Abstract
The expression of lipase from Pseudomonas sp. strain KWI-56 (recently reclassified as Burkholderia cepacia) had been found to be dependent on an activator gene (act) downstream of its structural gene (lip). In this work, the mature lipase was synthesized in an enzymatically active form with a cell-free Escherichia coli S30 coupled transcription-translation system by expressing a recombinant lipase gene (rlip) encoding the mature lipase in the presence of its purified activator or by coexpression of rlip and act. The in vitro expression systems were used for studying the folding process of the lipase. The addition of dithiothreitol in the expression systems decreased the activity dramatically without affecting the synthesis level of the lipase, whereas the in vitro-synthesized active lipase was relatively stable even in the presence of dithiothreitol. This phenomenon was further investigated by constructing mutant lipase genes only in vitro by PCR without gene cloning. Replacements of cysteine residues (Cys190 and Cys270) forming a sole putative disulfide bond to serine residues decreased the lipase activity greatly, suggesting that the disulfide bond was essential for the proper folding of the lipase. In addition, replacing Asp242 and Asp288, which were deduced to be part of a Ca2+ binding site, also greatly decreased the activities of the in vitro-synthesized lipases. The role of the Ca2+ binding site in the activation of the lipase is also discussed.
Extracellular lipase produced by members of the genus Pseudomonas has a wide range of potential applications in the hydrolysis, esterification, and transesterification of triglycerides or in the chiral selective synthesis of esters (11, 20). In recent years, a variety of Pseudomonas lipase genes have been cloned, sequenced, and characterized (5, 6, 8, 14, 21, 26). These lipases can be divided into three classes (designated classes I to III) depending on their amino acid sequence homology (34). The lipases of classes I and II have been found to require additional genes that are located downstream of the lipase genes for their active expression (1, 5, 9, 12, 15, 41). These genes were reported to direct the synthesis of chaperone-like proteins, named modulator, activator, or chaperone, to assist the correct folding of lipase specifically. The inactive lipases produced by recombinant Escherichia coli cells could be reactivated by an in vitro denaturation-renaturation method with the assistance of such chaperones (10, 13, 16, 33, 34).
The crystal structures of the lipases from Pseudomonas cepacia and Pseudomonas glumae have been determined previously (24, 31). It was found that a disulfide bond and a calcium binding site existed in the tertiary structures of the lipases. It is possible that they exist commonly among the Pseudomonas lipases of classes I and II because of the conservative amino acid sequence structures in the corresponding sites (39). The disulfide bond and the calcium binding site are thought to be important for the stability of lipase. However, their functions have not been fully examined experimentally.
The technology of cell-free protein synthesis has been improved by various methods to raise its productivity in a batch system since its establishment in the 1960s (22, 23, 27, 28, 40). Now, cell-free protein synthesis with various reactors from a cloned DNA fragment provides an alternative way to obtain large amounts of the desired protein without using living cells (29, 30, 37). Since it is not necessary to clone a mutated gene and to introduce a recombinant plasmid into host cells, this is an efficient method to get mutated proteins (4, 35). It is even more advantageous for the synthesis of proteins that are fatal to the host organism.
An extracellular lipase from Pseudomonas sp. strain KWI-56 (recently reclassified as Burkholderia cepacia) was reported to be a thermostable enzyme with potential industrial uses (17). It was purified, and its structural gene (lip) was cloned together with an activator gene (act), which was necessary for the active production of the lipase (18, 19). The expression of this lipase in recombinant E. coli resulted in only a small amount of active lipase, which was found in the membrane fraction of the E. coli cells. However, the processed site of its signal peptide was changed from the original one. Moreover, the recombinant plasmid was gradually lost in the E. coli hosts. These shortages greatly reduced available methods to investigate the activation and folding process.
In this work, a recombinant lipase gene (rlip) encoding the mature lipase without its N-terminal signal sequence and the activator gene (act) were subcloned into a high-expression plasmid under the control of the T7 RNA polymerase promoter. Active lipase was synthesized in a cell-free coupled transcription-translation system with E. coli S30 extract by coexpressing rlip and act or by expressing only rlip in the presence of partially purified activator protein which was obtained from activator-overproducing cells. Mutant lipases containing the site-directed replacements of amino acid residues were synthesized directly from templates prepared by PCR in the in vitro expression system. These methods greatly facilitated the analysis of the roles of the disulfide bond and the calcium binding site. The results suggest their important functions in the activation process of the lipase.
MATERIALS AND METHODS
Materials.
All restriction enzymes, T4 DNA ligase, and Ex Taq and LA Taq DNA polymerase were from TaKaRa Shuzo (Kyoto, Japan). DNA primers for PCR were synthesized by Nippon Flour Mills (Tokyo, Japan). p-Nitrophenyl octanoate was from Hayashi Kasei (Nagoya, Japan).
Bacterial strains, plasmids, and media.
The E. coli strain XL1-Blue (3) was used as a host for the manipulation of recombinant plasmids. E. coli BL21(DE3)/pLysS (38) was used as a host for overexpression of the recombinant lipase and the activator protein. A plasmid, pRSET, containing the T7 promoter and its terminator was purchased from Invitrogen (Groningen, The Netherlands). Luria-Bertani medium was used for the cultivation of E. coli, while ampicillin (50 μg/ml) and/or chloramphenicol (25 μg/ml) was added as a supplement if necessary.
Construction of plasmids.
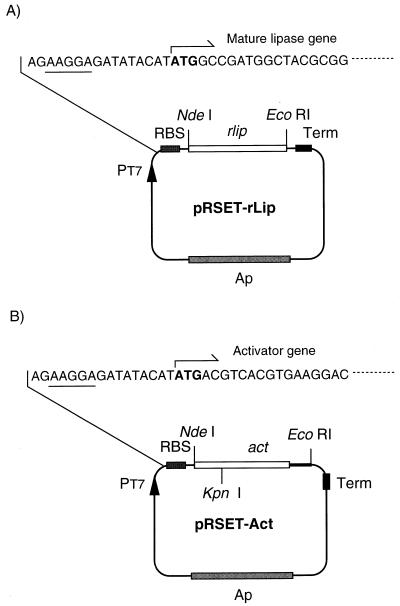
The lipase and activator genes of Pseudomonas sp. strain KWI-56 were subcloned by PCR from pLP64 (18), which contains a 2.9-kb fragment carrying both of the genes. A part of lip without its N-terminal signal-encoding region was amplified by PCR with primers Lip-F (GTCGGATCCATATGGCCGATGGCTACGCGGCGAC) and Lip-R (TATGAATTCATCGATTACACGCCCGCCAG), which contained NdeI and EcoRI sites, respectively. Then the fragment was ligated to NdeI-EcoRI-digested pRSET, resulting in pRSET-rLip (Fig. 1A). For cloning act, its 5′-end fragment was first amplified with primers Act1-F (AGGCCTATCATATGACGTCACGTGAAGGAC) and Act1-R (AACGGTACCGTCGAGCTGT), which contained NdeI and KpnI restriction sites, respectively. The amplified fragment was treated with NdeI and KpnI and ligated to pRSET. Then a KpnI-EcoRI fragment from pLP64 was inserted into this intermediate plasmid, yielding pRSET-Act (Fig. 1B). In both PCRs, DNA was amplified by Ex Taq DNA polymerase under the following conditions: 94°C for 5 min; 25 cycles of 94°C for 0.5 min, 60°C for 0.5 min, and 72°C for 1 min; and finally 72°C for 5 min. The subcloned genes were sequenced with a Taq Dye Deoxy cycle sequencing kit and an ABI Prism 310 genetic analyzer (Perkin-Elmer Corporation) in accordance with the instructions of the manufacturer. The nucleotide sequences of rlip and act were confirmed to be identical to the corresponding sequence of pLP64.
FIG. 1.
Schematic representation of structure of the expression plasmids. pRSET-rLip (A) and pRSET-Act (B) were constructed as described in the text. Underlined nucleotides are a ribosome binding site (RBS).
Expression of rlip and act in E. coli and partial purification of the activator.
E. coli BL21(DE3)/pLysS harboring pRSET-Act was cultivated in 800 ml of Luria-Bertani medium containing 1% glucose, 50 μg of ampicillin per ml, and 25 μg of chloramphenicol per ml at 37°C to achieve an optical density of about 0.6 at 660 nm. IPTG (isopropyl-β-d-thiogalactopyranoside) was added into the culture broth to a final concentration of 0.4 mM, and the culture was incubated at 37°C for another 2 h. The cells harvested by centrifugation were suspended in 35 ml of 50 mM Tris-HCl buffer (pH 8.0) and treated by ultrasonic disintegration, resulting in a crude cell extract. Following centrifugation (10,000 × g for 15 min at 4°C), the crude cell extract was precipitated by 20% saturated ammonium sulfate. The pellets, after centrifugation at 10,000 × g for 20 min, were resuspended in 20 ml of 50 mM Tris-HCl buffer (pH 7.4) and dialyzed against the same buffer containing 10 mM Mg(OAc)2. The purity of the activator was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) staining with Coomassie brilliant blue R-250.
The expression for rlip was carried out by the same method as that for act. Cells of E. coli BL21(DE3)/pLysS bearing pRSET-rLip harvested from the expression cultivation were suspended in 50 mM Tris-HCl buffer (pH 8.0) containing 30 mM NaCl. After ultrasonic disruption, the supernatant obtained by centrifugation at 3,000 × g for 10 min was collected as the crude cell extract. Then the soluble and insoluble proteins were separated by centrifugation at 8,000 × g for 20 min, and the distribution of the lipase protein was analyzed by SDS-PAGE.
In vitro coupled transcription-translation.
E. coli S30 extract in acetate buffer [9.9 mM Tris-acetate (pH 7.4) containing 14 mM Mg(OAc)2 and 60 mM KOAc] was prepared according to the procedure of Ellman et al. (7). The in vitro coupled transcription-translation was carried out by the method described by Ohuchi et al. (32) with some modifications. Three microliters of pRSET-rLip (100 μg/ml) or 3 μl of PCR-amplified DNA product was used as template in a total volume of 40 μl of reaction mixture containing the following: 42.3 mM Tris-acetate buffer, pH 7.4; 0.92 mM ATP; 0.64 mM GTP; 0.64 mM CTP; 0.64 mM UTP; 30 mM creatine phosphate; 0.11 mg of creatine kinase per ml; 0.24 mM (each) 20 kinds of unlabeled amino acid; 0.13 mg of E. coli tRNAs per ml; 26.0 μg of folinic acid per ml; 3% (wt/vol) polyethylene glycol 6000; 7.5 mM Mg(OAc)2; 112.5 mM KOAc; 26.9 mM NH4OAc; 7.5 μg of rifampin per ml; 10 μg of T7 RNA polymerase per ml; and 40% E. coli S30 extract. 14C-labeled leucine at 0.012 mM was included in the reaction system for scintillation counting and SDS-PAGE autoradiography.
Coexpression of rlip and act was carried out with both pRSET-rLip and pRSET-Act as transcription templates with a molar ratio of 1:1, while expression of rlip from pRSET-rLip with presynthesized activator was carried out by adding 0.3 mg of partially purified activator per ml.
After translation with [14C]leucine, 5 μl of the reaction solution was applied for measuring the incorporated [14C]leucine by the method of trichloroacetic acid precipitation. The amount of synthesized protein was deduced from the scintillation of radioactive leucine incorporated into the acid-insoluble fraction. Alternatively, 5 μl of reaction mixture was applied to SDS–12% polyacrylamide gels with rainbow 14C-methylated protein (molecular mass, 2,350 to 46,000 Da; Amersham Pharmacia Biotech Co., Tokyo, Japan) as molecular weight markers. The gels were dried and exposed to a Fuji imaging plate for 16 to 20 h and were analyzed with a Fujix BAStation (Fuji Photo Film Co. Ltd., Tokyo, Japan).
Introduction of mutation into lipase gene by PCR.
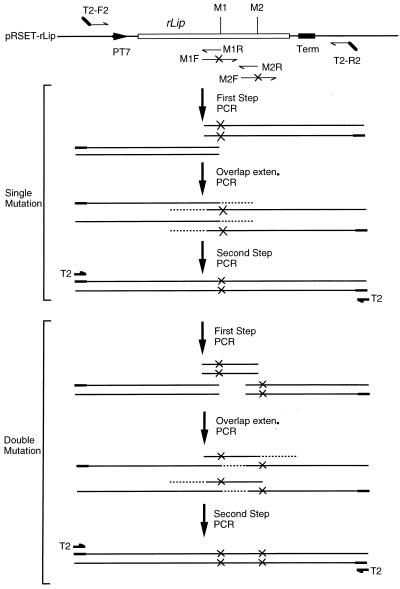
Mutations in rlip were generated by overlapping PCR (4) with some modifications as illustrated in Fig. 2. To introduce double mutations, for example, the whole sequence of the T7 promoter, rlip, and the T7 terminator was amplified as three fragments by using three pairs of primers. The standard conditions for the first-step PCR were as follows: 30 cycles of 10 s at 98°C, 30 s at 55°C, and 1 min at 72°C. The annealing temperatures varied depending on the sequences of primers. In the overlap extension PCR, the three independently amplified fragments of 1 μl each were mixed and subjected to five cycles of denaturation (10 s at 98°C), annealing (30 s at 55°C), and extension (1 min at 72°C) in 50 μl of reaction mixture without any primer. Finally, the reconstituted fragment was amplified by 25 cycles of 10 s at 98°C, 30 s at 55°C, and 1 min at 72°C after the addition of 0.5 μM T2 primer. By using the homoprimer T2 (2), amplification of the original sequence was greatly reduced. LA Taq polymerase was applied throughout this experiment.
FIG. 2.
Schematic representation of the strategy for overlapping PCR introducing mutations into rlip. M1 and M2 are mutation sites; M1R and M2R represent overlap primers; M1F and M2F represent primers containing mutation sites. In the first-step PCR, two fragments for single mutation or three fragments for double mutation are amplified. After overlap extension (exten.) PCR, the reconstituted mutant genes are further amplified in the second-step PCR by a homoprimer, T2. The sequences of the primers in the experiment are listed here: T2, GCGTACTAGCGTACCACGTG; F2-T2, GCGTACTAGCGTACCACGTGATCTCGATCCCGCGAAATTAATACGACTCAC; R2-T2, GCGTACTAG CGTACCACGTGGCCAGATCCGGATATAGTTCCTCCTTTCAG; C190R, ACTGCCCGGCGCACCCAGGC; C190SF, GCCTGGGTGCGCCGGGCAGTTCGCAGACCGGCGCGCCGACCGA; C270R, CTTCGACACGAGCCCGTCGTTCT; C270SF, AGAACGACGGGCTCGTGTCGAAGTCGAGTGCG CTGTACGGCAAGGTGC; D242R, GAGCACGTTCGCCAGATCAA; D242SF, TTGATCTGGCGAACGTGCTCGCGCCGTCGACGCTCGCGCTGTT; D288R, GAGGTGGTTCCACTTGTAGCTC; D288AF, GAGCTACAAGTGGAACCACCTCGCGGAGATCAACCAGCTGCTCGGCG.
Sets of primers used for each mutant were as follows: for C190S, T2-F2 and C190R and C190SF and T2-R2; for C270S, T2-F2 and C270R and C270SF and T2-R2; for C190/270S, T2-F2 and C190R, C190SF and C270R, and C270SF and T2-R2; for D242A, T2-F2 and D242R and D242AF and T2-R2; for D288A, T2-F2 and D288R and D288SF and T2-R2; for D242/288A, T2-F2 and D242R, D242SF and D288R, and D288SF and T2-R2; for wild type, T2-F2 and T2-R2.
Lipase activity assay.
p-Nitrophenyl octanoate was emulsified by ultrasonication at a final concentration of 25 mM in the presence of 0.5% Triton X-100 in 50 mM potassium phosphate buffer (pH 6.5). The substrate solution of 190 μl was preincubated at 37°C for 3 min. Properly diluted lipase sample (10 μl) was added and incubated for another 10 min. The reaction was stopped by adding 800 μl of ethanol. The mixtures were then clarified by centrifugation, and the absorbance at 400 nm was measured. One unit of lipase activity is defined as the amount of enzyme that liberates 1 μmol of p-nitrophenol in 1 min at 37°C.
RESULTS
Expression of the lipase and the activator in E. coli.
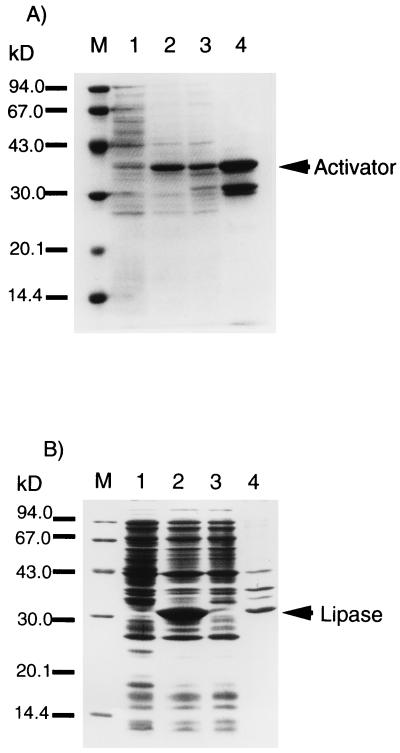
Both pRSET-rLip and pRSET-Act (Fig. 1) were transformed into E. coli BL21(DE3)/LysS. The SDS-PAGE analysis of the cell lysate of the recombinant E. coli cells showed that a 33-kDa band appeared in the cells bearing pRSET-rLip, while a 37-kDa band was observed in those bearing pRSET-Act (Fig. 3). The recombinant lipase was not soluble, suggesting that it was produced as an inclusion body, whereas the activator was recovered in the soluble fraction. Then the activator was further purified by precipitation with ammonium sulfate as described in Materials and Methods. The purity of the activator was over 50% as analyzed by SDS-PAGE.
FIG. 3.
SDS-PAGE analysis of in vivo expression of the lipase and the activator. (A) Expression of act encoded by E. coli BL21(DE3)/LysS bearing pRSET-Act. Lane M, marker proteins; lane 1, whole-cell proteins of E. coli BL21(DE3)/LysS bearing pRSET-Act before induction by IPTG; lane 2, whole-cell proteins of E. coli BL21(DE3)/LysS bearing pRSET-Act after induction by IPTG; lane 3, cell lysate of whole-cell proteins of lane 2; lane 4, activator partially purified by 20% saturated (NH4)2SO4 precipitation. (B) Expression of rlip by E. coli BL21(DE3)/LysS bearing pRSET-rLip. Lane M, marker proteins; lane 1, whole-cell proteins of E. coli BL21(DE3)/LysS; lane 2, whole-cell proteins of E. coli BL21(DE3)/LysS bearing pRSET-rLip induced by IPTG; lane 3, soluble proteins in whole-cell proteins of lane 2; lane 4, insoluble proteins in whole-cell proteins of lane 2.
Activation of the lipase synthesized in the in vitro coupled transcription-translation system by coexpression of rlip and act.
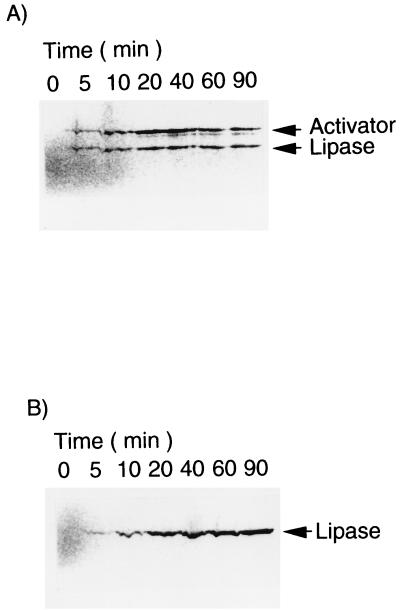
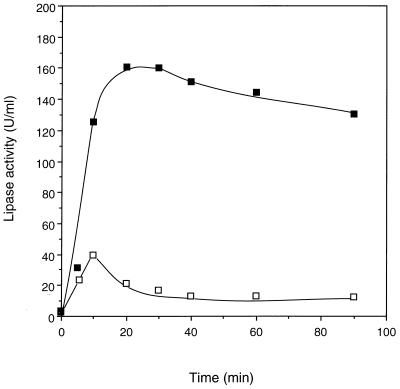
Simultaneous expression of rlip and act, encoded by pRSET-rLip and pRSET-Act, respectively, was carried out in the coupled transcription-translation system. As shown by autoradiography of SDS-PAGE (Fig. 4A), mature lipase and activator appeared in the reaction mixture with the molecular masses of 33 and 37 kDa, respectively. The total synthesized protein was also assayed by scintillation counting, revealing that it reached a plateau after about 20 min and kept nearly constant within a time course of 90 min (data not shown). On the other hand, the assay of the lipase activities of the coexpression reaction showed that the activity rose quickly after the translation started, reached a peak of 157 U/ml after 20 min, and decreased slightly afterward (Fig. 5). If the rlip gene was expressed without the act gene, no activity was detected in the reaction mixture, although the synthesis of lipase protein was not affected.
FIG. 4.
Autoradiography analysis of in vitro expression of rlip in the coupled transcription-translation system with the act gene (A) or partially purified activator (B). Five-microliter samples were analyzed by SDS-PAGE with subsequent autoradiography.
FIG. 5.
Time course of the lipase activity determined by coexpression of rlip and act without DTT (■) and with 2 mM DTT (□).
These data show that even in the coupled transcription-translation system, the expression of active lipase could be obtained and required the presence of the act gene. In addition, lipase thus produced was relatively stable compared with the lipase from Pseudomonas sp. strain 109 in the same expression system (42).
Activation of the lipase expressed from the rlip gene in the cell-free coupled transcription-translation system with partially purified activator.
The full length of the activator was successfully produced in the recombinant E. coli as a soluble protein. Its function in a partially purified form was investigated in the in vitro expression of rlip encoded by pRSET-rLip.
The synthesis of the lipase was analyzed by autoradiography of SDS-PAGE and scintillation counting. As shown in Fig. 4B, the lipase was synthesized as a single band of 33 kDa, which corresponded to the molecular mass of the mature lipase. The time course of radioactive leucine incorporation also showed that the translation of lipase reached a plateau after 20 min of reaction and leveled off at least up to 90 min (data not shown).
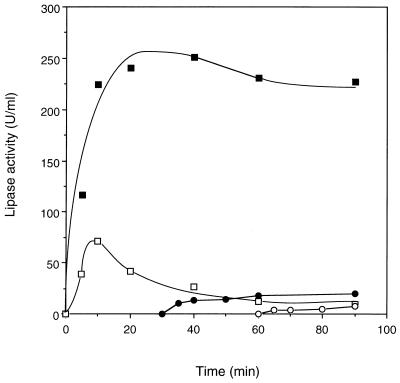
The enzyme activity in the reaction mixture with partially purified activator was assayed at 5, 10, 20, 40, 60, and 90 min. After 20 min of translation, the lipase activity also reached a peak and remained relatively stable afterward (Fig. 6). The specific activity of the lipase at 60 min of translation was 5,940 U/mg, calculated by dividing the activity by the protein concentration, which was deduced from scintillation counting. The activity level obtained with the presynthesized activator (Fig. 6) was higher than that obtained by the coexpression (Fig. 5). One reason might be that the amount of synthesized lipase protein generated by the former method (Fig. 4B) was higher than that by the latter (Fig. 4A).
FIG. 6.
Time course of the lipase activity determined by expressing rlip in the presence of the partially purified activator. The activator was added from the beginning (■), at 30 min (●), and at 60 min (○), without DTT. The reaction was also carried out with the activator in the presence of 2 mM DTT (□).
If the same amount of the activator was added into the expression mixture at 30 and 60 min after the translation started, only 20.1 and 7.3 U, respectively, of lipase activities per ml were recovered, although the protein synthesis did not change. They were 8.9 and 3.2%, respectively, of the activity obtained when the activator was present from the beginning (Fig. 6). Since the result of the radioautography of SDS-PAGE showed no serious degradation of the synthesized protein, the amount of the lipase protein remained relatively constant in the reaction mixture. These data suggest that without the activator the synthesized lipase protein would quickly fold to inactive forms that could not be easily accessed by the activator. And once the inactively folded lipase was produced, it would not be able to be quantitatively renatured even though the activator was added subsequently.
Effect of DTT on the synthesis of active lipase.
Usually, dithiothreitol (DTT) is included in the E. coli S30 expression system as a reducing agent. In the presence of a normal concentration (2 mM) of DTT, however, the lipase activity was much lower than that in its absence. In both cases of the coexpression of the two genes (Fig. 5) and the expression of rlip with the activator protein (Fig. 6), the activity of lipase decreased dramatically after 10 min of the translation reaction. But the synthesis of the lipase protein lasted longer, until a plateau was reached at about 20 min.
In the case of coexpression of the two genes, the activity obtained at 90 min in the presence of DTT was only 6.9% of that in the absence of it (Fig. 5). Similarly, with the presynthesized activator, the final product with DTT was only 4.3% as active as the lipase expressed without DTT (Fig. 6). It was also found that the translation rate and the yield of protein were almost unaffected by the addition of DTT (data not shown). Therefore, this suggests that DTT did not interfere with the protein synthesis of the lipase but affected its activation process or enzyme stability.
Effect of mutation at the disulfide bond-forming sites on the activation process of lipase.
By analyzing the amino acid sequence of the lipase from Pseudomonas sp. strain KWI-56 (18), two Cys residues were found at positions 190 and 270. It was reported that a disulfide bond was formed between the corresponding sites in the lipases of P. cepacia (24) and P. glumae (31). It is quite reasonable to deduce that a disulfide bond is formed between the two Cys residues of the lipase of Pseudomonas sp. strain KWI-56, because these two sites are highly conserved among many other Pseudomonas lipases.
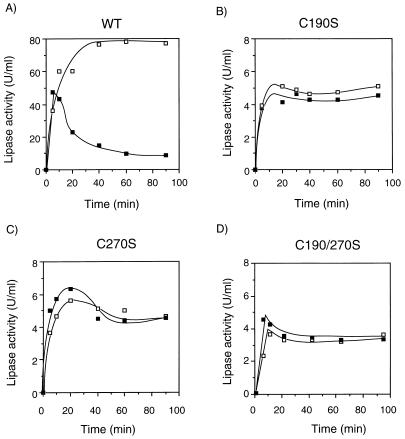
Therefore, the effect of DTT on the lipase was further investigated by making mutant lipases with amino acid replacements at these two sites. By means of overlapping PCR, Cys190 and Cys270 were replaced by Ser, yielding DNA fragments encoding mutant lipases of C190S, C270S, and C190/270S as illustrated in Fig. 2. The wild-type gene was also amplified by PCR, since the expression level from the linear template of PCR product was lower than that from the circular plasmid. The wild-type and mutant genes of rlip on PCR-amplified fragments were then coexpressed with act encoded by pRSET-Act. As a result, the time course of the wild type showed a great difference between the lipase activities with and without DTT (Fig. 7A). In its presence, a sharp peak appeared at an early stage of the time course and was followed by a rapid decrease before the activity reached a plateau gradually within 40 min. In contrast, all the mutant lipases showed a similar time course profile of the lipase activity in both the presence and the absence of DTT (Fig. 7B to D).
FIG. 7.
Expression of mutant lipases with cysteine replacements. PCR-amplified lipase genes of wild type (WT) (A), C190S (B), C270S (C), and C190/270S (D) were coexpressed with the act gene. In each panel, solid and open squares indicate the lipase activities with and without 2 mM DTT, respectively.
In order to compare the specific activities of the wild-type and the mutant lipases, the genes of the wild type, C190S, C270S, and C190/270S were expressed from their PCR templates with the presynthesized activator. The protein concentrations were deduced from the radioactive leucine by scintillation counting. The results show that the specific activities of the mutant lipases were about 11 to 27 times lower than that of the wild type (Table 1).
TABLE 1.
Specific activities of wild-type and mutant lipasesa
| Lipase | Activity (U/ml) | Protein concn (10−3 mg/ml) | Sp act (103 U/mg) |
|---|---|---|---|
| WT | 101.4 ± 3.5 | 6.9 ± 0.35 | 14.7 ± 1.3 |
| C190S | 2.20 ± 0.14 | 4.0 ± 0.22 | 0.55 ± 0.07 |
| C270S | 3.66 ± 0.11 | 3.8 ± 0.13 | 0.96 ± 0.07 |
| C190/270S | 3.05 ± 0.26 | 2.4 ± 0.05 | 1.27 ± 0.14 |
| D242A | 5.49 ± 0.08 | 2.4 ± 0.02 | 2.29 ± 0.05 |
| D288A | 7.08 ± 0.32 | 6.4 ± 0.29 | 1.11 ± 0.10 |
| D242/288A | 3.17 ± 0.13 | 2.8 ± 0.06 | 1.13 ± 0.07 |
Wild-type (WT) and mutant lipases were expressed by the coupled transcription-translation system with the PCR-amplified DNA as templates in the presence of presynthesized activator. Lipase activities and protein concentrations of the samples from 60 min of translation reaction were analyzed. Protein concentration was deduced from the radioactivity of [14C]leucine incorporated into the synthesized lipase.
DTT (2 mM) was added into the mixture of wild-type lipase, which was synthesized freshly by the cell-free system. The lipase activity remained 92% after the mixture was incubated at 37°C for 2 h. So the low activities of the wild-type lipase synthesized in the presence of DTT and the mutant lipases should be due to the folding process instead of enzyme stability.
The role of a calcium binding site in the activation of the lipase.
A calcium binding site was found in the crystal structure of the lipase from P. cepacia and P. glumae (24, 31). The calcium ion ligands in the P. cepacia lipase include two carboxylate groups of Asp242 and Asp288, two carbonyl groups of Gln292 and Val 296, and two water molecules (24). The same primary structure was also found in the lipase of Pseudomonas sp. strain KWI-56. Because of the high similarity among the three lipases, it can be deduced that the calcium binding site also exists in this lipase.
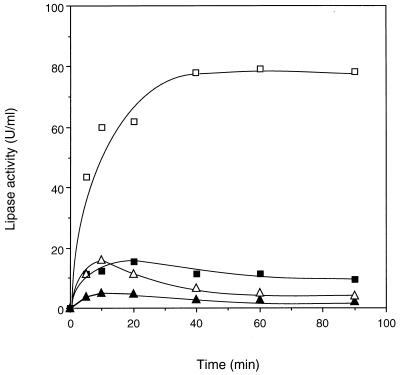
In order to investigate the role of calcium ion in the activation process of the lipase, the sole calcium binding sites of Asp242 and Asp288 were replaced by Ala. Three mutant lipase genes were constructed by overlapping PCR, yielding D242A, D288A, and D242/288A. These three genes and the wild-type gene amplified by PCR were coexpressed with the act gene in the coupled transcription-translation system. The activity levels of the single mutants D242A and D288A were about 12 and 5.3% of the wild type, respectively, while that of the double mutant D242/288A was only 2.8%. The time courses of the mutant gene expression showed a decrease in activity after 10 or 20 min, while the wild type showed a stable state after the plateau was reached (Fig. 8).
FIG. 8.
Expression of lipases mutated at the calcium binding site. PCR-amplified lipase genes from wild type (□), D242A (■), D288A (▵), and D242/288A (▴) were coexpressed with the act gene.
Because it was difficult to calculate accurately the concentration of the lipase in the coexpression system, the mutant genes were also expressed in the presence of the presynthesized activator. The specific activities of D242A, D288A, and D242/288A were 15.6, 7.6, and 7.7% of that of the wild-type lipase, respectively (Table 1).
The wild-type lipase synthesized in vitro was incubated with 20 mM EDTA at 37°C for 2 h, and no activity loss was observed. Since the wild-type lipase was relatively stable even in the presence of the chelator, the low activities of the mutated lipases were possibly owing to their ineffectiveness in the activation process without calcium binding.
DISCUSSION
Pseudomonas lipases classified as classes I and II need a kind of helper protein, named chaperone, modulator, or activator by different authors, which helps in correct folding for their active production. The activation process of the lipases has been studied in detail in vitro by the denaturation-renaturation method that refolds denatured lipase obtained from inclusion body by dialysis in the presence of the chaperone proteins. A 1:1 complex between the lipase and its modulator was reported to be formed during the refolding process (1, 13, 33). The formation of complex was proposed to weaken the activity of lipase, thus preventing membrane degradation. Furthermore, the proper folding of lipase in the periplasm of Pseudomonas cells appears to be essential for its Xcp-mediated translocation across the outer membrane (25).
The in vitro refolding, however, may have some differences from the authentic folding process in living organisms. In the denaturation-renaturation system, the refolding of the polypeptide starts from a random coil. But in living organisms, it is possible for lipase to fold partially from its N terminus during translation before it forms a complex with its specific chaperone. The intermediate structure of the lipase during the refolding process by denaturation-renaturation might be different from that of the native process.
In this work, we have succeeded in expressing the active lipase of Pseudomonas sp. strain KWI-56 in an E. coli coupled transcription-translation system. The activator protein of the lipase was added in the expression mixture after partial purification or expressed in situ from its act gene. In both cases, active lipases were synthesized, whereas no activity was detected without the activator protein or act gene. Therefore, it is confirmed that the activator is necessary for activation of the lipase in the in vitro expression system. In addition, compared with the denaturation-renaturation system, we believe that the activation process of our system is closer to that of living organisms, especially for the coexpression of rlip and act.
The timing of addition of the activator did affect protein synthesis of the lipase but did not affect the activation greatly. When the activator was added after 30 min of the protein translation, the activity was only 8.9% of that obtained by adding the activator from the beginning. When the activator was added after 60 min of the reaction, only 3.2% of the activity was recovered. Therefore, it seems that the freshly synthesized lipase will fold into an inactive form soon after being synthesized if there is no activator in the reaction mixture. It can be concluded that if an incorrect conformation of the lipase is once formed, it is difficult for it to be refolded into the active form even by the activator in a natural environment.
The crystal structure of the lipase of P. cepacia (PcL) contains a disulfide bond between Cys190 and Cys270, which was determined by X-ray crystallography (24). Because the lipase of Pseudomonas sp. strain KWI-56 has a primary structure identity of 97% with PcL, the same disulfide bond is likely to be formed. This disulfide bond has been believed to contribute to the stability of the lipase. However, Hobson et al. reported that in the presence of DTT, no lipase activity could be recovered by renaturation from the denatured lipase of P. cepacia DSM3959 even if the modulator LimA was added (13). This report suggests that the disulfide bond may play an important role during the folding of the lipase.
The role of the disulfide bond in the production of active lipase was investigated in detail with a cell-free protein synthesis system in this work. If 2 mM DTT was added into the expression system, the activity decreased dramatically after a peak at 10 min, while without DTT the activity kept rising for an additional 10 to 20 min (Fig. 5 and 6). Once the active lipase was finally formed in the absence of DTT, it was relatively stable at 37°C even with DTT being added subsequently. In the presence of DTT, active lipase was synthesized, but at a much lower activity level than that without DTT. A further analysis of the protein synthesis showed little difference between the two cases. One possible explanation of this phenomenon is that the activator binds to the freshly synthesized lipase and assists in its correct folding to yield a transient active state, and this step does not depend on the formation of the disulfide bridge. The transient active state, however, is not stable without the disulfide bond, which is also possibly important for the correct folding steps following its formation. Finally, a stable and active tertiary structure is reached when the disulfide bond is formed between the corresponding Cys residues. In contrast, if this disulfide bond is not formed because of DTT, the lipase will change to a much less active form.
This phenomenon was further investigated by constructing mutant lipases by PCR in vitro. Cysteine residues forming the sole putative disulfide bridge were replaced with serine because they have quite similar structures except for the difference between the thiol and the hydroxyl group. Both mutant lipases with a single replacement (C190S and C270S) and that with a double replacement (C190/270S) showed a similar time course of the activity in the presence and in the absence of DTT, and their activity level was less than 10% of the wild-type activity obtained in the absence of DTT. In contrast, the time course of the wild-type lipase expression had a sharp peak in the presence of DTT but not in its absence (Fig. 7). Since none of the mutant lipases had a DTT effect, it can be confirmed that DTT directly reduced the disulfide bond, causing a failure in the activation of the synthesized lipase. But in the time courses of mutant lipase expression, no activity peak was found in the early stage of translation as expected. Therefore, another explanation could be that at the start of the reaction some disulfide bonds were still formed, resulting in transiently active lipase. The disulfide bonds were then disrupted by DTT. But the SDS-PAGE experiment to discriminate between lipase molecules with and those without disulfide bonds failed to show any difference in electrophoretic mobility for lipases synthesized at 5, 10, 20, and 60 min of reaction (data not shown). This transient state remains to be further investigated for a correct explanation.
The specific activity levels of the mutant lipases were 10 times lower than that of the wild type (Table 1). Also, it was found that the stability of the wild-type lipase at 37°C changed little even with the addition of DTT, although its stability at 70°C was decreased by DTT (data not shown). Therefore, in in vivo conditions the role of the disulfide bond between C190 and C270 should be first to make the correct folding and second to stabilize the final structure of the lipase.
Another characteristic of the structure of the lipase from Pseudomonas sp. strain KWI-56 is that it may contain a calcium binding site like the lipases from P. cepacia and P. glumae (24, 31). Because of the distance between this Ca2+ binding site and the active site, a direct involvement of calcium ion in catalysis seems unlikely; therefore, a structural role of the metal ion is proposed. It has been reported that the active conformation of Pseudomonas lipase is stabilized by calcium ion (39). Shibata et al. (36) found that Ca2+ was essential for the formation of the lipase-modulator complex from Pseudomonas aeruginosa in the denaturation-renaturation process, and the addition of EDTA rapidly caused inactivation of the reactivated lipase. Mn2+ could also play the role of Ca2+, although it was not so effective (36). In our work, however, the stability of the in vitro-synthesized lipase was not affected by addition of 20 mM EDTA while it was incubated at 37°C for 2 h. This result suggests the independence of the stability of the lipase at 37°C from free divalent cations. It is possible that for this heat-resistant lipase, its stability at low temperatures does not depend so much on the metal ion as does that of other lipases.
If 20 mM EDTA was added to the expression system of the lipase, its synthesis was inhibited completely by the removal of Mg2+, which is necessary for the protein synthesis reaction. So the effect of Ca2+ on lipase activation could not be examined by the method of adding EDTA to the cell-free lipase synthesis system. Further investigation was carried out by mutant lipases with calcium-binding-site-forming Asp residues being replaced by Ala. The coexpression of the mutant rlip genes (D242A, D288A, and D242/288A) with the act gene did not show any high level of activity during the time course (Fig. 8). Also, the specific activity of each mutated lipase was much lower than that of the wild type (Table 1). It suggests the importance of this Ca2+ binding site in the production of active lipase. Considering the relative stability of the wild-type lipase in the presence of EDTA, the binding of divalent ions to this site seems to be important for the activation process of the lipase.
ACKNOWLEDGMENTS
The genes of lipase from KWI-56 encoded by pLP64 were kindly provided by Kurita Water Industries Ltd.
This work was financially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, and the “Research for the Future” program of The Japan Society for the Promotion of Science (JSPS-RFTF96100306).
REFERENCES
- 1.Aamand J L, Hobson A H, Buckley C M, Jørgensen S T, Diderichsen B, McConnell D J. Chaperone-mediated activation in vivo of a Pseudomonas cepacia lipase. Mol Gen Genet. 1994;245:556–564. doi: 10.1007/BF00282218. [DOI] [PubMed] [Google Scholar]
- 2.Brownie J, Shawcross S, Theaker J, Whitcombe D, Ferrie R, Newton C, Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullok W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 4.Burks E A, Chen G, Georgiou G, Iverson B L. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc Natl Acad Sci USA. 1997;94:412–417. doi: 10.1073/pnas.94.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chihara-Siomi M, Yoshikawa K, Oshima-Hirayama N, Yamamoto K, Sogabe Y, Nakatani T, Nishioka T, Oda J. Purification, molecular cloning, and expression of lipase from Pseudomonas aeruginosa. Arch Biochem Biophys. 1992;296:505–513. doi: 10.1016/0003-9861(92)90604-u. [DOI] [PubMed] [Google Scholar]
- 6.Chung G H, Lee Y P, Jeohn G H, Yoo O J, Rhee J S. Cloning and nucleotide sequence of thermostable lipase gene from Pseudomonas fluorescens SIK W1. Agric Biol Chem. 1991;55:2359–2365. [PubMed] [Google Scholar]
- 7.Ellman J, Mendel D, Anthony-Cahill S, Noren C J, Schultz P G. Biosynthetic method for introducing unnatural amino acids site-specifically into proteins. Methods Enzymol. 1991;202:301–337. doi: 10.1016/0076-6879(91)02017-4. [DOI] [PubMed] [Google Scholar]
- 8.Frenken L G J, Egmond M R, Batenburg A M, Bos J W, Visser C, Verrips C T. Cloning of the Pseudomonas glumae lipase gene and determination of the active site residues. Appl Environ Microbiol. 1992;58:3787–3791. doi: 10.1128/aem.58.12.3787-3791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frenken L G J, Bos J W, Visser C, Müller W, Tommassen J, Verrips C T. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol Microbiol. 1993;9:579–589. doi: 10.1111/j.1365-2958.1993.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 10.Frenken L G J, Groot A, Tommassen J, Verrips C T. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol Microbiol. 1993;9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 11.Harwood J. The versatility of lipases for industrial uses. Trends Biochem Sci. 1989;14:125–126. doi: 10.1016/0968-0004(89)90140-0. [DOI] [PubMed] [Google Scholar]
- 12.Hobson A H, Buckley C M, Aamand J L, Jørgensen S T, Diderichsen B, McConnell D J. Activation of a bacterial lipase by its chaperone. Proc Natl Acad Sci USA. 1993;90:5682–5686. doi: 10.1073/pnas.90.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobson A H, Buckley C M, Jørgensen S T, Diderichsen B, McConnell D J. Interaction of the Pseudomonas cepacia DSM3959 lipase with its chaperone, LimA. J Biochem. 1995;118:575–581. doi: 10.1093/oxfordjournals.jbchem.a124948. [DOI] [PubMed] [Google Scholar]
- 14.Ihara F, Kageyama Y, Hirata M, Nihira T, Yamada Y. Purification, characterization, and molecular cloning of lactonizing lipase from Pseudomonas species. J Biol Chem. 1991;266:18135–18140. [PubMed] [Google Scholar]
- 15.Ihara F, Okamoto I, Nihira T, Yamada Y. Requirement in trans of the downstream limL gene for activation of lactonizing lipase from Pseudomonas sp. 109. J Ferment Bioeng. 1992;73:337–342. [Google Scholar]
- 16.Ihara F, Okamoto I, Akao K, Nihira T, Yamada Y. Lipase modulator protein (LimL) of Pseudomonas sp. strain 109. J Bacteriol. 1995;177:1254–1258. doi: 10.1128/jb.177.5.1254-1258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizumi T, Nakamura K, Fukase T. Purification and characterization of a thermostable lipase from newly isolated Pseudomonas sp. KWI-56. Agric Biol Chem. 1990;54:1253–1258. [Google Scholar]
- 18.Iizumi T, Nakamura K, Shimada Y, Sugihara A, Tominaka Y, Fukase T. Cloning, nucleotide sequencing, and expression in Escherichia coli of a lipase and its activator genes from Pseudomonas sp. KWI-56. Agric Biol Chem. 1991;55:2349–2357. [PubMed] [Google Scholar]
- 19.Iizumi T, Fukase T. The role of the gene encoding lipase activator from Pseudomonas sp. KWI-56 in in vitro activation of lipase. Biosci Biotechnol Biochem. 1994;58:1023–1027. doi: 10.1271/bbb.58.1023. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki M, Hiratake J, Nishioka T, Oda J. Lipase-catalyzed stereoselective acylation of [1,1′-binaphthyl]-2,2′-diol and deacylation of its esters in an organic solvent. Agric Biol Chem. 1989;53:1879–1884. [Google Scholar]
- 21.Jørgensen S, Skov K W, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in heterologous hosts requires two Pseudomonas genes. J Bacteriol. 1991;173:559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawarasaki Y, Nakano H, Yamane T. Prolonged cell-free protein synthesis in a batch system using wheat germ extract. Biosci Biotechnol Biochem. 1994;58:1911–1913. doi: 10.1271/bbb.58.1911. [DOI] [PubMed] [Google Scholar]
- 23.Kawarasaki Y, Kawai T, Nakano H, Yamane T. A long-lived batch reaction system of cell-free protein synthesis. Anal Biochem. 1995;226:320–324. doi: 10.1006/abio.1995.1231. [DOI] [PubMed] [Google Scholar]
- 24.Kim K K, Song H K, Shin D H, Hwang K Y, Suh W S. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure. 1997;5:173–185. doi: 10.1016/s0969-2126(97)00177-9. [DOI] [PubMed] [Google Scholar]
- 25.Kok R G, Thor J J, Nugteren-Roodzant I M, Vosman B, Hellingwerf K J. Characterization of lipase-deficient mutants of Acinetobactor calcoaceticus BD413: identification of a periplasmic lipase chaperone essential for the production of extracellular lipase. J Bacteriol. 1995;177:3295–3307. doi: 10.1128/jb.177.11.3295-3307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordel M, Hofmann B, Schomburg D, Schmid R D. Extracellular lipase of Pseudomonas sp. strain ATCC 21808: purification, characterization, crystallization, and preliminary X-ray diffraction data. J Bacteriol. 1991;173:4836–4841. doi: 10.1128/jb.173.15.4836-4841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano H, Tanaka T, Kawarasaki Y, Yamane T. An increased rate of cell-free protein synthesis by condensing wheat-germ extract with ultrafiltration membranes. Biosci Biotechnol Biochem. 1994;58:631–634. doi: 10.1271/bbb.58.631. [DOI] [PubMed] [Google Scholar]
- 28.Nakano H, Tanaka T, Kawarasaki Y, Yamane T. Highly productive cell-free protein synthesis system using condensed wheat-germ extract. J Biotechnol. 1996;46:275–282. [Google Scholar]
- 29.Nakano H, Yamane T. Cell-free protein synthesis systems. Biotechnol Adv. 1998;16:367–384. doi: 10.1016/s0734-9750(97)00082-7. [DOI] [PubMed] [Google Scholar]
- 30.Nakano H, Shinbata T, Okumura R, Sekiguchi S, Fujishiro M, Yamane T. Efficient coupled transcription/translation from PCR template by a hollow-fiber membrane bioreactor. Biotechnol Bioeng. 1999;64:194–199. doi: 10.1002/(sici)1097-0290(19990720)64:2<194::aid-bit8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Nobel M E M, Cleasby A, Johnson L N, Egmond M R, Frenken L G J. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 1993;331:123–128. doi: 10.1016/0014-5793(93)80310-q. [DOI] [PubMed] [Google Scholar]
- 32.Ohuchi S, Nakano H, Yamane T. In vitro method for the generation of protein libraries using PCR amplification of a single DNA molecule and coupled transcription/translation. Nucleic Acids Res. 1998;26:4339–4346. doi: 10.1093/nar/26.19.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshima-Hirayama N, Yoshikawa K, Nishioka T, Oda J. Lipase from Pseudomonas aeruginosa production in Escherichia coli and activation in vitro with a protein from the downstream gene. Eur J Biochem. 1993;215:239–246. doi: 10.1111/j.1432-1033.1993.tb18028.x. [DOI] [PubMed] [Google Scholar]
- 34.Quyen D T, Schmidt-Dannert C, Schmid R D. High-level formation of active Pseudomonas cepacia lipase after heterologous expression of the encoding gene and its modified chaperone in Escherichia coli and rapid in vitro refolding. Appl Environ Microbiol. 1999;65:787–794. doi: 10.1128/aem.65.2.787-794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryabova L A, Desplancq D, Spirin A S, Plückthun A. Functional antibody production using cell-free translation: effects of protein disulfide isomerase and chaperones. Nat Biotechnol. 1997;15:79–84. doi: 10.1038/nbt0197-79. [DOI] [PubMed] [Google Scholar]
- 36.Shibata H, Kato H, Oda J. Calcium ion-dependent reactivation of a Pseudomonas lipase by its specific modulating protein, LipB. J Biochem. 1998;123:136–141. doi: 10.1093/oxfordjournals.jbchem.a021900. [DOI] [PubMed] [Google Scholar]
- 37.Spirin A S, Baranov V I, Ryabova L A, Ovodov S Y, Alakhov Y B. A continuous cell-free translation system capable of producing polypeptide in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 38.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Svendsen A, Borch K, Barfoed M, Nielsen T B, Gormsen E, Patkar S A. Biochemical properties of cloned lipases from Pseudomonas family. Biochim Biophys Acta. 1995;1259:9–17. doi: 10.1016/0005-2760(95)00117-u. [DOI] [PubMed] [Google Scholar]
- 40.Ueda T, Tohoda H, Chikazumi N, Eckstein F, Watanabe K. Phosphorothioate-containing RNAs show mRNA activity in the prokaryotic translation system in vitro. Nucleic Acids Res. 1991;19:547–552. doi: 10.1093/nar/19.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wohlfarth S, Hoesche C, Strunk C, Winkler U K. Molecular genetics of the extracellular lipase of Pseudomonas aeruginosa PAO1. J Gen Microbiol. 1992;138:1325–1335. doi: 10.1099/00221287-138-7-1325. [DOI] [PubMed] [Google Scholar]
- 42.Yang J H, Kobayashi K, Nakano H, Tanaka J, Nihira T, Yamada Y, Yamane T. Modulator-mediated synthesis of highly active lipase of Pseudomonas sp. 109 by Escherichia coli cell-free coupled transcription/translation system. J Biosci Bioeng. 1999;88:605–609. doi: 10.1016/s1389-1723(00)87087-5. [DOI] [PubMed] [Google Scholar]