Abstract
In the phosphorelay signal transduction system for sporulation initiation in Bacillus subtilis, the opposing activities of histidine kinases and aspartyl phosphate phosphatases determine the cell's decision whether to continue with vegetative growth or to initiate the differentiation process. Regulated dephosphorylation of the Spo0A and Spo0F response regulators allows a variety of negative signals from physiological processes that are antithetical to sporulation to impact on the activation level of the phosphorelay. Spo0F∼P is the known target of two related phosphatases, RapA and RapB. In addition to RapA and RapB, a third member of the Rap family of phosphatases, RapE, specifically dephosphorylated the Spo0F∼P intermediate in response to competence development. RapE phosphatase activity was found to be controlled by a pentapeptide (SRNVT) generated from within the carboxy-terminal domain of the phrE gene product. A synthetic PhrE pentapeptide could (i) complement the sporulation deficiency caused by deregulated RapE activity of a phrE mutant and (ii) inhibit RapE-dependent dephosphorylation of Spo0F∼P in in vitro experiments. The PhrE pentapeptide did not inhibit the phosphatase activity of RapA and RapB. These results confirm previous conclusions that the specificity for recognition of the target phosphatase is contained within the amino acid sequence of the pentapeptide inhibitor.
Reversible protein phosphorylation mediated by kinases and phosphatases plays a cardinal role in regulating essentially all aspects of eukaryotic cell physiology (12). Similarly, protein phosphorylation in prokaryotes is a common mechanism utilized in signal transduction as a means of information transfer. The two-component signal transduction system is a widespread mechanism that couples a large variety of stimuli to a diverse array of adaptive responses through a signal-stimulated phosphotransfer pathway between two proteins: a histidine protein kinase and a response regulator (11, 22, 35). Moreover, it is now appreciated that in prokaryotes, as well as in eukaryotes, protein phosphatases with distinct specificities exist to counteract histidine kinase activities (3). Thus signal transduction must be viewed as a competitive process in which kinases and phosphatases are the instruments of positive and negative signals on the system. A complex example of such interplay is provided by the phosphorelay signal transduction system that governs the initiation of the developmental process of sporulation in Bacillus subtilis.
The phosphorelay is a more complex version of the typical two-component system. Since its original discovery in B. subtilis (4), phosphorelays have been described as regulating important and complex pathways such as pathogenesis in Bordetella pertussis (41), osmosensing in Saccharomyces cerevisiae (29), and anaerobic gene expression in Escherichia coli (6), among others. In the B. subtilis phosphorelay, multiple kinases provide signal input into the system through an autophosphorylation reaction with subsequent transfer of the phosphoryl group to the Spo0A transcription factor via the Spo0F response regulator and the Spo0B phosphotransferase intermediates. The use of a multicomponent system, in place of the classic two-component system, was proposed to provide multiple entry levels to negative regulators for controlling the flow of phosphoryl groups in the system and the ultimate production of Spo0A∼P (4). Negative regulation is carried out through controlled dephosphorylation at the level of Spo0F∼P and Spo0A∼P response regulators. The phosphorylation level of Spo0A is specifically and directly modulated by the Spo0E phosphatase in response to signals that remain unknown (21). Spo0F∼P is the target for the RapA and RapB phosphatases (26). These response regulator aspartyl phosphate phosphatases provide access for negative signals to influence the cell's decision of whether to initiate the sporulation process or to continue with vegetative growth.
The expression of RapA and RapB phosphatases is known to be differentially activated by physiological processes alternative to sporulation, such as competence and growth (17, 26), thereby allowing the recognition of a variety of negative signals and providing a means to impact on the phosphorelay and its output product Spo0A∼P. A further level of complexity is brought into the system by the mechanism modulating the Rap phosphase activities. The RapA gene is transcriptionally coupled to a second gene, phrA, which encodes the phosphatase regulator protein PhrA. The Phr family of phosphatase regulators is comprised of seven members (PhrA, -C, -E, -F, -G, -I, and -K), each of which is associated with a corresponding Rap phosphatase (13, 25). The 44-amino-acid (aa) product of phrA is subject to a series of proteolytic events through an export-import control circuit that results in an active pentapeptide (ARNQT). This PhrA pentapeptide specifically and directly inhibits the phosphatase activity of RapA (24). The series of events that characterize the formation of the active PhrA pentapeptide, through export by the SecA-dependent system (5, 32) and reimportation by the oligopeptide permease (27, 30, 31), may be subject to a series of temporal and spatial regulatory mechanisms. Therefore, the production of the active Phr pentapeptides was postulated to be a regulatory mechanism required for timing coordination of alternative physiological events such as growth, competence, and sporulation (24).
In this communication, we characterized the RapE protein as the third member of the Rap family of phosphatases that specifically dephosphorylates the Spo0F∼P response regulator of the phosphorelay. We showed that the phosphatase activity of RapE is specifically modulated by a pentapeptide generated from within the carboxy-terminal domain of the PhrE protein, which suggests a processing event distinct from the one postulated to produce the PhrA active pentapeptide.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this study are listed in Table 1. Sporulation assays were carried out in Schaeffer's sporulation medium or in Sterlini-Mandelstam resuspension medium (19). Cells were grown for the time indicated in the figure or tables and then treated with CHCl3 before plating on Schaeffer's sporulation agar plates. Cultures for β-galactosidase assays were grown in Schaeffer's sporulation medium as previously described. β-Galactosidase activity was expressed in Miller units (15).
TABLE 1.
Bacillus subtilis strains used in this study
| Straina | Relevant genotype | Source or reference(s)b |
|---|---|---|
| JH642 | trpC2 phe-1 | Laboratory stock |
| JH703 | spo0AΔ204 | Laboratory stock |
| JH12474 | Δspo0H::cat | BH-1→JH642; 10 |
| JH12546 | spo0A12 abrB::Tn917::mls | 23a |
| JH12575 | abrB::Tn917::mls | 23a |
| JH12834 | rapA::Tn917::mls | 26 |
| JH12954 | ΔphrA::cat | 28 |
| JH11125 | rapE::cat | pSK28→JH642 |
| JH11435 | spo0FY13S | 26 |
| JH11450 | ΔphrE::cat | pSK34→JH642 |
| JH11455 | ΔphrE::erm | pCm::Erm→JH11450; 34 |
| JH11499 | ΔphrA::cat ΔphrE::erm | JH12954→JH11455 |
| JH11505 | amyE::(rapE-lacZ cat) | pSK35→JH642 |
| JH11506 | amyE::(phrE-lacZ cat) | pSK36→JH642 |
| JH11534 | rapE::tet | pCm::Tet→JH11125; 34 |
| JH11542 | rapE::tet phrE::cat | pSK34→JH11534 |
| JH11751 | rapA::Tn917::mls rapE::cat | JH12834→JH11125 |
| JH11760 | spo0FY13S ΔphrE::cat | pSK34→JH11435 |
| JH22141 | comA::cat amyE::(rapE-lacZ spc) | BD1626→JH11505 Spc; 8c |
| JH22143 | spo0A Δ204 amyE::(rapE-lacZ spc) | 11505 Spc→JH703c |
| JH22144 | comA::cat amyE::(phrE-lacZ spc) | BD1626→JH11506 Spcc |
| JH22145 | spo0H::cat amyE::(phrE-lacZ spc) | JH12474→JH11506 Spcc |
| JH22146 | spo0A Δ204 amyE::(phrE-lacZ spc) | JH11506 Spc→JH703; 8 |
| JH22152 | spo0A Δ204 abrB::Tn917::mls amyE::(rapE-lacZ spc) | JH12575→JH22143 |
| JH22154 | spo0A Δ204 abrB::Tn917::mls amyE::(phrE-lacZ spc) | JH12575→JH22146 |
All JH strains are derivatives of JH642 and therefore carry the trpC2 phe-1 auxotrophic markers.
→, construction by transformation.
These strains were obtained by replacement of the chloramphenicol resistance gene with the spectinomycin resistance gene by means of plasmid pCm::Spc (34).
Antibiotics were used at the following concentrations: chloramphenicol, 5 μg/ml; spectinomycin, 50 μg/ml; erythromycin, 25 μg/ml (for strains carrying pHT315 and its derivatives) or 1 μg/ml (for strains carrying the macrolide-lincosamide-streptogramin B resistance gene from the Tn917 transposon). E. coli DH5α was used for plasmid construction and propagation.
DNA manipulations.
The construction of the chromosomal library in the multicopy vector pHT315 was described previously (42). Plasmid pRM17 was subject to nucleotide sequence analysis at the 5′ and 3′ terminal ends. Plasmids pSK28 and pSK44 were derived from pRM17 by subcloning fragments in pJM103 and pHT315, respectively. The fragment carried by pSK38 was generated by PCR amplification of JH642 chromosomal DNA with oligonucleotides that introduced a KpnI site at the 5′ end and a BamHI site at the 3′ end. The fragment was first cloned in pJM103 (pSK31) and subjected to full-length sequence analysis. This revealed three nucleotide mismatches, only one of which resulted in an amino acid change (from G to E) at position 278 of the published sequence of the RapE protein (GenBank accession no. D32216) (36). The fragment from pSK31 was then transferred to pHT315, producing pSK38, and also digested and subcloned, producing the multicopy plasmids pSK39 and pSK43. The fragments carried by plasmids pSK33, pSK34, pSK35, and pSK36 were also generated by PCR amplification and subjected to sequence analysis. The fragment carried by pSK33 was transferred to the pHT315 multicopy vector, producing plasmid pSK40. The vectors used in this study were the integrative vector pJM103 (23); the multicopy vector pHT315 (2); the lacZ transcriptional fusion vectors pDH32 and pJM783 (23); the antibiotic cassette exchange plasmids pCm::Erm, pCm::Tet, and pCm::Spc (34); and the Cm cassette vector pJM105A, used for the construction of pSK34 (23).
Protein expression and purification.
A fragment carrying the RapE coding sequence was generated by PCR amplification from JH642 chromosomal DNA with oligonucleotides that introduced a BamHI site at both the 5′ and 3′ ends. The fragment was cloned in the pET16b expression vector (Novagen) and verified by sequence analysis. This cloning generated an extension of 10 histidine codons to the 5′ end of the rapE gene. Protein expression was obtained in E. coli BL21(DE3) pLysS (Novagen) by induction at an optical density at 600 nm of 0.7 with 2 mM isopropyl-β-d-thiogalactopyranoside. Cells were grown for 2 h at 37°C and the protein was purified by affinity chromatography on Ni-nitrilotriacetic acid agarose (Qiagen) as previously described (7). Purification of RapA, RapB, KinA, Spo0F and Spo0A was performed as previously described (7, 24). Spo0B∼P was produced in a reaction mixture containing KinA, Spo0F, and [γ-32P]ATP and then purified as previously described (40).
In vitro assay conditions.
RapE-dependent dephosphorylation of Spo0F∼P was tested in a reaction mixture containing 0.1 μM KinA, 5 μM Spo0F, 1.0 mM ATP, and 1.8 mCi of [γ-32P]ATP (6,000 Ci/mmol; NEN) per ml. The reaction buffer was 50 mM N-(2-hydroxyethyl)piperazine-N′-(3-propanesulfonic acid) (EPPS) (pH 8.5), 20 mM MgCl2, 100 μM EDTA, and 5% glycerol. The reaction mixture was allowed to equilibrate for 30 min at room temperature. RapE was then added at a 2.5 μM final concentration. Time points were taken at the indicated times and the reactions were stopped by addition of sodium dodecyl sulfate (SDS) loading buffer. The Rap-Phr in vitro assays were carried out under the buffer conditions described above. KinA and Spo0F at the concentrations indicated in the figures were incubated for 1 h prior to the addition of Rap phosphatases or premixed Rap phosphatases and Phr peptides. The reactions were allowed to proceed for an additional 30 min and then stopped with SDS loading buffer. Purified Spo0B∼P (1 μM) and Spo0B∼P with Spo0A (2 μM) were incubated in the presence or absence of RapE (2 μM) for 30 min at room temperature in the reaction buffer described above. The reactions were run on SDS–glycine–15% polyacrylamide gels at constant current (25 mA) for 1.5 h. The gels were immediately exposed to Kodak X-Omat RP films at −80°C and then exposed to a Molecular Dynamics PhosphorImager and analyzed with ImageQuant software. The concentration of Phr peptides was determined by amino acid analysis.
RESULTS
The product of rapE is a negative regulator of sporulation initiation.
Many negative regulators of the phosphorelay were found by their property of inhibiting sporulation when overexpressed on a multicopy plasmid. The KipI histidine kinase inhibitor was identified by screening a B. subtilis chromosomal library constructed in the shuttle vector pHT315 (42). This library yielded a series of sporulation-deficient clones, and plasmids isolated from these clones were subject to nucleotide sequence analysis. Among the plasmids isolated, pRM17 contained a 1,802-bp fragment of the B. subtilis genome from nucleotide 40974 to nucleotide 42776 (GenBank accession no. D32216) on the skin excisable element (36). An open reading frame (ORF) was identified on this fragment, ORF5, and the high level of similarity between the product of ORF5 and RapA (47% identity; 66% similarity) suggested this may be an additional member of the Rap family of phosphatases. ORF5 was renamed RapE.
The presence of plasmid pRM17 in the wild-type strain JH642 resulted in a sporulation-deficient phenotype (≃3-fold-fewer spores than in the wild-type strain carrying the vector pHT315) (Table 2). The sporulation deficiency was associated with the presence of an intact RapE coding sequence, since plasmids pSK43 and pSK44, which carried only the rapE promoter region and upstream sequences, did not inhibit sporulation (Fig. 1A and Table 2).
TABLE 2.
Sporulation efficiency of B. subtilis strains carrying multicopy plasmidsa
| Plasmidb | Insert | Viable cell count | Spore count | % Efficiency of sporulation |
|---|---|---|---|---|
| pRM17 | rapE | 2.2 × 108 | 2.3 × 107 | 10.4 |
| pSK43 | rapE promoter | 4.2 × 108 | 1.8 × 108 | 42.8 |
| pSK38 | rapE phrE | 3.9 × 108 | 1.2 × 108 | 30.7 |
| pSK39 | phrE | 3.0 × 108 | 1.1 × 108 | 36.6 |
| pSK40 | phrE promoter | 4.0 × 108 | 1.3 × 108 | 32.5 |
| pHT315 | Vector | 3.2 × 108 | 1.2 × 108 | 37.5 |
Representative of several independent experiments.
JH642 derivatives harboring the multicopy plasmids were grown for 46 h at 37°C in Schaeffer's sporulation medium with the addition of erythromycin at a concentration of 5 μg/ml.
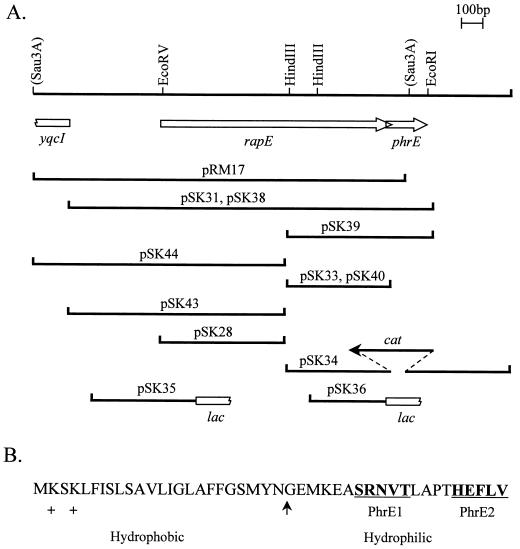
FIG. 1.
(A) Restriction map of the chromosomal region containing the rapE and phrE loci. Fragments cloned in plasmids used in this study are indicated by lines. The fragments in plasmids pSK34, -35, -36, -38, and -40 were generated by PCR amplification from JH642 chromosomal DNA with oligonucleotides carrying restriction sites suitable for cloning. Restriction sites in parentheses are not unique. (B) Amino acid sequence of the PhrE protein. The arrow denotes the putative type I signal peptidase cleavage site, as determined by the SignalP program (20). The PhrE-1 and PhrE-2 pentapeptides are in boldface and underlined. +, positively charged residues.
The phrE gene.
Analysis of the nucleotide sequence of the region downstream of the rapE gene revealed the presence of an overlapping small ORF, phrE (Fig. 1). phrE encodes a 44-aa peptide with low primary sequence homology to the PhrA peptide regulating RapA but with similar structural features, i.e., a positively charged amino-terminal hydrophobic domain separated from a hydrophilic carboxy-terminal domain by a putative signal peptidase cleavage site (Fig. 1B) (20). Nucleotide sequence analysis also revealed the presence of a putative sigma H promoter region buried within the rapE coding sequence and immediately upstream of the phrE ribosome binding site (not shown).
To test the possibility that the product of phrE regulated the activity of RapE, as is the case of PhrA with RapA, we constructed a multicopy plasmid carrying the entire rapE phrE operon. As shown in Table 2, the strain harboring plasmid pSK38 sporulated as efficiently as the control strain, indicating that the presence of phrE overcame the sporulation defect caused by rapE overexpression. Multicopy plasmids carrying the phrE gene (pSK39) or the phrE promoter alone (pSK40) did not significantly affect the efficiency of sporulation (Fig. 1A and Table 2).
When a chromosomal inactivation of the phrE gene was obtained by the insertion of a chloramphenicol resistance cassette within the gene, the resulting strain, JH11450, showed a reduction in sporulation efficiency (Table 3). The sporulation defect of strain JH11450 was suppressed by the concurrent inactivation of the rapE gene (strain JH11542) (Table 3). The sporulation efficiency of strain JH11542 was comparable to the efficiency of strain JH11125 carrying the inactivated rapE alone; both strains sporulated at a higher level than the wild-type strain JH642. Furthermore, the sporulation defect of the phrE mutant was totally overcome by an spo0F mutation, Y13S, which renders the Spo0F response regulator insensitive to the activity of both RapA and RapB phosphatases (strain JH11760) (Table 3) (26). These observations strongly suggested that the PhrE peptide acts as a modulator of RapE activity and the target of RapE is the Spo0F∼P response regulator intermediate of the phosphorelay.
TABLE 3.
Sporulation efficiency of rap and/or phr mutant strainsa
| Strainb | Relevant phenotype | Viable cell count | Spore count | % of sporulationc |
|---|---|---|---|---|
| JH642 | WT | 2.3 × 108 | 1.35 × 108 | 58.7 |
| JH11450 | PhrE− | 4.8 × 108 | 1.1 × 108 | 22.9 |
| JH11542 | RapE− PhrE− | 2.2 × 108 | 1.5 × 108 | 68.2 |
| JH11125 | RapE− | 2.3 × 108 | 1.5 × 108 | 65.2 |
| JH11435 | Spo0F (Y13S) | 1.5 × 108 | 1.3 × 108 | 86.6 |
| JH11760 | Spo0F (Y13S) PhrE− | 1.2 × 108 | 1.0 × 108 | 83.3 |
| JH12954 | PhrA− | 4.4 × 108 | 4.0 × 107 | 9.1 |
| JH11499 | PhrA− PhrE− | 4.9 × 108 | 3.6 × 107 | 7.3 |
| JH12834 | RapA− | 1.8 × 108 | 1.4 × 108 | 77.7 |
| JH11751 | RapA− RapE− | 1.6 × 108 | 1.2 × 108 | 75.0 |
Representative of three independent experiments. WT, wild type.
Strains were grown for 30 h at 37°C in Schaeffer's sporulation medium.
Percentage of sporulation is expressed as the ratio between spore counts and viable cell counts. The significant differences observed in percentages, despite the limited differences in absolute spore counts, are due to the highly reproducible differences in viable counts between sporulating and nonsporulating cells.
An exogenously provided synthetic PhrE pentapeptide complements the phrE mutant.
The structural features of the phrE gene product are reminiscent of the phrA and phrC gene products. Therefore, we investigated whether the 44-aa PhrE protein was subject to the same maturation process through the export-import control circuit that results in the formation of an active pentapeptide. It has recently been shown that the PhrA carboxy-terminal pentapeptide ARNQT is specifically active on RapA while the PhrC carboxy-terminal pentapeptide ERGMT weakly, but specifically, inhibits RapB (24). Although there is a very limited amino acid sequence homology among the members of the Phr family, there is a highly conserved arginine residue at position 2 and a threonine residue at position 5 of active carboxy-terminal pentapeptides. Analysis of the amino acid sequence of PhrE (Fig. 1) revealed that while the carboxy-terminal pentapeptide HEFLV (PhrE-2) did not contain any of the characteristic R or T residues, a pentapeptide corresponding to the sequence SRNVT (PhrE-1) was located 9 aa from the carboxy-terminal end. Thus, experiments were designed to determine which peptide was the active inhibitor.
Peptides corresponding to the sequences of PhrE-1 and PhrE-2 were chemically synthesized and used to determine their ability to complement the phrE deficiency in vivo. Strain JH11450 (PhrE−) was grown in Sterlini-Mandelstam medium, as described in Materials and Methods. At resuspension time, the synthetic PhrE-1 and PhrE-2 peptides were added at increasing concentrations and the cells were allowed to sporulate for 12 h. The results of the sporulation assay (Fig. 2) indicated that the PhrE-1 peptide (SRNVT) restored the sporulation capability of the phrE mutant strain JH11450 when used at a 1 μM concentration, while the PhrE-2 peptide (HEFLV) was totally inactive even at the highest concentration tested (10 μM). The assay also showed that at low concentrations, the PhrA and PhrC synthetic pentapeptides did not complement the phrE defect, confirming the observation that no significant cross-reactivity exists in vivo among Phr peptides (24).
FIG. 2.
In vivo complementation of the phrE mutant by synthetic Phr pentapeptides. The assay was carried out by the Sterlini-Mandelstam resuspension method, as described in Materials and Methods. Cells were grown for 12 h at 37°C. The efficiency of sporulation of strains JH642 (wild type) (■) and JH11450 (phrE) (●) is indicated. □, PhrE-1; ○, PhrE-2; ▵, PhrA; ◊, PhrC.
RapE dephosphorylates Spo0F∼P and PhrE-1 inhibits its activity.
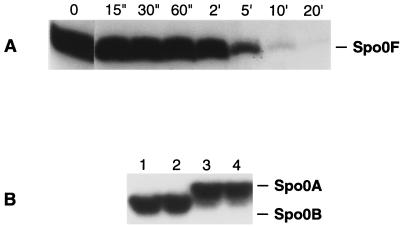
The results of the genetic analysis prompted us to carry out in vitro biochemical assays in order to confirm the target of RapE activity and the role of the PhrE pentapeptides. The RapE protein was purified from an overexpressing E. coli strain and tested in vitro. The results of a time course of RapE-dependent dephosphorylation of Spo0F∼P are shown in Fig. 3A. RapE is specifically active on Spo0F∼P and it does not directly affect the phosphorylation level of Spo0A∼P or Spo0B∼P, as previously observed for RapA and RapB (Fig. 3B).
FIG. 3.
(A) Time course of Spo0F∼P dephosphorylation by RapE. The reaction mixture containing KinA (0.1 μM), Spo0F (5 μM), and RapE (2.5 μM) was incubated in the presence of [γ-32P]ATP as described in Materials and Methods, and aliquots were taken at the indicated times. (B) RapE does not dephosphorylate Spo0B∼P or Spo0A∼P. Purified Spo0B∼P (1 μM) (lanes 1 and 2) or Spo0B∼P and Spo0A (2 μM) (lanes 3 and 4) were incubated without (lanes 1 and 3) or with (lanes 2 and 4) RapE (2 μM) for 30 min at room temperature.
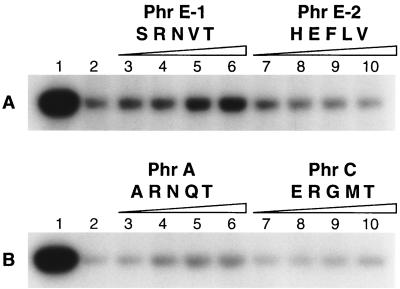
The ability of the synthetic PhrE-1 peptide to inhibit RapE activity, compared to the in vivo-inactive PhrE-2 peptide, was then tested. PhrE pentapeptide concentrations ranging from 50 to 400 μM were utilized and the results of the in vitro assays are reported in Fig. 4A. Increasing concentrations of the PhrE-1 peptide (SRNVT) inhibited the RapE-dependent dephosphorylation of Spo0F∼P, while the PhrE-2 peptide (HEFLV) was totally inactive and actually seemed to stimulate phosphatase activity. These results corroborate the in vivo properties of the PhrE-1 and PhrE-2 peptides.
FIG. 4.
Inhibition of RapE activity by Phr peptides. The reaction mixture containing KinA (0.1 μM), Spo0F (5 μM), and [γ-32P]ATP was incubated for 1 h at room temperature. Aliquots were then incubated with RapE (2.5 μM) (lanes 2) or RapE premixed with PhrE-1 and PhrE-2 (A) or PhrA and PhrC (B) synthetic pentapeptides at 50, 100, 200, and 400 μM (lanes 3 to 6 and 7 to 10, respectively). The control level of Spo0F phosphorylation is shown in lanes 1.
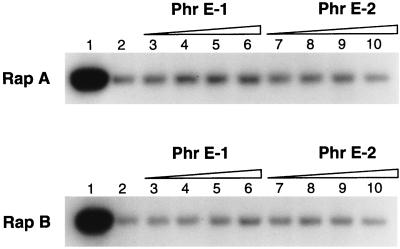
In order to test the specificity of RapE inhibition by Phr peptides, a RapE-dependent dephosphorylation assay of Spo0F∼P was carried out in the presence of PhrA (ARNQT) or PhrC (ERGMT). PhrA showed some inhibitory activity (a threefold lower level of activity than PhrE-1 at the highest concentration used) while PhrC was totally inactive (Fig. 4B). Despite the weak inhibitory activity observed with PhrA, the in vitro and in vivo data indicate that RapE is most likely inhibited specifically by the PhrE-1 peptide. Furthermore, PhrE-1 and PhrE-2 did not show any inhibitory activity toward RapA or RapB (Fig. 5). Therefore, we concluded that the PhrE-1 pentapeptide specifically inhibits the RapE phosphatase activity on Spo0F∼P both in vivo and in vitro.
FIG. 5.
PhrE peptides do not inhibit RapA or RapB. PhrE-1 (lanes 3 to 6) or PhrE-2 (lanes 7 to 10) at 50, 100, 200, and 400 μM was incubated with RapA or RapB at 2.5 μM and the reaction mixture containing KinA (0.1 μM), Spo0F (5 μM), and [γ-32P]ATP. Lanes 1 contain the control level of Spo0F phosphorylation. Lanes 2 show the control level of Spo0F∼P dephosphorylation by RapA or RapB.
Transcription regulation of rapE and phrE.
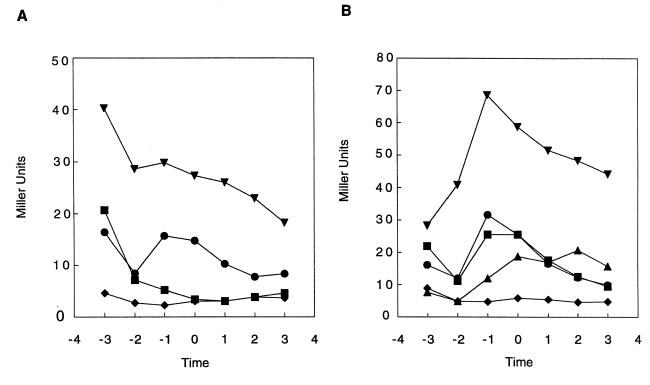
The RapA and RapB proteins are known to be phosphorelay regulators that each prevent sporulation in response to specific and unique physiological conditions (26). Transcription of rapA is dependent upon the ComA-ComP two-component system for competence development (17), whereas rapB is under control of the AbrB transition state regulator and is induced by vegetative growth conditions (26; our unpublished data). Examination of the nucleotide sequence of the rapE promoter region revealed the presence of a putative ComA binding site (18) followed by −35 and −10 consensus sequences for ςA-containing RNA polymerase. A rapE promoter fusion to the E. coli lacZ gene was constructed in the transcriptional fusion vector pJM115 and β-galactosidase assays were carried out in various genetic backgrounds. As shown in Fig. 6A, when wild-type cells were grown under sporulation conditions, transcription from the rapE promoter was induced approximately 2 h before the transition from vegetative growth to sporulation. The rapE gene was transcribed at a very low level (10 to 15 Miller units) and the induction was dependent upon an active ComA protein. The transcription of rapE was also inhibited by an spo0A mutation and this inhibition was released by inactivation of the abrB gene. The effect of the spo0A and abrB mutations, however, was most likely indirect and a result of the Spo0A and AbrB regulatory role in ComA-ComP activation and competence development (9).
FIG. 6.
Time course of β-galactosidase activity of rapE-lacZ (A) and phrE-lacZ (B) fusion constructs integrated at the amyE locus. Time points were taken hourly before and after the transition (To) from exponential to stationary phase. Cells were grown in Schaeffer's sporulation medium. Symbols: ●, wild-type; ⧫, spo0A; ▾, spo0A, abrB; ■, comA; ▴, spo0H.
The genetic organization of the rapE and phrE genes (Fig. 1) was suggestive of an operon structure in which phrE is transcriptionally coupled to rapE in a manner similar to that in the rapA-phrA operon. However, examination of the nucleotide sequence within the rapE coding region that immediately precedes the phrE gene revealed the presence of −35 and −10 consensus sequences for ςA- and ςH-containing RNA polymerase (16). This suggested that in addition to being coupled to rapE transcription, phrE could be independently transcribed from its own promoter. A phrE promoter-lacZ fusion was constructed and integrated in the amyE locus of wild-type strain JH642. β-Galactosidase assays (Fig. 6B) confirmed that phrE is transcribed independently of rapE. The transcription of phrE is also induced approximately 2 h before the transition time (To), as observed for rapE transcription, but at a slightly and reproducibly higher level than rapE. Induction of phrE, however, is not dependent upon ComA and is totally inhibited in a spo0A background, owing to repression by AbrB. When phrE transcription was analyzed in a spo0H background, the level of induction was approximately 50% lower than that in the wild-type strain, suggesting that the putative ςH promoter might have a limited, if any, role in phrE transcription.
β-Galactosidase assays were also carried out on JH642 derivative strains carrying the same rapE-lacZ and phrE-lacZ fusions integrated at the rapE and phrE loci. The results (data not shown) indicated that, although the patterns of transcription were comparable to the ones observed in the amyE locus, the levels of transcription at the isotopic position were 50% lower than the ones obtained at the ectopic integration site.
Relative contribution of RapA-PhrA and RapE-PhrE to the modulation of phosphorelay activity.
Transcription of rapE and rapA is similarly controlled by the ComA-ComP signal transduction system. However, rapA is transcribed at a very high level (approximately 1,500 Miller units at To) (our unpublished data) (17) while rapE expression never exceeds 15 Miller units, as determined by measuring the β-galactosidase activity of rapA-lacZ and rapE-lacZ fusion constructs. In order to assess the relative contribution of RapA and RapE in modulating the level of Spo0F∼P in the phosphorelay in vivo, we carried out a sporulation efficiency test on various rap or phr mutant strains. As shown in Table 3, a deletion of phrE that results in the deregulation of RapE had a minor effect on sporulation efficiency, compared to the deletion of phrA (40% of residual sporulation versus 15%). The double mutant phrA phrE did not display a significantly additive phenotype. Furthermore, while the deletion of the RapA coding gene resulted in an increase of sporulation efficiency (35% more spores than in the wild type), a deletion of rapE only resulted in a 12% increase in spore formation. Once again, the double mutant rapA rapE did not exhibit an additive phenotype. Furthermore, the sporulation defect of an oligopeptide transport mutant which is unable to transport either the phrA or phrE peptide is overcome by a deletion of rapA, but it is not significantly suppressed by a deletion of rapE (Table 4).
TABLE 4.
Suppression of the oppD sporulation phenotype by rapA and rapE mutationsa
| Strainb | Relevant phenotype | % of sporesc |
|---|---|---|
| JH642 | WT | 65.3 |
| JH12795 | OppD− | 5.3 |
| JH11110 | OppD− RapE− | 6.5 |
| JH11053 | OppD− RapA− | 66.0 |
| JH11108 | OppD− RapA− RapE− | 70.0 |
The results are the average of two independent experiments. WT, wild type.
Strains were grown in Schaeffer's sporulation medium for 30 h.
Calculated as the ratio between viable cells and CHCl3-resistant spores.
Altogether, these results suggest that the major role in modulating the phosphate flow through the phosphorelay by the dephosphorylation of Spo0F∼P is played by the RapA phosphatase, while RapE has an accessory role. Indeed, the location of the rapE and phrE genes on the B. subtilis skin excisable element supports this view. The skin element is seen as a cryptic remnant of an ancestral temperate phage and it is reportedly absent from other B. subtilis stock strains at the University of Tokyo (Y. Kobayashi, personal communication) (36) or from closely related bacilli such as Bacillus thuringiensis (1). We analyzed various natural isolates of B. subtilis for the presence of the rapE gene with the rapA gene as a control. PCR amplifications were carried out on chromosomal DNA isolated from the following sporulating Bacillus strains: B. subtilis “Polish” (J. A. Hoch strain collection), Bacillus natto, B. subtilis 23SR, ATCC 10783, ATCC 12139, ATC 14593, and ATCC 10774. The results showed that a fragment of the size of the rapA coding sequence (1.1 kb) was generated by the rapA oligonucleotides in all the strains tested, while a rapE fragment was detected only in the JH642 laboratory strain and in B. subtilis “Polish” (data not shown).
These observations confirm the hypothesis that RapE and PhrE play a dispensable role in the overall context of sporulation initiation and support previous findings about the nonubiquitous presence of the skin element in Bacillus strains (1, 36).
DISCUSSION
The RapE member of the Rap family of response regulator aspartyl phosphate phosphatases was found to promote the dephosphorylation of Spo0F∼P, the response regulator intermediate of the phosphorelay. RapE contributes, with RapA and RapB, to the integration of negative signals into the phosphorelay for sporulation initiation, in response to physiological conditions antithetical to the developmental process. Transcription of the RapE-coding gene is under control of the ComA-ComP two-component system for competence development, which also activates transcription of rapA (17). Expression of the rapB gene, on the contrary, is induced by conditions that favor vegetative growth. Since vegetative growth and competence are processes that cannot occur in a sporulating cell, the induction of rap phosphatases prevents sporulation from interfering with these processes.
Inhibition of RapE phosphatase activity both in vivo and in vitro occurs by action of a pentapeptide, SRNVT, generated from the central portion of the C-terminal half of the phrE gene product. Transcription of phrE occurs independently of the rapE promoter and is controlled by the Spo0A-AbrB pair of transcription regulators. This may represent a mechanism ensuring sufficient production of PhrE peptide to inhibit RapE activity when the level of phosphorylated Spo0A in the cells is high enough to prevent abrB transcription, therefore allowing the initiation of the transition phase to sporulation.
PhrE, like PhrA, has the characteristics of a protein exported by the SecA-dependent system (5, 28). Key features are the positively charged amino end followed by a stretch of hydrophobic residues, a putative type I signal peptidase cleavage site, and a slightly hydrophilic carboxy-terminal portion. Buried within this latter region is the PhrE-1 pentapeptide (SRNVT) that specifically inhibits RapE activity. The C-terminal PhrE-2 pentapeptide (HEFLV) was inactive both in vivo and in vitro. The PhrE-1 pentapeptide does not act in vitro or in vivo on RapA or RapB. Similarly, synthetic PhrA or PhrC pentapeptides do not inhibit RapE activity in vitro when used at low concentrations. These results support previous observations on the highly specific recognition of phosphatase targets by Phr peptides (24). It was reported that single amino acid substitutions within a pentapeptide can severely affect its inhibitory activity and/or its target specificity. Thus, it is not surprising that, despite the fact that the PhrA pentapeptide shares 3 aa with PhrE-1, no cross-reactivity is observed in vivo and very little in vitro. Moreover, the PhrC pentapeptide is inactive on RapE in vitro and the partial complementation in vivo is most likely due to inhibition of RapB phosphatase activity, which would result in increased sporulation frequency (24, 33).
Production of active Phr pentapeptide inhibitors was postulated to occur through an export-import control circuit (24). The major unanswered question is the identity of the proteases responsible for the liberation of the active Phr pentapeptides from the phr gene products. The first modification of the primary gene product is a SecA-dependent export process associated with proteolytic cleavage by type I signal peptidases, as suggested by the primary structure and organization of Phr proteins (5, 28). Five type I signal peptidase-coding genes (sipS, -T, -U, -V and -W) have been identified by the B. subtilis genome sequencing project (13, 38). However, none of them seems to be specifically involved in processing Phr peptides, since single inactivation of the sip genes does not result in a sporulation defective phenotype (M. Jiang and M. Perego, unpublished data). Such phenotype would be expected if the control circuit leading to the production of the active PhrA and PhrE pentapeptides were interrupted.
It has been reported that a sipS-sipT double deletion is lethal to the cells (39) and we have observed that a sipV-sipT double deletion results in a severe sporulation defect that cannot be rescued by deletion of Rap phosphatases (our unpublished work). All this suggests that some processing specificity must exist in signal peptidases and more than one signal peptidase must be involved in processing the Phr proteins, unless a still-unidentified processing enzyme exists with signal peptidase enzymatic activity. However, the sporulation-deficient phenotype of the sipV-sipT mutant is the result of a more severe defect than the inability to process Phr peptides.
After the initial signal peptidase cleavage of Phr proteins, a second proteolytic event was postulated to occur in the extra cytoplasmic compartment in order to generate the active inhibitor pentapeptide from the inactive proinhibitor of presumably 19 aa. This step is necessary to generate a peptide of a size (5 aa) suitable for reimportation by the oligopeptide transport system (37). If one proteolytic event was required to generate the PhrA and PhrC pentapeptides from the C-terminal ends of their respective precursors, two processing events are needed to generate the PhrE-1 peptide from within its precursor. These differential processing events among Phr peptide maturation processes raise intriguing questions about how the specificity of proteolytic events is achieved. Are the determinants for protease recognition embedded within the sequence of the pentapeptides or do they extend within the inactive propeptide inhibitor? Are the conserved R and T residues at positions 2 and 5 of active pentapeptides involved in recognition by proteolytic enzymes? The R and T residues were previously proposed to define the sites of Rap-Phr interaction by providing the correct orientation for binding, while the remaining amino acids may determine specificity through the interactions established by their side chains (24).
A remarkable feature of Rap phosphatases is their specificity for target recognition. RapA, RapB, and RapE are specifically active on Spo0F∼P and do not promote the dephosphorylation of Spo0A∼P or other response regulators tested (our unpublished data). Likewise, Spo0F∼P is not dephosphorylated by RapF or by RapC (our unpublished data); the latter phosphatase is known from genetic studies to affect competence development (14, 33). Nevertheless, RapC and RapF residues share 44 and 41% identity, respectively, and 62% identity with RapA. Furthermore, the remaining chromosomally coded Rap phosphatases all share between 25 to 45% identity with RapA but none of them seem to affect sporulation, based on genetic analysis (our unpublished data). The stringent substrate specificity, despite the high level of homology of Rap phosphatases, and the conserved structural features of response regulators raise challenging questions about molecular recognition. Answering these questions will significantly move forward our understanding of the mechanisms governing signal transduction.
ACKNOWLEDGMENTS
This research was supported, in part, by Public Health Service grants GM55594 and GM19416 from the National Institute of General Medical Sciences, National Institutes of Health.
Oligonucleotides were provided, in part, by the Stein Beneficial Trust.
Footnotes
Publication 12191-MEM of the Department of Molecular and Experimental Medicine, The Scripps Research Institute.
REFERENCES
- 1.Adams L F, Brown K L, Whiteley H R. Molecular cloning and characterization of two genes encoding sigma factors that direct transcription from a Bacillus thuringiensis crystal protein gene promoter. J Bacteriol. 1991;173:3846–3854. doi: 10.1128/jb.173.12.3846-3854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Brautigan D L. Signaling by kinase cascade. In: Corbin J, Francis S, editors. Phosphatases as partners in signaling networks. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 113–124. [Google Scholar]
- 4.Burbulys D, Trach K A, Hoch J A. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 5.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgellis D, Lynch A S, Lin E C C. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimsley J K, Tjalkens R B, Strauch M A, Bird T H, Spiegelman G B, Hostomsky Z, Whiteley J M, Hoch J A. Subunit composition and domain structure of the Spo0A sporulation transcription factor of Bacillus subtilis. J Biol Chem. 1994;269:16977–16982. [PubMed] [Google Scholar]
- 8.Guillen N, Weinrauch Y, Dubnau D A. Cloning and characterization of the regulatory Bacillus subtilis competence genes comA and comB. J Bacteriol. 1989;171:5354–5361. doi: 10.1128/jb.171.10.5354-5361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn J, Roggiani M, Dubnau D. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J Bacteriol. 1995;177:3601–3605. doi: 10.1128/jb.177.12.3601-3605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Healy J, Weir J, Smith I, Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor sigma H in Bacillus subtilis. Mol Microbiol. 1991;5:477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 12.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 13.Kunst F, et al. The complete genome sequence of the Gram-positive model organism Bacillus subtilis (strain 168) Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 14.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 16.Moran C P. RNA polymerase and transcription factors. In: Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 17.Mueller J P, Bukusoglu G, Sonenshein A L. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M M, Xia L, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. pp. 391–450. [Google Scholar]
- 20.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Ohlsen K L, Grimsley J K, Hoch J A. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 23.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 23a.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 24.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci USA. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perego M, Glaser P, Hoch J A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 26.Perego M, Hanstein C G, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 27.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 28.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito J. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 30.Quentin Y, Fichant G, Denizot F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 31.Rudner D Z, Ladeaux J R, Breton K, Grossman A D. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 34.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 35.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive response in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemaru K-I, Mizuno M, Sato T, Takeuchi M, Kobayaski Y. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 37.Tame J R H, Murshudov G N, Dodson E J, Neil T K, Dodson G G, Higgins C F, Wilkinson A J. The structural basis of sequence-independent peptide binding by OppA protein. Science. 1994;264:1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 38.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W J, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tjalsma H, Noback M A, Bron S, Venema G, Yamane K, van Dijl J M. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- 40.Tzeng Y-L, Hoch J A. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 41.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Grau R, Perego M, Hoch J A. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]