Abstract
Paracoccidioidomycosis is a systemic and endemic mycosis, restricted to tropical and subtropical areas of Latin America. The infection is caused by the thermal dimorphic fungus Paracoccidioides brasiliensis and Paracoccidioides lutzii. The diagnosis of paracoccidioidomycosis is usually performed by microscopic examination, culture and immunodiagnostic tests to respiratory specimens, body fluids and/or biopsies; however these methods require laboratory personnel with experience and several days to produce a result. In the present study, we have validated and evaluated a nested PCR assay targeting the gene encoding the Paracoccidioides gp43 membrane protein in 191 clinical samples: 115 samples from patients with proven infections other than paracoccidioidomycosis, 51 samples as negative controls, and 25 samples from patients diagnosed with paracoccidioidomycosis. Additionally, the specificity of the nested PCR assay was also evaluated using purified DNA isolated from cultures of different microorganisms (n = 35) previously identified by culture and/or sequencing. The results showed that in our hands, this nested PCR assay for gp43 protein showed specificity and sensitivity rates of 100%. The optimized nested PCR conditions in our laboratory allowed detection down to 1 fg of P. brasiliensis DNA.
Keywords: Paracoccidioides brasiliensis, Nested PCR, Paracoccidioidomycosis, Molecular diagnosis
Introduction
Paracoccidioidomycosis (PCM) is an important systemic granulomatous disease restricted to tropical and subtropical endemic areas in Latin America, from Mexico to northern Argentina caused by the thermal dimorphic fungi of Paracoccidioides genus.1
Until 2006, it was considered that this genus consisted of a single species, Paracoccidioides brasiliensis, however, molecular studies have revealed four distinct phylogenetic species: S1 (present in Brazil, Argentina, Paraguay, Peru and Venezuela), PS2 (found in Brazil and Venezuela), PS3 (restricted exclusively to Colombia), and PS4 (recently identified clinical isolates in Venezuela).2, 3, 4, 5 However, a new monophyletic species, Paracoccidioides lutzii formerly known as ‘Pb01-like’ strains in the P. brasiliensis complex, has been proposed as a new species based on phylogenetic and comparative genomics data, recombination analysis, and morphological characteristics.6 There is little information about the prevalence or incidence of this new species; the few cases of PCM caused by P. Lutzii have been reported predominately in the central west region of Brazil, principally at Mato Grosso and Goias states.7, 8, 9 However, there is evidence of some reported cases outside this area10 and outside Brazil.11 In respect to Colombia, there have been no reported cases of PCM caused by P. lutzii; so far, all cases of PCM reported in Colombia were caused by P. brasiliensis, although these cases were diagnosed by conventional laboratory tests.
Both species grow in the mycelial form at room temperature or in the budding yeast form in host tissue or in culture at 36 °C. It is well accepted that the infection is acquired through inhalation of infectious particles (conidia) produced by the fungus mycelia form present in its as yet unknown natural habitat.12, 13, 14 In this infectious disease, the fungus can remain restricted to the lungs, the primary focus of infection, or spread to other organs and tissues, resulting in diverse clinical manifestations.
Different clinical forms of the disease are recognized and the severity depends on the gender, age, and immune status of the host, as well as on the size of the inhaled fungal inoculum. The infection could be asymptomatic or may give rise to different forms of disease as follows: regressive sub-clinical infection, progressive disease that may be either chronic (adult type) or acute/sub-acute (juvenile type), and the residual form.13
PCM shows a broad geographic distribution of cases from Mexico to Argentina; Brazil has the highest incidence but in Venezuela, Colombia, Ecuador, and Argentina many cases of PCM have also been reported.14, 15, 16 In Brazil areas where PCM is highly endemic, the estimated incidence ranges from 10 to 30 cases/million inhabitants and a mortality rate of 1.4 deaths/million inhabitants.17
The diagnosis of PCM is usually made by microscopic detection of rounded yeast with peripheral budding giving a “ship's wheel” appearance, which are observed directly on patient's clinical samples, especially in sputum and in skin or mucous membrane biopsies.13, 14, 18 Depending on the histopathological pattern it is possible that small forms of Paracoccidioides may be mistaken with other fungi.14, 18, 19, 20 The gold standard to diagnosis of PCM is still the microbiological culture with a disadvantage that the fungus growing can take up to six weeks to yield some result.20, 21, 22 In this sense, diagnostic immunodiagnostic techniques based on detection of antibodies or fungal antigens are of special value.15, 18, 20, 23 Nevertheless, these techniques yield many cases of cross-reactivity with other fungi and false-negative results in patients with some type of immunodeficiency.8, 18, 20, 24
In the last decade, several molecular methods such as polymerase chain reaction (PCR) have been developed for DNA detection of P. brasiliensis in clinical samples, providing both high sensitivity and specificity rates.25, 26, 27, 28 The most frequently used target sequences for molecular detection of P. brasiliensis have been the gp4323 and ribosomal DNA genes.26, 29 These PCR assays have been used to detect P. brasiliensis in the sera and tissues of infected mice,23, 26, 27 in artificially contaminated soil and in environmental samples.26, 30 However, molecular diagnosis of PCM is not currently used in clinical routine.
In the present study, our aim was to assess PCM the diagnostic accuracy of a nested PCR technique targeting the gene coding for the specific gp43 protein of P. brasiliensis using biological samples from in a cohort of patients with suspected or clinically diagnosed PCM. In addition, to implement this molecular assay as an integral component of the diagnostic tests regularly used in our laboratory for the diagnosis of PCM in patients suspected of fungal infection.
Materials and methods
Over the 17-month period, from January 2011 to June 2012, 25 clinical samples from 25 patients with confirmed PCM were collected. The clinical specimens included: bronchoalveolar lavage (BAL, n = 2), biopsies (n = 15), and sputum (n = 8). All of the specimens were collected at hospitals in Medellín, Colombia, and sent to the Medical and Experimental Mycology Unit of the Corporación para Investigaciones Biológicas (CIB), for mycological diagnosis. Of the 25 samples positives for P. brasiliensis, 15 (60%) were diagnosed by culture and 10 (40%) by microscopic direct examination using KOH; all then where subsequently analyzed by a gp43 nested PCR.
To assess the specificity of the nested PCR, we analyzed 115 clinical samples from patients suffering from the following infections confirmed by culture or direct examination: aspergillosis (n = 19), candidiasis (n = 16), pneumocystosis (n = 6), tuberculosis (n = 20), cryptococcosis (n = 44) and histoplasmosis (n = 10). As negative controls we used 30 respiratory negative samples for P. brasiliensis or any of the most common respiratory infectious pathogens diagnosed by culture and/or specific stains as Gomori methenamine silver in the Pneumocystis cases and 21 peripheral blood samples from healthy individuals (Table 1). Additionally, the specificity of the nested-PCR was also evaluated using purified DNA isolated from cultures of different microorganisms (n = 35) previously identified by culture and/or sequencing (Table 2).
Table 1.
Samples used to evaluate specificity during the standardization of the nested PCR assay for the diagnosis of Paracoccidioides brasiliensis.
| Kind of patient | Type of sample | Diagnosis | ||
|---|---|---|---|---|
| Negative controls (n:51) | Respiratory symptomatic (n:30) | BAL (25) | ||
| BL (4) | ||||
| Sputum (1) | ||||
| Healthy individuals (n:21) | Whole blood (21) | |||
| Positive controls (n:25) | Respiratory symptomatic (n:25) | BAL (2) | Paracoccidioides brasiliensis (n:25) | |
| Biopsies (15) | ||||
| Sputum (8) | ||||
| Clinical samples (n:191) | Samples used to evaluate the specificity (n:115) | Respiratory symptomatic | Body fluids (4) | Aspergillus fumigatus (n:1) |
| Candida albicans (n:1) | ||||
| Candida tropicalis (n:2) | ||||
| Tracheal aspirates (9) | Aspergillus fumigatus (n:1) | |||
| Candida albicans (n:6) | ||||
| Candida tropicalis (n:1) | ||||
| Mycobacterium tuberculosis (n:1) | ||||
| CSF (24) | Cryptococcus neoformans (n:24) | |||
| Biopsies (22) | Aspergillus fumigatus (n:4) | |||
| Aspergillus flavus (n:2) | ||||
| Candida spp (n:1) | ||||
| Candida albicans (n:3) | ||||
| Candida parasilopsis (n:1) | ||||
| Candida guillermondii (n:1) | ||||
| Cryptococcus neoformans (4) | ||||
| Histoplasma capsulatum (n:5) | ||||
| Mycobacterium tuberculosis (n:1) | ||||
| Sputum (9) | Aspergillus fumigatus (n:2) | |||
| Mycobacterium tuberculosis (n:7) | ||||
| BL (16) | Aspergillus fumigatus (n:3) | |||
| Mycobacterium tuberculosis (n:7) | ||||
| Cryptococcus neoformans (6) | ||||
| BAL (31) | Aspergillus fumigatus (n:4) | |||
| Aspergillus flavus (n:1) | ||||
| Aspergillus versicolor (n:1) | ||||
| Mycobacterium tuberculosis (n:4) | ||||
| Cryptococcus neoformans (10) | ||||
| Histoplasma capsulatum (n:5) | ||||
| Pneumocystis jirovecii (n:6) | ||||
BAL, bronco alveolar lavage; BL, bronchial lavage; CSF, cerebrospinal fluid.
Table 2.
Results of the Paracoccidioides brasiliensis nested PCR assay to several fungi and related microorganisms.
| Molecular analysis | ||
|---|---|---|
| Isolates | Strain source | Nested PCR gp43 |
| Aspergillus fumigatus | CIB | − |
| Aspergillus terreus | CDC | − |
| Aspergillus flavus | CIB | − |
| Aspergillus niger | CDC | − |
| Cryptococcus neoformans | CIB | − |
| Cryptococcus gattii | CIB | − |
| Candida albicans | CIB | − |
| Candida guillermondii | CIB | − |
| Candida tropicalis | CIB | − |
| Candida parasilopsis | CIB | − |
| Candida dubliniensis | CIB | − |
| Candida glabrata | CIB | − |
| Candida krusei | CIB | − |
| Candida lusitanae | CIB | − |
| Candida bracariensis | CIB | − |
| Candida ohmeri | CIB | − |
| Candida famata | CIB | − |
| Candida ortopsilosis | CIB | − |
| Candida metapsilosis | CIB | − |
| Paracoccidioides brasiliensis | ATCC 60855 | + |
| Paracoccidioides brasiliensis | CIB/Pb339 | + |
| Paracoccidioides brasiliensis | CIB/Pb18 | + |
| Paracoccidioides lutzii | CIB | − |
| Coccidioides immitis | ATCC 28868 | − |
| Blastomyces dermatitidis | CDC 2008011573 | − |
| Blastomyces dermatitidis | ATCC 26199 | − |
| Histoplasma capsulatum var. capsulatum | CDC/Thon | − |
| Histoplasma capsulatum var. capsulatum | G217B | − |
| Histoplasma capsulatum var. capsulatum | G184B | − |
| Histoplasma capsulatum var. capsulatum | CDC/3670 | − |
| Histoplasma capsulatum var. duboisii | CDC/5822 | − |
| Histoplasma capsulatum var. duboisii | CDC/5823 | − |
| Schizophyllum commune | CIB | − |
| Mycobacterium tuberculosis | CIB | − |
| Mycobacterium avium | CIB | − |
CIB, Corporación para Investigaciones Biológicas, Medellín, Colombia; CDC, Centers for Disease Control and Prevention, Atlanta, USA; −, PCR negative result; +, PCR positive result.
Processing of clinical samples
Respiratory tract specimens (BAL, BL fluid and sputum) and body fluids, including peritoneal fluid, pleural fluid and cerebrospinal fluids (CSF) were collected in 50 mL sterile Falcon tubes (Becton Dickinson) and centrifuged at 1550 × g for 30 min (Centra MP4R, IEC). The fresh tissues (biopsies) were manually homogenized in 3 mL of sterile saline solution. The pelleted samples and the homogenate obtained were used for culture and Gomori methenamine stain, and 0.6 mL of each sample was stored at −20 °C for subsequent DNA extraction. Processed clinical samples were cultured on Sabouraud dextrose agar and Mycosel (Becton Dickinson) by incubation at room temperature (18–22 °C) for 20–30 days and were examined weekly macroscopically and microscopically. These procedures were carried out in a biosafety laboratory level 3 (BSL3).
DNA extraction
Two hundred microliters of each previously processed clinical sample or yeast suspension were used for DNA extraction and purification. The QIAamp® DNA Mini kit (Qiagen, Hildenberg, Germany) was used with some modifications: the initial incubation with lysis buffer was performed at 65 °C for one hour, followed by AL buffer incubation at 90 °C for 10 min and additional incubation with recombinant lyticase (1 UI/μL) at 37 °C for 45 min. For filamentous fungal isolates, DNA extraction was performed using the phenol–chloroform method or a commercial kit with Genomic G-100 columns (Qiagen Inc., CA).31 DNA extraction from whole blood was performed using a protocol described by Einsele et al.,32 with some modifications. The relative concentrations of DNA extracted were determined using a NanoDrop ND2000 (Thermo Scientific).
P. brasiliensis nested PCR assay
Specific primers for P. brasiliensis that target the gene encoding the gp43 membrane protein, were used in a nested PCR reaction as described by Bialek et al.,23 with minor modifications. The master mix for the first PCR consisted of 5 μL of purified DNA in a total PCR volume of 50 μL with final concentrations of 3 mM MgCl2 (Invitrogen), 0.2 μM of each outer and inner primer (Invitrogen) and 0.03 U of Taq polymerase (Invitrogen). The mixture was incubated at 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 5 min. For the second (nested) PCR, the mix was similar to the first, except that 2 μL of the first PCR product was used as template DNA and the reaction mixture was incubated at 94 °C for 5 min; 30 cycles of 94 °C for 30 s and 72 °C for 1 min; with final extension at 72 °C for 5 min. The final product of the nested PCR is a 196 bp fragment that indicates the presence of P. brasiliensis DNA in the samples analyzed. As a positive control, 10 μL containing 10 ng of purified P. brasiliensis DNA was used in all PCR assays. In order to ensure validity and reliability of our results, positive and negative internal controls were always included. Additionally, sample preparation, DNA extraction, mix and primer preparation as well as the agarose gel electrophoresis were performed in separated laboratory areas using different sets of pipettes for each step and in separate air-flow hoods regularly decontaminated using UV light and DNAZap solution.
As a control to verify amplifiable DNA or to detect the presence of PCR inhibitors in the clinical samples, a PCR designed to amplify the human gene for β-globin was carried out as described by Bialek et al.33 All of the PCR reactions were run on a Peltier Thermal Cycler PT100 (MJ Research, USA). The PCR products were visualized by electrophoresis on 2% agarose gels (Sigma Chemical Co., St. Louis, MO, USA), using red gel and a UV trans illuminator (Molecular imager® Gel DocTMXR+BIORAD). All of the nested PCR products were sequenced to verify that the amplified DNA fragment corresponded to the gp43 membrane protein of P. brasiliensis.
Detection limit
To establish the detection limit of the nested PCR assay, we extracted and quantified DNA from a P. brasiliensis yeasts suspension and performed serial dilutions (1:10) ranging from 100 ng to 1 fg units. Each of these dilutions was used for a specific PCR, to determine the amount of DNA at the assay's detection limit.
Data analysis
The sequences obtained were properly edited and aligned using Sequencher software (version 4.8) and homology searches of all sequences were carried out using the BLASTn program from the National Center for Biotechnology Information, Washington, DC. The sequences were categorized according to E-values (error probability) as provided by BLASTn, using values lower than 1 × 10−40.
Sensitivity and specificity rates of the P. brasiliensis nested PCR were calculated using those samples diagnosed positively by microbiological culture or microscopic observation of the fungus on direct exam or histopathology and according to the method of Galen and Gambino.34
Results
Detection of P. brasiliensis gp 43 DNA in clinical samples
A total of 25 clinical samples from patients with paracoccidioidomycosis that had been diagnosed by culture or Gomori methenamine stain were analyzed through nested PCR. Fifteen samples (60%) turned out positive by culture and 10 were positive by KOH direct examination (40%) and all of the 25 specimens tested positive for gp43 nested PCR assay. To assess the specificity of this nested PCR assay, 115 clinical samples collected from patients with other diagnosed infections by culture and/or specific stains (Gomori methenamine silver) and 51 negative controls (30 respiratory negative samples and 21 peripheral blood samples from healthy individuals) were analyzed. Our P. brasiliensis gp43 nested PCR exhibited 100% specificity using both negative controls as well as those clinical samples diagnosed with other infections (Table 3).
Table 3.
Nested PCR results obtained on human clinical samples from patients with paracoccidioidomycosis (A) or other respiratory diseases different from paracoccidioidomycosis as well as healthy individuals taken as controls (B).
| Specimen type | Proven disease | Number of samples |
|
|---|---|---|---|
| Total | Positive PCR | ||
| (A) | |||
| Bronco alveolar lavage | Paracoccidioidomycosis | 2 | 2 |
| Sputum | Paracoccidioidomycosis | 8 | 8 |
| Biopsy | Paracoccidioidomycosis | 15 | 15 |
| Total | 25 | 25 | |
| (B) | |||
| Bronco alveolar lavage | Cryptococcosis | 10 | 0 |
| Pneumocystis pneumonia | 6 | 0 | |
| Histoplasmosis | 5 | 0 | |
| Aspergillosis | 6 | 0 | |
| Tuberculosis | 4 | 0 | |
| No disease | 25 | 0 | |
| Bronchial lavage | Cryptococcosis | 6 | 0 |
| Aspergillosis | 3 | 0 | |
| Tuberculosis | 7 | 0 | |
| No disease | 4 | 0 | |
| Sputum | Aspergillosis | 2 | 0 |
| Tuberculosis | 7 | 0 | |
| No disease | 1 | 0 | |
| Tracheal aspirate | Aspergillosis | 1 | 0 |
| Candidiasis | 7 | 0 | |
| Tuberculosis | 1 | 0 | |
| Biopsy | Candidiasis | 6 | 0 |
| Aspergillosis | 6 | 0 | |
| Cryptococcosis | 4 | 0 | |
| Tuberculosis | 1 | 0 | |
| Histoplasmosis | 5 | 0 | |
| Cerebrospinal fluid | Cryptococcosis | 24 | 0 |
| Pleural fluid | Aspergillosis | 1 | 0 |
| Peritoneal fluid | Candidiasis | 3 | 0 |
| Whole blood | No disease | 21 | 0 |
| Total | 166 | 0 | |
The presence of PCR inhibitors was ruled out because all of the clinical samples with negative result in the P. brasiliensis gp43 nested PCR assay allowed amplification of a specific fragment of the human β-globin gene.
The entire purified DNA from the three P. brasiliensis isolates used (ATCC 60855, Pb339 and Pb18), tested positive in the nested PCR assay; however, the DNA isolated from culture of P. lutzii (Pb 01 Like) yielded a negative result in the nested PCR assay. In addition, none of the DNA isolated from cultures of related microorganisms tested positive in the P. brasiliensis gp43 nested PCR assay (Table 2).
Cross-contamination during the extraction procedure was not evident. All of the DNA extraction controls tested negative in the nested PCR assays.
Detection limits
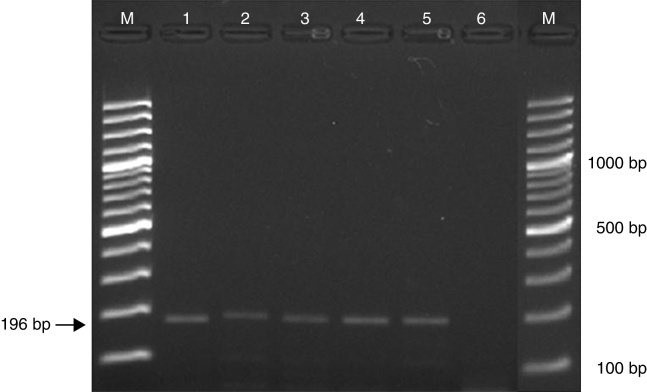
The optimized nested PCR conditions in our laboratory allowed detection down to a 1 fg of P. brasiliensis DNA (Fig. 1).
Fig. 1.
Detection limit of the Paracoccidioides brasiliensis nested PCR. M: molecular weight marker, 1: 40 ng/μL, 2: 400 pg/μL, 3: 4 pg/μL, 4: 40 fg/μL, 5: 1 fg/μL, 6: empty.
Controls
All of the DNA extraction controls tested negative in the nested PCR assays; therefore any possible cross-contamination during the extraction procedure was ruled out.
Discussion
Usually the diagnosis of PCM is based mainly on direct observation of multi budding yeast cells in clinical samples; microbiological growth in culture is too slow and a specific diagnosis can be obtained only on late stages of fungal growth. Additionally, the culture must be kept for at least two months before reporting a negative result. Although PCM immunodiagnostic is also widely employed, these tests may lead to cross-reactivity with other fungi or yield false-negative results.14, 20, 26, 27 Due to the above reasons, molecular techniques such as PCR have been, although it is still at early stages of being incorporated as routine diagnostic test for PCM.20, 23, 24, 27
Regarding the design of specific primers for fungal molecular diagnosis, it is clear that within the gene chosen as target must exist very conserved regions that could serve as primer binding sites. Additionally, such regions must flank sequences with sufficient variability to permit discrimination at genus level, at least. Therefore ribosomal genes particularly those related with 18S RNA are constantly used as they exist in all organisms in high amounts, which favors high sensitivity.20, 35, 36 Furthermore the rRNA gene contains both conserved (28S, 5.8S and 18S) and specific (ITS1 and ITS2) regions, making this gene a valuable option for molecular identification in the majority of microorganisms.
However, for the molecular diagnosis of PCM, several assays with the 18S rRNA genes of P. brasiliensis have evidenced high homology between this fungus and other similar fungi like Histoplasma capsulatum or Blastomyces dermatitidis.20 As a result other associated specific genes have been used, such as the P. brasiliensis gp43 gene, which enhances the specificity of PCM diagnosis.23 Gp43 is a 43-kDa glycoprotein that participates in the microbial interaction with the host cells, its initial interaction helping fungal internalization, incoming successful infection and posterior dissemination.37, 38
After complete cloning, sequencing and characterization of the gene encoding the gp43 region, numerous primer sets have been designed for molecular diagnosis of P. brasiliensis.23, 28, 39, 40
In this study we amplified a specific fragment of the sequence of gp43 P. brasiliensis protein deposited in the GenBank (U26160) using previously described primers in 2000 by Bialek et al.23; additionally, and in order to increase the disruption efficiency and liberate the P. brasiliensis DNA from tissue homogenates, the DNA extraction protocol used an additional incubation period with recombinant lyticase (1 UI/μL) at 37 °C for 45 min.
Given that the DNA isolated from cultures of P. lutzii yielded negative results with our P. brasiliensis nested PCR assay, we decided to confirm these results aligning different sequences of gp43 protein of P. lutzii, obtained from the web-page of the Fungal Genomics Group at the Broad Institute, to run an in-silico PCR using the sequences of our primers. Effectively we found that the primers used in our nested PCR did not match P. lutzii sequences.
Polymorphisms in the sequence of the gene encoding for the glycoprotein gp43 in P. lutzii in contrast to P. brasiliensis have been reported; these polymorphisms cause significant and decisive changes on some motifs of the protein, interfering directly with its structure and function. The most significant differences between both gp43 sequences of P. lutzii and P. brasiliensis are: the homology rate in the gp43 sequence between those Paracoccidioides species is only 81%; the gp43 protein in P. lutzii is not glycosylated due to a change in the amino acid sequence from NRT to KRT in the N-glycosylation site. P. lutzii gp43 protein has an amino acid change from NKP to NEP in the glucanase active site. However, this protein retain full glucanase activity,36 in contrast to that seen in other studies with P. brasiliensis gp43 protein37; in respect to the role of gp43 protein in the adhesion of yeast to components of the extracellular matrix, the gp43 protein in both Paracoccidioides species maintain similar ability to bind to laminin and fibronectin proteins. Finally, it is suggested that the gp43 protein is expressed at low amounts by P. lutzii both in vivo and in vitro.36 Although both Paracoccidioides species express gp43 glycoprotein, there are some genetic differences in P. lutzii which modify structural properties that possibly affect functionality and protein expression, therefore, its recognition by the primers used in our nested PCR.41, 42
We found that the P. brasiliensis nested PCR assay tested positive for all 25 clinical samples from patients diagnosed with PCM. None of the 115 clinical samples from patients diagnosed with infections other than PCM tested positive for P. brasiliensis in the nested PCR assay, translating 100% specificity.
In conclusion and in line with the report by Bialek et al.,23 P. brasiliensis nested PCR assay is a sensitive, specific, and reproducible diagnostic method to be used in different clinical samples. We confirmed a high sensitivity for detecting P. brasiliensis DNA in concentrations down to 1 fg. The nested PCR may be a useful tool not only for the rapid diagnosis of acute or chronic PCM but also for monitoring parasite clearance during therapy and follow-up. Although currently in Colombia there has been no reported case of PCM by P. lutzii, as perspective of this study, we suggest the necessity of future studies focusing in the design of primers able to detect molecular targets more conserved in both species in order to detect both P. brasiliensis and P. lutzii and be able to make a timely diagnosis of PCM at any endemic area for this mycosis.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by: COLCIENCIAS, Bogotá, Colombia (Project No. 2213-519-28916), the Corporación para Investigaciones Biológicas (CIB), the Universidad de Antioquia (UdeA), the Universidad Pontificia Bolivariana (UPB), and the Research Committee (CODI) of the Universidad de Antioquia through the Sustainability Strategy for Groups A1 and A Program 2013–2014. Medellín, Colombia. Convocatoria Nacional Jóvenes Investigadores e Innovadores of Colciencias, Number 501, year 2010 (supporting Marcela Gaviria). Convocatoria Nacional Jóvenes Investigadores e Innovadores of Colciencias, Number 645, year 2014 (supporting Vanessa Rivera). Mycotic Research Branch, Centers for Disease Control and Prevention (CDC), for providing purified DNAs (see Table 2).
References
- 1.Nucci M., Colombo A.L., Queiroz Telles F. Paracoccidioidomycosis. Curr Fung Infect. 2009;3:15–20. [Google Scholar]
- 2.Bagagli E., Sano A., Coelho K.I., et al. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus noveminctus) captured in an endemic area of paracoccidioidomycosis. Am J Trop Med Hyg. 1998;58:505–512. doi: 10.4269/ajtmh.1998.58.505. [DOI] [PubMed] [Google Scholar]
- 3.Matute D.R., McEwen J.G., Puccia R., et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol. 2006;23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- 4.Matute D.R., Sepulveda V.E., Quesada L.M., et al. Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J Clin Microbiol. 2006;44:2153–2157. doi: 10.1128/JCM.02540-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira M.D.M., Cordeiro Theodoro R., Derengowski L.D.S., Nicola A.M., Bagagli E., Felipe M.S. Molecular and morphological data support the existence of a sexual cycle in species of the genus Paracoccidioides. Eukaryot Cell. 2013;12:380–389. doi: 10.1128/EC.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira M.M., Theodoro R.C., Nino Vega G., Bagagli E., Felipe M.S.S. Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 2014;10:e1004397. doi: 10.1371/journal.ppat.1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordeiro Theodoro R., Teixeira M.D.M., Soares Felipe M.S., et al. Genus paracoccidioides: species recognition and biogeographic aspects. PLoS ONE. 2012;7:e37694. doi: 10.1371/journal.pone.0037694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bocca A.L., Amaral A.C., Teixeira M.M., et al. Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiol. 2013;8:1177–1191. doi: 10.2217/fmb.13.68. [DOI] [PubMed] [Google Scholar]
- 9.Gegembauer G., Araujo L.M., Pereira E.F., et al. Serology of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl Trop Dis. 2014;8:e2986. doi: 10.1371/journal.pntd.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marques da Silva S.H., Rodrigues A.M., Sybren de Hoog G., Silveira Gomes F., Pires De Camargo Z. Occurrence of Paracoccidioides lutzii in the Amazon region: description of two cases. Am J Trop Med Hyg. 2012;87:710–714. doi: 10.4269/ajtmh.2012.12-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixeira M.M., Theodoro R.C., de Carvalho M.J., et al. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol. 2009;52:273–283. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Restrepo A., McEwen J.G., Castañeda E. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med Mycol. 2001;39:233–241. doi: 10.1080/mmy.39.3.233.241. [DOI] [PubMed] [Google Scholar]
- 13.Restrepo A., Benard G., de Castro C., Agudelo C.A., Tobo A.M. Pulmonary Paracoccidioidomycosis. Semin Respir Crit Care Med. 2008;29:182–197. doi: 10.1055/s-2008-1063857. [DOI] [PubMed] [Google Scholar]
- 14.Restrepo A., Tobón A.M., Cano L.E. In: Principles and practice of infectious diseases. 8th ed. Mandell G.L., Bennett J.E., Raphael D., editors. Philadelphia; 2014. Paracoccidioidomycosis; pp. 2995–3002. [Google Scholar]
- 15.Brummer E., Castañeda E., Restrepo A. Paracoccidioidomycosis: an update. Clin Microbiol rev. 1993;6:89–117. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes Colombo A., Tobón A., Restrepo A., Queiroz Telles F., Nucci M. Epidemiology of endemic systemic fungal infections in Latin America. Med Mycol. 2011;49:785–798. doi: 10.3109/13693786.2011.577821. [DOI] [PubMed] [Google Scholar]
- 17.Bellissimo Rodrigues F., Machado A.A., Martinez R. Paracoccidioidomycosis epidemiological features of a 1,000-cases series from a hyperendemic area on the southeast of Brazil. Am J Trop Med Hyg. 2011;85:546–550. doi: 10.4269/ajtmh.2011.11-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Queiroz Telles F., Escuissato D.L. Pulmonary paracoccidioidomycosis. Semin Respir Crit Care Med. 2011;32:764–774. doi: 10.1055/s-0031-1295724. [DOI] [PubMed] [Google Scholar]
- 19.Graybill J., Anstead G., Queiroz Telles F. In: Diagnosis of fungal infections. Maertens J., Marr K., editors. Informa Healthcare; New York: 2007. The diagnosis of endemic Mycoses; pp. 291–353. [Google Scholar]
- 20.Teles F.R.R., Martins M.L. Laboratorial diagnosis of paracoccidioidomycosis and new insights for the future of fungal diagnosis. Talanta. 2011;85:2254–2264. doi: 10.1016/j.talanta.2011.07.099. [DOI] [PubMed] [Google Scholar]
- 21.Ayats J., Martín Mazuelos E., Pemán J., et al. Recomendaciones sobre el diagnóstico de la enfermedad fúngica invasora de la Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC). Actualización 2010. Enferm Infecc Microbiol Clin. 2011;29:39e1–415e. doi: 10.1016/j.eimc.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Sifuentes Osornio J., Corzo León D.E., Ponce de León L.A. Epidemiology of invasive fungal infections in Latin America. Curr Fungal Infect Rep. 2012;6:23–34. doi: 10.1007/s12281-011-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bialek R., Ibricevic A., Aepinus C., et al. Detection of Paracoccidioides brasiliensis in tissue samples by a nested PCR assay. J Clin Microbiol. 2000;38:2940–2942. doi: 10.1128/jcm.38.8.2940-2942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Faveri Pitz A., Koishi A.C., Tavares E.R., et al. An optimized one-tube, semi-nested PCR assay for Paracoccidioides brasiliensis detection. Rev Soc Bras Med Trop. 2013;46:783–785. doi: 10.1590/0037-8682-1625-2013. [DOI] [PubMed] [Google Scholar]
- 25.Buitrago M.J., Merino P., Puente S., et al. Utility of real-time PCR for the detection of Paracoccidioides brasiliensis DNA in the diagnosis of imported paracoccidioidomycosis. Med Mycol. 2009;47:879–882. doi: 10.3109/13693780802713208. [DOI] [PubMed] [Google Scholar]
- 26.Koishi A.C., Fonseca Vituri D., Dionízio Filho P.S.R., Sasaki A.A., Soares Felipe M.S., Venancio E.J. A semi-nested PCR assay for molecular detection of Paracoccidioides brasiliensis in tissue samples. Rev Soc Bras Med Trop. 2010;43:728–730. doi: 10.1590/s0037-86822010000600026. [DOI] [PubMed] [Google Scholar]
- 27.Dias L., de Carvalho L.F., Romano C.C. Application of PCR in serum samples for diagnosis of paracoccidioidomycosis in the southern Bahia-Brazil. PLoS Negl Trop Dis. 2012;6:e1909. doi: 10.1371/journal.pntd.0001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatibana B.T., Sano A., Uno J., et al. Detection of Paracoccidioides brasiliensis gp43 gene in sputa by loop-mediated isothermal amplification method. J Clin Lab Anal. 2009;23:139–143. doi: 10.1002/jcla.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai T., Sano A., Mikami Y., et al. A new PCR primer for the identification of Paracoccidioides brasiliensis based on rRNA sequences coding the internal transcribed spacers (ITS) and 5 × 8S regions. Med Mycol. 2000;38:323–326. doi: 10.1080/mmy.38.4.323.326. [DOI] [PubMed] [Google Scholar]
- 30.Theodoro R.C., Candeias J.M.G., Araújo J.P., et al. Molecular detection of Paracoccidioides brasiliensis in soil. Med Mycol. 2005;43:725–729. doi: 10.1080/13693780500129418. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J., Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2001. Commonly used techniques in molecular cloning; p. 2344. [Google Scholar]
- 32.Einsele H., Hebart H., Roller G., et al. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bialek R., Konrad F., Kern J., et al. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J Clin Pathol. 2005;58:1180–1184. doi: 10.1136/jcp.2004.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galen R., Gambino S. J. Willey and Sons; New York: 1975. Beyond normality: the predictive value and efficiency of medical diagnosis. [Google Scholar]
- 35.Lindsley M.D., Hurst S.F., Iqbal N.J., Morrison C.J. Rapid identification of dimorphic and yeast-like fungal pathogens using specific DNA probes. J Clin Microbiol. 2001;39:3505–3511. doi: 10.1128/JCM.39.10.3505-3511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richini Pereira V.B., Gimenes Bosco S.D.M., Cordeiro Theodoro R., Da Graça Macoris S.A., Bagagli E. Molecular approaches for eco-epidemiological studies of Paracoccidioides brasiliensis. Mem Inst Oswaldo Cruz. 2009;104:636–643. doi: 10.1590/s0074-02762009000400018. [DOI] [PubMed] [Google Scholar]
- 37.Popi A.F., Lopes J.D., Mariano M. GP43 from Paracoccidioides brasiliensis inhibits macrophage functions. An evasion mechanism of the fungus. Cell Immunol. 2002;218:87–94. doi: 10.1016/s0008-8749(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 38.Konno F.T.C., Maricato J., Konno A.Y.C., et al. Paracoccidioides brasiliensis GP43-derived peptides are potent modulators of local and systemic inflammatory response. Microbes Infect. 2012;14:517–527. doi: 10.1016/j.micinf.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 39.Semighini C.P., de Camargo Z.P., Puccia R., Goldman M.H.S., Goldman G.H. Molecular identification of Paracoccidioides brasiliensis by 5′ nuclease assay. Diagn Microbiol Infect Dis. 2002;44:383–386. doi: 10.1016/s0732-8893(02)00472-8. [DOI] [PubMed] [Google Scholar]
- 40.Gomes G.M., Cisalpino P.S., Taborda C.P., de Camargo Z.P. PCR for diagnosis of paracoccidioidomycosis. J Clin Microbiol. 2000;38:3478–3480. doi: 10.1128/jcm.38.9.3478-3480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leitão N.P., Vallejo M.C., Conceição P.M., Camargo Z.P., Hahn R., Puccia R. Paracoccidioides lutzii Plp43 is an active glucanase with partial antigenic identity with P. brasiliensis gp43. PLoS Negl Trop Dis. 2014;8:e3111. doi: 10.1371/journal.pntd.0003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cisalpino P.S., Puccia R., Yamauchi L.M., Cano M.I.N., da Silveira F., Travassos L.R. Cloning, characterization, and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J Biol Chem. 1996;271:4553–4560. doi: 10.1074/jbc.271.8.4553. [DOI] [PubMed] [Google Scholar]