenylacetyl-CoA: acceptor oxidoreductase, a membrane-bound molybdenum-iron-sulfur enzyme involved in anaerobic metabolism of phenylalanine in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1999;262:1–10. doi: 10.1046/j.1432-1327.1999.00399.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schmitz A, Gartemann K-H, Fiedler J, Grund E, Eichenlaub R. Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU. Appl Environ Microbiol. 1992;58:4068–4071. doi: 10.1128/aem.58.12.4068-4071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider S, Fuchs G. Phenylacetyl-CoA: acceptor oxidoreductase, a new α-oxidizing enzyme that produces phenylglyoxylate. Assay, membrane localization, and differential production in Thauera aromatica. Arch Microbiol. 1998;169:509–516. doi: 10.1007/s002030050604. [DOI] [PubMed] [Google Scholar]
- 39.Schneider S, Mohamed M E, Fuchs G. Anaerobic metabolism of l-phenylalanine via benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Arch Microbiol. 1997;168:310–320. doi: 10.1007/s002030050504. [DOI] [PubMed] [Google Scholar]
- 40.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Alerdge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Oiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sparnins V L, Chapman J, Dagley S. Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J Bacteriol. 1974;120:159–167. doi: 10.1128/jb.120.1.159-167.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takami H, Nakasone K, Ogasawara N, Hirama C, Nakamura Y, Masui N, Fuji F, Takaki Y, Inoue A, Horikoshi K. Sequencing of three lambda clones from the genome of alkaliphilic Bacillus sp. strain C-125. Extremophiles. 1999;3:29–34. doi: 10.1007/s007920050096. [DOI] [PubMed] [Google Scholar]
- 43.van den Tweel W J J, Smits J P, de Bont J A M. Catabolism of dl-α-phenylhydracrylic, phenylacetic and 4-hydroxyphenylacetic acid via homogentisic acid in a Flavobacterium sp. Arch Microbiol. 1988;149:207–213. [Google Scholar]
- 44.Velasco A, Alonso S, Garcia J, Perera J, Diaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vitovski S. Phenylacetate-coenzyme A ligase is induced during growth on phenylacetic acid in different bacteria of several genera. FEMS Microbiol Lett. 1993;108:1–6. doi: 10.1016/0378-1097(93)90477-j. [DOI] [PubMed] [Google Scholar]