Abstract
Introduction:
POU2F3 is a recent marker of a small cell lung carcinoma (SCLC) subtype related to chemosensory tuft cells (SCLC-P). The characteristics of SCLC-P have not been fully defined, and the data on POU2F3 expression in other lung tumors are scarce.
Methods:
We screened 254 SCLC for POU2F3 expression, and comprehensively analyzed histopathologic, genomic, and clinical characteristics of POU2F3-positive tumors. We also explored POU2F3 expression in other major lung cancer types (n=433) and a targeted set of potential diagnostic mimics of SCLC (n=123).
Results:
POU2F3 was expressed in 30 of 254 (12%) SCLC, and was strongly associated with low expression of standard neuroendocrine markers (synaptophysin, chromogranin A, CD56, INSM1). Notably, POU2F3 was expressed in 75% of SCLC with entirely negative or minimal neuroendocrine marker expression (n=20) and was helpful in supporting the diagnosis of SCLC in such cases. Broad targeted next-generation sequencing revealed that SCLC-P (n=12) exhibited enrichment in several alterations, including PTEN inactivation, MYC amplifications, and 20q13 amplifications, but similar rates of RB1 and TP53 alterations as other SCLC (n=155). Beyond SCLC, POU2F3 expression was exclusively limited to large cell neuroendocrine carcinoma (12%) and basaloid squamous cell carcinoma (22%).
Conclusions:
This is the largest cohort of SCLC-P clinical samples to-date, where we describe the diagnostic utility of POU2F3 in a challenging subset of SCLC with low or absent expression of standard neuroendocrine markers. The distinct genomic alterations in SCLC-P may offer a novel avenue for therapeutic targeting. The role of POU2F3 in a narrow subset of other lung cancer types warrants further study.
Keywords: POU2F3, small cell lung carcinoma, SCLC-P, neuroendocrine-low
Introduction
POU2F3 is a master transcriptional regulator of tuft cells – a rare cell type thought to have a chemosensory and immunomodulatory function1, 2. These cells are sparsely present in a wide variety of epithelia and are alternatively known as brush cells in the lung airways 3. Tuft cell-like variant of small cell lung cancer (SCLC-P) was first described by Huang et al. as tumors showing gene expression signature of tuft cells, with POU2F3 representing a lineage oncogene essential for tumor cell survival4. This initial and several subsequent studies have described the highly distinctive transcriptomic and epigenetic profiles of SCLC-P, with POU2F3 expression found to be mutually exclusive of other major SCLC transcriptional subtype markers, ASCL1 and NEUROD14–8. SCLC-P tumors were suggested to have distinct therapeutic vulnerabilities in model systems4, 9–11 and recently in patients5, nominating POU2F3 as an attractive marker for potential personalized treatment of SCLC. Given its recent definition and relatively low prevalence among SCLC (7–14%), there are only limited data on clinicopathologic and genomic characteristics of SCLC-P.
A notable characteristic of SCLC-P identified in prior studies was the low or absent expression of neuroendocrine (NE) phenotype markers (so-called NE-negative or NE-low SCLC). This was initially observed by mRNA.4 In a recent IHC-based study, we found that these tumors were also associated with low protein expression of all 4 standard NE markers used in diagnostic pathology (synaptophysin, chromogranin A, CD56/NCAM, INSM1)12. Significant variation in the extent of NE marker expression in SCLC has long been documented both at protein and mRNA levels13–15. By mRNA, ~20% of SCLC are regarded as NE-low/negative15, 16, and a similar proportion of SCLC exhibit low expression of NE markers by IHC14. The biological characteristics of NE-low SCLC have largely remained a mystery until the identification of POU2F3 expression as a defining characteristic in many of these tumors. In practice, SCLC with extremely low or negative NE markers can present a diagnostic challenge, and identification of a novel diagnostic marker to define this subtype would represent a significant diagnostic advance. Recent identification of POU2F3 as a marker associated with NE-low subset of SCLC has therefore prompted us to consider whether POU2F3 could have a practical application as an additional marker in the diagnosis of SCLC.
In this study, using the largest cohort of SCLC-P to date (n=30) we set out to comprehensively analyze the clinicopathologic and genomic characteristics of this SCLC subtype. To evaluate potential diagnostic utility of POU2F3, we specifically focused on SCLC with minimal or negative NE marker expression, some of which presented a diagnostic challenge. To explore the specificity of POU2F3, we surveyed the expression of POU2F3 by IHC in a large set of other lung cancer types (n=433 tumors total), including lung adenocarcinoma, squamous cell carcinoma (SCC), large cell neuroendocrine carcinoma (LCNEC), lung carcinoids and SMARCA4-deficient undifferentiated tumors (SMARCA4-UT), and examined POU2F3 expression in various tumors that may mimic SCLC pathologically (n=123 total). Notably, given the recently proposed close developmental relationship of tuft cells and basal cells in lung airways17–20, we included in the analysis a rare lung carcinoma variant with presumed relationship to basal cells known as basaloid SCC. Lastly, we analyzed expression of POU2F3 in conjunction with standard NE markers in normal lung tissue.
Methods
Sample selection and study design
The study was performed with the approval of the institutional review board of Memorial Sloan Kettering Cancer Center (MSKCC). SCLC specimens included in the analysis comprised 254 consecutive clinical samples of SCLC reviewed at MSKCC primarily between January 2017 and October 2021. Of these, 218 were from whole-tissue sections and 36 were from a previously constructed TMA12. The set contained 140 cases (up to January 2020) that were included in a prior study12.
Other lung cancer types included in the analysis comprised 100 lung adenocarcinomas, 63 SCC, and 167 carcinoids (136 typical and 31 atypical); these were analyzed using previously constructed tissue microarrays (TMAs)21, 22. In addition, 52 large cell neuroendocrine carcinomas (35 from whole-tissue sections and 17 from a previously constructed TMA23), and 32 whole-tissue sections of basaloid SCC were analyzed.
Potential diagnostic mimics of SCLC included in the study comprised 29 thoracic lymphomas (20 pulmonary mucosa associated lymphoid tissue [MALT] lymphomas and 9 primary mediastinal B-cell lymphomas [PMBCL]), 25 melanomas, 49 Merkel cell carcinomas, and 20 round cell sarcomas. Lymphomas and sarcomas were analyzed in whole-tissue sections, while melanomas and Merkel cell carcinomas were analyzed using previously constructed multi-tissue arrays24 and TMAs25, respectively.
All histologic diagnoses were made using the WHO 2021 criteria13, which for SCLC vs LCNEC relied largely on morphologic differences (larger cell size, more abundant cytoplasm, and more prominent nucleoli for LCNEC), and for SCLC vs basaloid SCC relied on morphologic features (such as abrupt keratinization and basement membrane material deposition) and/or labeling for squamous marker p40 for the latter.
POU2F3 immunohistochemistry (IHC) was performed on all the above tumors (n total = 810). All POU2F3-positive SCLC and a control group of 142 POU2F3-negative SCLC were further analyzed for the expression of the four standard NE markers (synaptophysin, chromogranin A, CD56/NCAM, and INSM1), SCLC transcriptional subtype markers (ASCL1, NEUROD1), retinoblastoma protein (Rb), and if sufficient tissue was available, TTF-1, Ki-67, and MYC. Pan-keratin (AE1/AE3 and/or Cam5.2) and p40 expression was also examined in a subset of SCLC.
IHC methods and scoring criteria
For POU2F3, epitope retrieval was performed using Leica Bond III ER2 for 40 minutes on the stainer. The sections were incubated with the mouse monoclonal antibody to POU2F3 (Santa Cruz, clone 6D1; 1:500 dilution) for 30 minutes, and detection was carried out using Bond Polymer Refine DAB IHC detection kit (Supplementary Table 1). Detailed IHC protocols and scoring criteria for other markers are summarized in Supplementary Table 1. Briefly, labeling for POU2F3, NE markers, ASCL1, NEUROD1 and MYC was scored using the intensity of labeling (1 = weak, 2 = moderate, 3 = strong) and percentage of positive tumor cells (1–100%), with the product of these two parameters yielding a histoscore (H-score). When tumors were dichotomized as positive vs negative, tumors with H-score ≤10 were regarded as negative, unless stated otherwise. Ki-67 proliferative index was expressed as a percentage of positive tumor cells, and TTF-1 was recorded as positive (any amount of nuclear labeling) or negative. For Rb, only complete lack of nuclear staining in the presence of a positive internal control was interpreted as the loss of expression. The extent of NE marker expression (NE-score) was derived as an average H-score of the four standard NE markers. For descriptive purposes, tumors were grouped as NE-low (NE-score ≤150) vs NE-high (NE-score >150)12. Within the NE-low group, tumors with NE-score of 0–50 were defined as a NE-extremely low/negative subgroup.
Next-generation sequencing
A total of 167 SCLC samples, including 12 POU2F3-positive and 155 POU2F3-negative, were analyzed using MSK-IMPACT (hybrid capture-based NGS platform) for somatic mutations in up to 505 cancer genes, as previously described26.
Statistical analysis
JMP version 14.0 software (SAS Institute Inc., Cary, NC) was used for statistical evaluation of the IHC data. Two-tailed t-test was used for analysis of continuous variables, while the likelihood-ratio χ2 and Fisher exact tests were used for analysis of categorical data. Survival analysis was performed using IBM SPSS Statistics for Windows, Version 27.0 software (IBM Core Inc., Armonk, NY). Univariate Kaplan-Meier methodology was used to estimate the probability of overall survival (OS) in each group, for all stages, and stratified by extensive vs limited stage disease.
Results
Prevalence and extent of POU2F3 expression in SCLC
The screen of 254 SCLC revealed POU2F3 expression in 30 cases (12%). Of 30 POU2F3-positive SCLC, the vast majority (n=28) demonstrated robust nuclear staining in >50% of tumor cells (mean 82% of nuclei labeling) and 2–3+ intensity was seen in 90% of cases (Supplementary Table 2). The mean H-score of POU2F3 expression was 177 (range 40–300). The presence of rare scattered POU2F3-positive tumor cells was noted in three SCLC cases; those cases were considered negative for further analysis.
Immunohistochemical characteristics of SCLC-P
Comparison of POU2F3-positive SCLC (n=30) and a control group of 142 POU2F3-negative SCLC confirmed prior observation of the markedly lower level of NE marker expression in these tumors (Figure 1A, Supplementary Table 3). Both the number of NE markers expressed and extent of each NE marker reactivity were significantly lower in SCLC-P compared to other SCLC (p<0.0001 for all comparisons; Figure 1A). The NE-scores (average H-score for all four NE markers combined) for SCLC-P vs other SCLC were 56 and 184, respectively (p<0.0001). Similarly, TTF-1 expression was substantially lower in SCLC-P compared to other SCLC (7% vs 82%, respectively; p<0.0001). Conversely, Ki-67 proliferation was comparably high in SCLC-P and other SCLC. Pan-keratin expression was also similar in SCLC-P (93% positive [14/15], 13% weakly or focally; Supplementary Table 2) and other SCLC (94% positive [34/36], 19% weakly or focally). All SCLC-P were negative for squamous marker p40 (with the exception of squamous components in combined SCLC-P; Supplementary Table 2).
Figure 1. Immunohistochemical characteristics of SCLC-P.
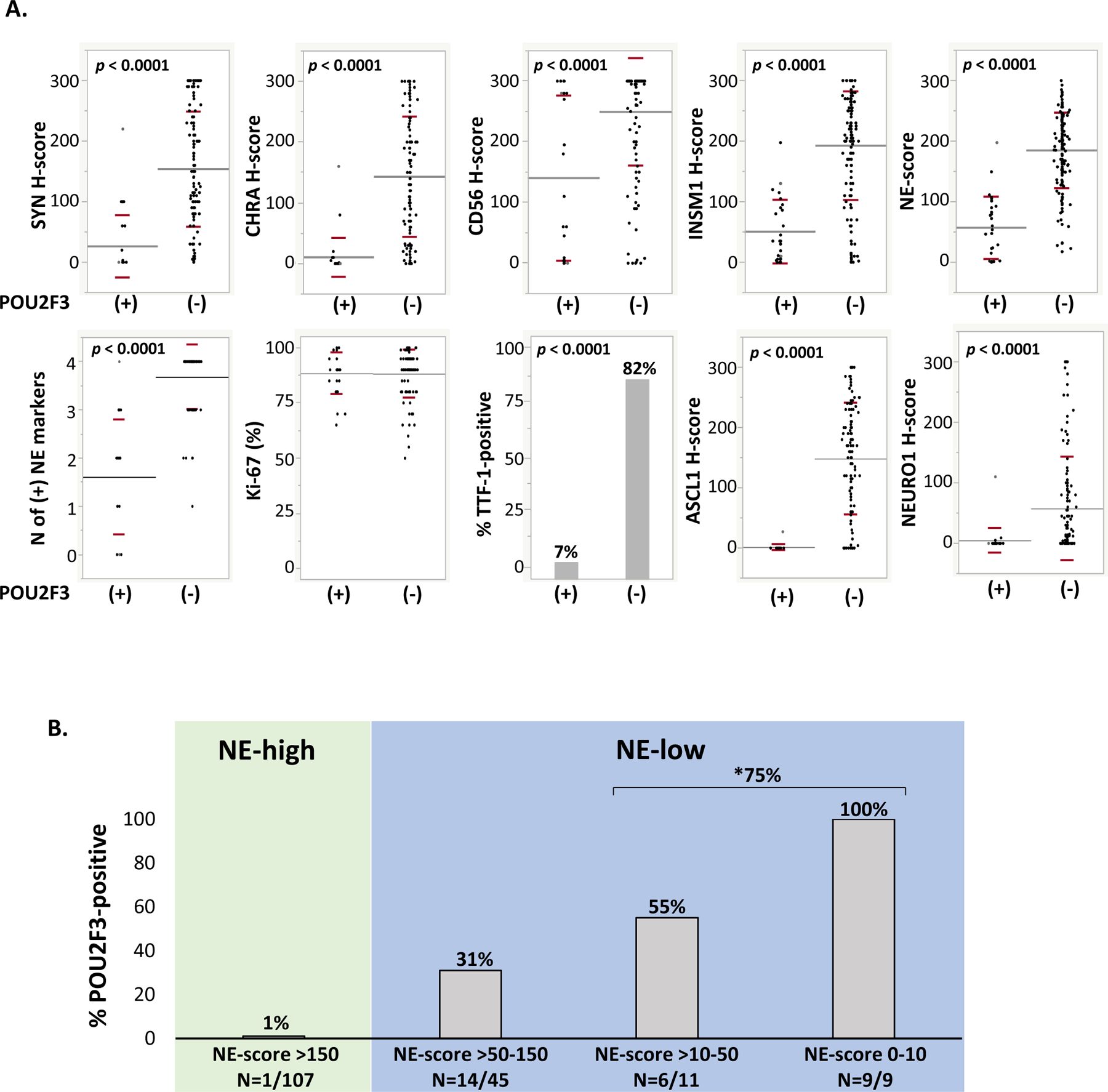
(A) Dot plots depicting expression of various markers in POU2F3-positive (n=30) versus POU2F3-negative SCLC (n=142 for all markers, except n=127 for Ki67 and n=122 for TTF-1). Mean and standard deviation for each comparison are graphically depicted by grey and red lines, respectively (see Supplementary Table 3 for details). (B) Bar graph depicting the proportion of POU2F3-positive cases among SCLC groups defined by extent of NE marker labeling, as expressed by the NE-score (average H-score for all four conventional NE markers: synaptophysin, chromogranin A, CD56, and INSM1). *75% refers to proportion of POU2F3-positive cases among all NE-extremely low/negative SCLC (H-score 0–50).
Abbreviations: CHRA chromogranin A, NE neuroendocrine, SYN synaptophysin
In line with prior studies, expression of POU2F3 was mutually exclusive of ASCL1 and NEUROD1 (Figure 1A), except for two cases containing immunophenotypically distinct tumor cell populations (subclones) with separate areas positive for POU2F3, ASCL1, or NEUROD1; one of these cases was described in our prior study12.
Detailed IHC results for all 30 SCLC-P are summarized in Supplementary Table 2.
POU2F3 in SCLC grouped by the extent of NE marker expression
To further explore the relationship between POU2F3 and NE marker expression, we divided SCLC into NE-high (n=107) vs NE-low (n=65) based on the NE-score of >150 or ≤150, respectively. Of the 30 SCLC-P, only one case was NE-high, whereas all other tumors were NE-low (Figure 1B). These NE-low SCLC generally lacked synaptophysin and chromogranin A expression, and were only positive for CD56 and/or INSM1 (Supplementary Table 2), in line with prior studies on the relative sensitivities of these markers in SCLC14.
Within the 29 NE-low SCLC-P, 20 cases had extremely low or negative NE marker expression (NE-score 0–50); of these, 9 cases were either entirely negative or nearly negative (labeling in only rare cells, generally with only one NE marker) [Figure 1B]. Of 20 NE-extremely low/negative cases, 75% were positive for POU2F3. Notably, within the subgroup with entirely absent (or nearly absent) NE markers, all 9 cases were POU2F3-positive (Figure 1B, Figure 2).
Figure 2. Illustration of preferential POU2F3 expression in SCLC with low or absent expression of standard NE markers.
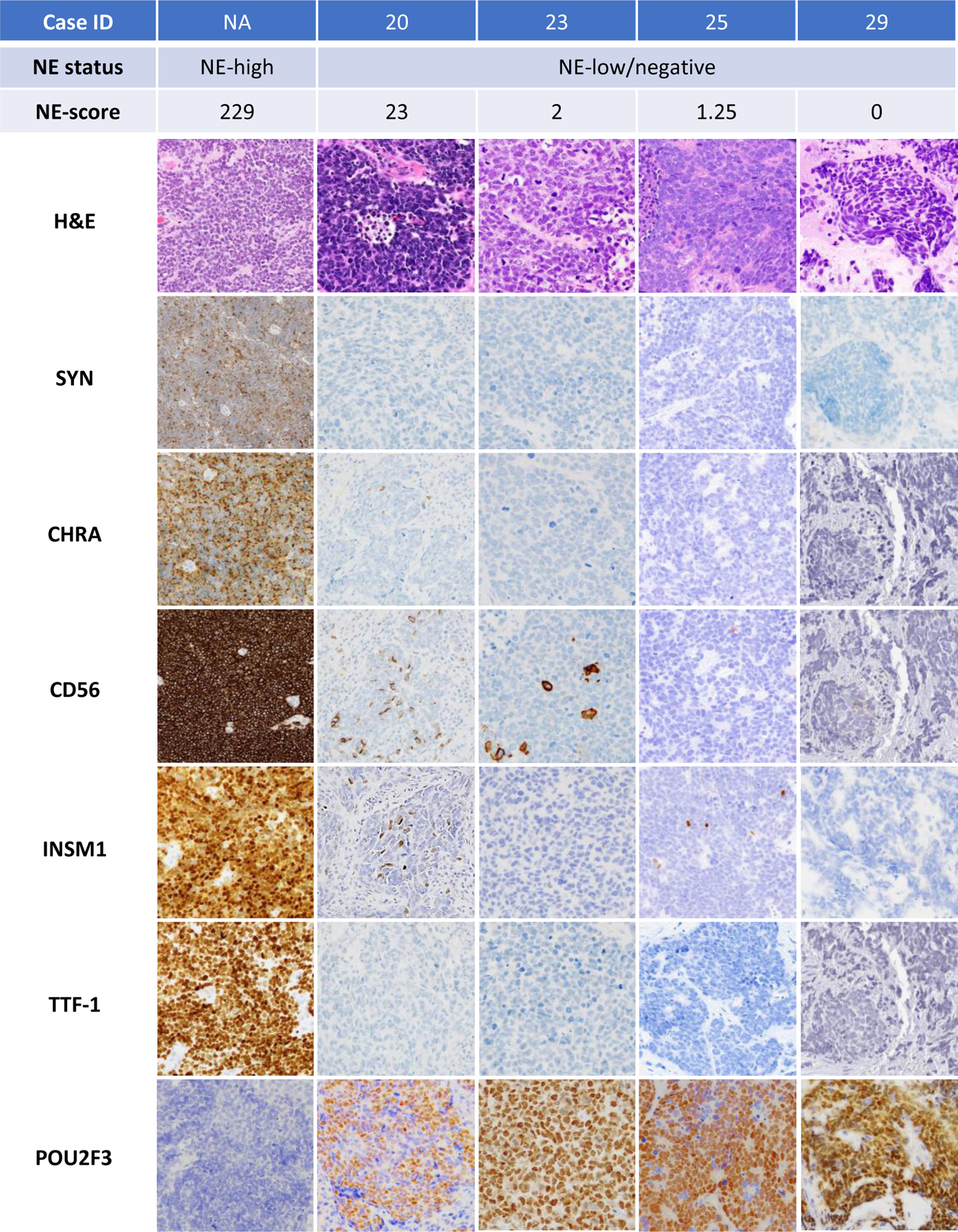
Case in the left panel illustrates a NE-high SCLC (NE-score = 229) showing strong diffuse expression of all conventional NE markers and TTF-1, and negative POU2F3. Other cases are examples of NE-low (Case IDs 23 and 25) or entirely negative (Case ID 29) SCLC with robust expression of POU2F3.
Abbreviations: CHRA chromogranin A, NE neuroendocrine, SYN synaptophysin
POU2F3 as a novel diagnostic marker in NE-low/negative SCLC
During the course of the study, we encountered three tumors that presented a diagnostic challenge (Case IDs 28–30, Supplementary Table 2). These tumors had either entirely absent labeling for NE markers (1 case) or minimal labeling in rare cells (2 cases). Two of the cases (Case IDs 28 and 30) were biopsies obtained at outside institutions, where they were both diagnosed as “poorly differentiated carcinomas” with reports indicating insufficient IHC support for definitive tumor classification despite extensive IHC panels (15 and 12 stains, respectively). In all 3 cases, the presence of robust POU2F3 staining provided direct support for the diagnosis of SCLC despite the complete or near-complete absence of NE marker expression. Case 29 is illustrated in Figure 2 (right panel) and case 28 is illustrated in Figure 3.
Figure 3. Illustration of a case in which POU2F3 supported the diagnosis of SCLC (Case ID 28).
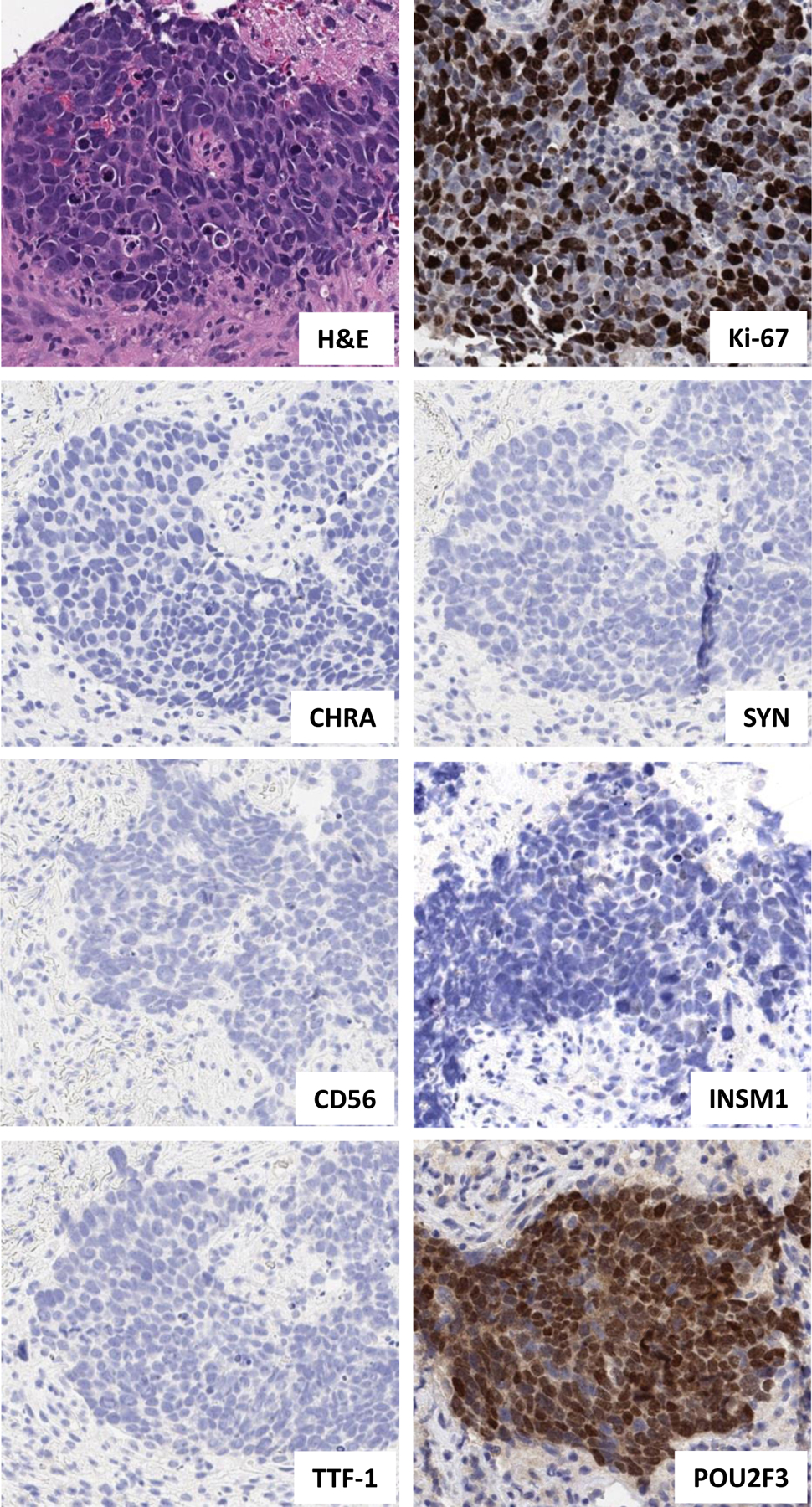
High-grade tumor showing histologic features of SCLC on H&E (high N:C ratio, nuclear molding, and finely granular chromatin) and high Ki-67 proliferative index (70%). However, all NE markers (with the exception of rare cells labeling weakly with INSM1) and TTF-1 are negative. Various other markers were initially evaluated to exclude an alternative diagnosis (including p40 to exclude squamous cell carcinoma), and were negative. Subsequently performed POU2F3 provides direct support for the diagnosis of SCLC.
Abbreviations: CHRA chromogranin A, H&E hematoxylin and eosin, IHC immunohistochemistry, N:C nuclear-to-cytoplasmic, NE neuroendocrine, SYN synaptophysin
Clinicopathologic characteristics
As summarized in Figure 4A, patient characteristics (age, gender, smoking history) and stage at presentation were comparable for SCLC-P vs other SCLC. Specimen characteristics (sampling type and site) were also similar (Supplementary Table 4).
Figure 4. Clinicopathologic and genomic characteristic of SCLC-P.
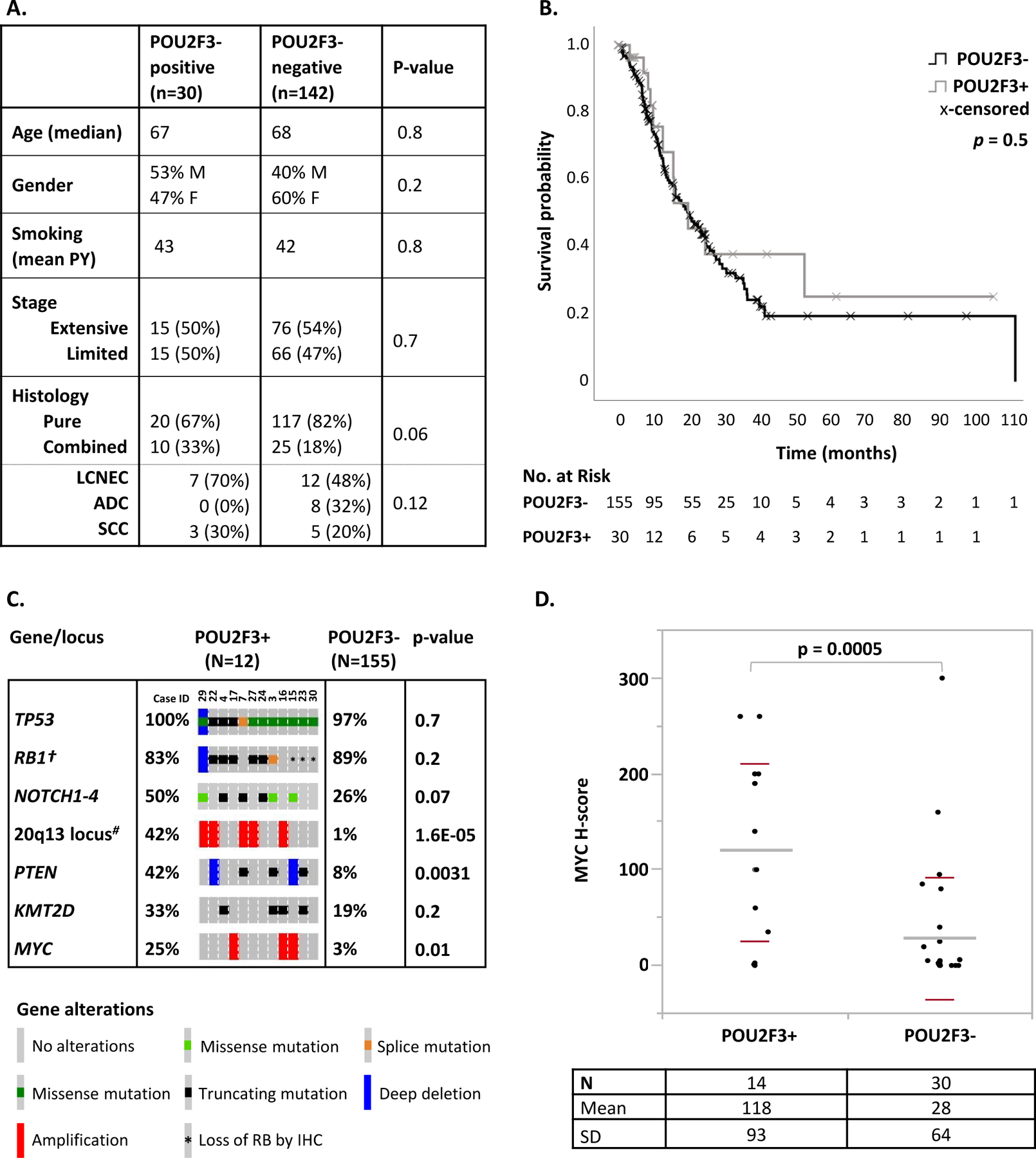
(A) Table comparing clinical and histologic features of SCLC-P (POU2F3-positive) vs other SCLC (POU2F3-negative). (B) Univariate Kaplan-Meyer analysis comparing overall survival in patients with SCLC-P vs other SCLC. (C) Summary of molecular characteristics of SCLC-P. The most prevalent (≥33%) genomic alterations in SCLC-P are shown, as well as genes with divergent distribution in SCLC-P compared to other SCLC. †Rb IHC was performed on all SCLC cases, and the rate of shown RB1 alterations incorporates genomic and IHC findings. #Amplifications of the 20q13 locus involved several genes, including NCOA3 in all 5 cases, and AURKA and GNAS in 3 of 5 cases (Case ID 29, 22 and 16). (D) MYC expression by IHC in SCLC-P vs other SCLC.
Abbreviations: ADC adenocarcinoma, IHC immunohistochemistry, LCNEC large cell neuroendocrine carcinoma, PY pack years, SCC squamous cell carcinoma
SCLC-P showed a trend toward higher rate of combined histology with NSCLC compared to other SCLC (33% vs 18%, respectively; p=0.06; Figure 4A). In all combined carcinomas, POU2F3 expression was seen exclusively or predominantly in SCLC components (Supplementary Figure 1). Morphology of SCLC-P in pure and combined carcinomas was that of typical SCLC.
Clinical follow-up was available for all 30 patients with SCLC-P. Kaplan-Meier analysis showed no significant difference in the overall survival (OS) between SCLC-P and a control group of 155 patients with other SCLC for all stages combined (median OS 19.5 vs 19.8 months, respectively; p=0.5; Figure 4B) and stratified by stage (Supplementary Figure 2).
Genomic characteristics: exploratory analysis
Genomic analysis was performed using targeted next-generation sequencing (MSK-IMPACT) on 12 SCLC-P in comparison to 155 other SCLC (Figure 4C; Supplementary Table 5). SCLC-P harbored a mean of 22 non-synonymous mutations per sample (range 8–49) with a mean coverage of 691x (range: 340–1005x). Tumor mutation burden was similar in SCLC-P vs other SCLC (median 7.0 vs 7.9, respectively), as was microsatellite instability score27 (median 2.3 vs 1.5, respectively) [Supplementary Figure 3].
The most prevalent alterations in SCLC-P were TP53 mutations, present in 100% of cases. RB1 alterations, defined as genomic alterations and/or loss of expression by IHC, were present in 83% of cases. The frequencies of TP53 and RB1 alterations were similar to those seen in other SCLC, as was the rate of mutations in NOTCH family genes (50%) and KMT2D chromatin modifier (33%). Notably, SCLC-P exhibited significantly higher rate of PTEN truncating mutations/losses (42% vs 8%, respectively; p=0.0031) and MYC amplifications (25% vs 3%, respectively; p=0.01) compared to other SCLC. In addition, amplification of 20q13 locus (which harbors AURKA, GNAS and NCOA3 oncogenes) was seen in 42% of SCLC-P compared to only 1% of other SCLC (p<0.0001).
In an exploratory analysis, we assessed MYC protein expression by IHC in 14 SCLC-P vs 30 other SCLC, and found that it was significantly higher in SCLC-P than in other SCLC (mean H-score 118 vs 28, respectively; p=0.0005; Figure 4D). Only 9 SCLC-P had data for both MYC amplification and expression (Supplementary Table 2). Of those, 2/2 MYC-amplified SCLC-P had high levels of MYC expression (H scores 260 and 140), but SCLC-P without MYC amplification (n=7) showed a range of MYC expression (H scores 1–260; mean 115) with 5 cases showing MYC H-scores of ≥100.
POU2F3 expression in other major lung cancer types and SCLC mimickers
To examine POU2F3 expression in lung tumors other than SCLC, POU2F3 immunoreactivity was surveyed in other major lung cancer types (433 total tumors tested; Figure 5A). All major conventional lung cancer types (adenocarcinoma n=100, SCC n=63, and carcinoids n=167) were completely negative for POU2F3. We also tested 19 thoracic SMARCA4-deficient undifferentiated tumors (SMARCA4-UT) – the primitive undifferentiated tumors that can mimic SCLC14, 28; all of those were completely POU2F3-negative. Conversely, POU2F3 was expressed in 12% (6/52) of LCNEC and 22% (7/32) of basaloid SCC (Figure 5A–B). Expression of standard NE markers in POU2F3-postive LCNEC was lower than in other LCNEC (not shown). For POU2F3-positive basaloid SCC, the diagnosis was confirmed by diffuse p40 immunoreactivity in all cases. In both LCNEC and basaloid SCC, POU2F3 immunoreactivity was strong and diffuse (mean H-scores 153 and 204, respectively).
Figure 5. POU2F3 expression in other major lung cancer types and histologic mimics of SCLC.
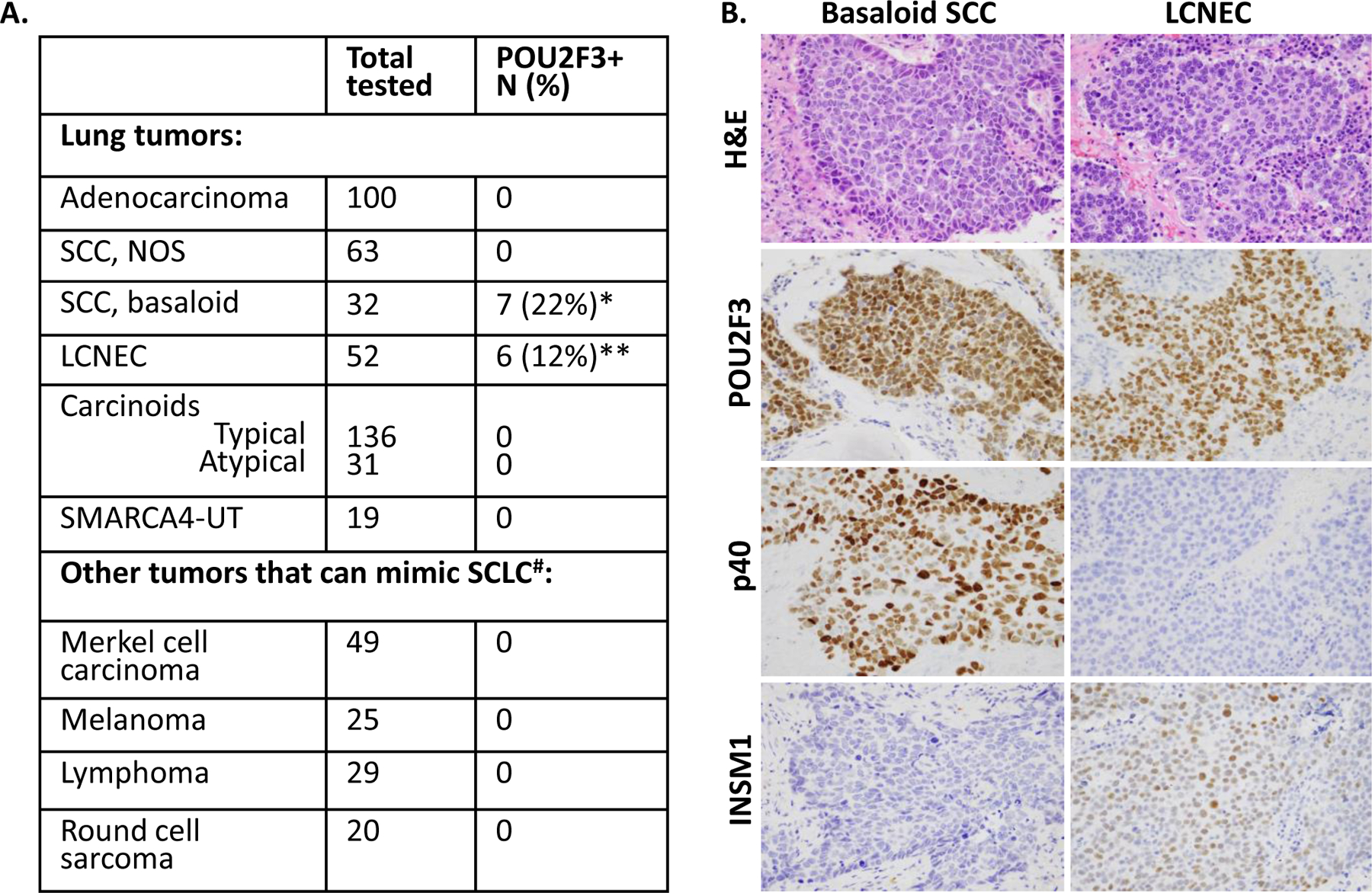
(A) Summary of the frequency of POU2F3 expression in other major lung cancer types. *Mean (range) of POU2F3 H-score in basaloid SCC: 153 (15–275). **Mean (range) of POU2F3 H-score in LCNEC: 204 (85–300). #Lymphomas included 20 MALT lymphomas and 9 primary mediastinal B-cell lymphomas. Round cell sarcomas included 10 Ewing/Ewing family sarcomas, 7 CIC-rearranged sarcomas, 1 NCOA2-rearranged sarcoma, 1 BCOR-CCNB3 sarcoma, and 1 desmoplastic round cell tumor. (B) Illustration of representative cases of basaloid SCC (left panel) and LCNEC (right panel) with POU2F3 expression. Confirming the diagnosis of basaloid SCC is diffuse expression of p40 and the lack of NE markers. LCNEC is distinguished from SCLC based on standard morphologic criteria.
Abbreviations: LCNEC large cell neuroendocrine carcinoma, NOS, not otherwise specified, SCC squamous cell carcinoma, SMARCA4-UT SMARCA4-deficient undifferentiated tumor
In addition, we explored the expression of POU2F3 in various tumors that can pathologically mimic SCLC, including Merkel cell carcinomas, lymphomas, round cell sarcomas, and melanomas (n total=123; Figure 5A). All these tumors were POU2F3 negative, although one Ewing sarcoma showed weak nuclear staining in rare tumor cells (H-score = 7).
POU2F3 expression in normal lung
Lastly, POU2F3 expression was examined in randomly selected sections of the benign lung tissue containing airways from 5 lobectomy specimens for tumor resections. Nuclear staining for POU2F3 was detected in rare cells within airways but not in alveolar lung tissue. POU2F3-positive cells were located primarily in the distal airways, whereas the proximal lobar and segmental airways were largely negative. On manual quantification, we found that the number of POU2F3-positive cells ranged from 0 to 3 cells per distal bronchiolar cross-section, with the majority of bronchioles having none (average <1). Triple-staining for POU2F3, synaptophysin and p40 highlighted that POU2F3-positive cells were located suprabasally, and were synaptophysin-negative. Instead, synaptophysin highlighted separate scattered intrabronchial neuroendocrine cells that lacked the expression of POU2F3 (Figure 6).
Figure 6. POU2F3 expression in normal airways.
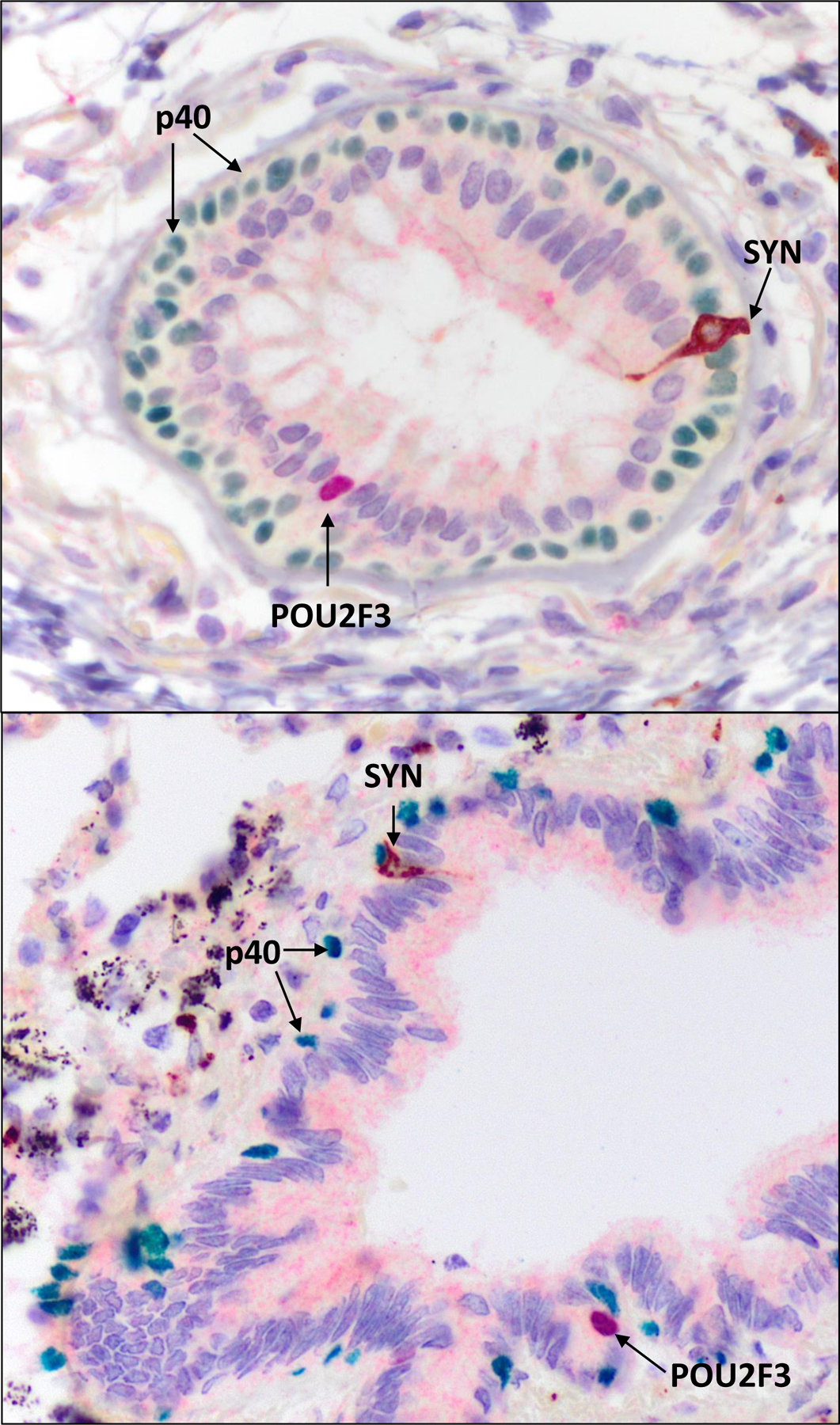
Triple synaptophysin (brown chromogen), p40 (green chromogen), and POU2F3 (red chromogen) IHC in representative bronchiolar cross-sections shows nuclear POU2F3 expression in rare cells that are distinct from the flask-shaped synaptophysin-positive resident pulmonary neuroendocrine cells, with POU2F3-positive nuclei overlying a discretely separate continuous p40-positive basal cell layer and located at or above the level of ciliated cell nuclei.
Abbreviations: IHC immunohistochemistry, SYN synaptophysin
Discussion
POU2F3 expression and tuft-cell-like phenotype in SCLC has been described in several prior studies following the initial description in 20184–8, 12. This is the largest study to-date on this distinct subtype of SCLC, significantly expanding the clinicopathologic and genomic characteristics of this subset, and providing first evidence for the utility of POU2F3 as an ancillary marker for the diagnosis of NE-low SCLC. This is also the first study to perform a survey of POU2F3 expression by IHC in large cohorts of all other major lung cancer types and several histologic mimics of SCLC (n=556 tumors in total), confirming a very narrow and specific expression pattern for POU2F3.
We and others have previously found that SCLC-P are associated with a low “NE program”, characterized by low expression of various standard markers of neuroendocrine differentiation4–8, 12. In our prior study, which examined the distribution of ASCL1, NEURDO1, POU2F3 and YAP1 by IHC in SCLC12, only 12 tumors were of SCLC-P type, whereas in the current study, this group was expanded to 30 cases, allowing a more granular examination of the characteristics of these tumors. In this expanded set, we have confirmed the strict exclusivity of POU2F3 with ASLC1 and NEUROD1 (except for rare SCLC with regional subclones exhibiting distinct transcriptional regulators, as noted previously12) and strong association of POU2F3 with a NE-low program. In addition, we specifically examined the tumors with extremely low or negative NE marker expression (NE-score 0–50; n=20). We found that 75% of tumors in this subgroup expressed POU2F3, with the rate of POU2F3 expression reaching 100% in tumors with negative or nearly negative labeling for NE markers. Therefore, the likelihood of POU2F3 expression in SCLC is exquisitely and quantitatively linked with the level of NE marker expression, although it can also be rarely seen in SCLC with NE-high phenotype. These observations provide a rationale for including POU2F3 as a potential additional diagnostic marker in SCLC lacking or exhibiting minimal levels of standard NE marker expression.
Although SCLC has been traditionally regarded as primarily a morphologic diagnosis that can be made on H&E-stained slides alone, in recent years, with the increasing recognition of various poorly differentiated tumors that can mimic SCLC, the diagnosis is increasingly supported by IHC13, 14. The incorporation of IHC was shown to increase accuracy and reproducibility of the SCLC diagnosis29, with labeling for NE markers representing the key feature of SCLC. However, even with the utilization of the full panel of NE markers, some SCLC have minimal or entirely negative expression of NE markers14. Because of the lack of positive markers, such tumors may cause a diagnostic challenge and/or require an extensive IHC work-up to exclude other entities. Having POU2F3 as a positive marker for these NE-minimal/negative SCLC constitutes a valuable addition to the IHC marker armamentarium for the diagnosis of SCLC. Notably, we illustrate here several cases with NE-negative or NE-minimal profiles, where robust expression of POU2F3 served as supporting evidence for SCLC diagnosis.
In addition to proposing the diagnostic utility of POU2F3 in SCLC, in this study we also demonstrated the pattern of POU2F3 expression in other major lung cancer types (n=433) and histologic mimics of SCLC (n=123). We found that POU2F3 expression was consistently negative in all tested lung adenocarcinomas, conventional SCC, carcinoid tumors, and SMARCA4-UT, as well as lymphomas, melanomas, Merkel cell carcinomas, and various round cell sarcomas. As expected, based on the overlap in the gene expression and genomic profiles between SCLC and LCNEC15, 30, 31, we found that the frequency of POU2F3 expression in LCNEC was similar to that of SCLC (12% in both). The characterization of POU2F3-expressing LCNEC warrants further study. From the diagnostic perspective, the distinction of POU3F3-positive SCLC vs LCNEC was made based on the standard morphologic criteria (see Methods)13.
An unexpected and intriguing finding in this study was that of expression of POU2F3 in 22% of basaloid SCC – primitive carcinomas of presumed basal/squamous cell lineage. This finding may be understood in the context of the recent data from in vivo lineage tracing studies that demonstrated basal cell origin of pulmonary tuft cells18, 20. Furthermore, a similarity between a NE-low subset of SCLC (SQ-P) and “primitive phenotype lung squamous cell carcinomas” was suggested in prior epigenetic studies based on the shared methylation profiles32. Generally, there may be a closer biologic relationship between SCLC and basaloid SCC than currently recognized. Notably, in a recent study, it was demonstrated that basaloid SCC exhibit upregulated expression of various NE markers compared to conventional poorly differentiated SCC33. Therefore, POU2F3 expression in basaloid SCC could represent activation of chemosensory pathways, analogous to increased rate of expression of NE markers in these tumors. From the diagnostic perspective, POU2F3-positive basaloid SCC were distinct from SCLC by morphologic features (see Methods) and consistently diffuse expression of squamous marker p40, whereas SCLC-P were consistently p40-negative. Furthermore, in an exploratory analysis, the rate of Rb loss by IHC was significantly lower in POU2F3-positive basaloid SCC than in POU2F3-positive SCLC (not shown), suggesting their likely distinct genomic features despite the putatively related histogenesis. Currently, comprehensive genomic studies dedicated to basaloid SCC are lacking. Overall, further studies with side-by-side comparison of genomic, transcriptomic, epigenetic and proteomic profiles of POU2F3-expressing tumors with NE (SCLC and LCNEC) and non-NE histologies are needed to more clearly define the relationship between these tumors.
To our knowledge, there is only one study to date - by Yamada et al. - that has examined POU2F3 expression in tumor types other than SCLC34. That study identified POU2F3 expression and tuft cell-like signature in 72% of thymic carcinomas – an interesting observation suggesting a wider distribution of this phenotype than previously recognized. In addition, the authors analyzed publicly available databases from The Cancer Genome Atlas (TCGA) and other studies, and identified POU2F3 mRNA expression and tuft-cell-like signature in 2% of lung SCC, all of which were poorly differentiated, and <1% of adenocarcinomas. It was noted, however, that a thorough pathologic evaluation of tumors was not possible due to the nature of publicly available datasets. In fact, our re-review of virtual images of the three purported POU2F3-positive adenocarcinomas suggests a possible morphologic overlap with LCNEC (Supplementary Figure 4).
Genomic characteristics of SCLC-P are not well established, although prior studies documented that these tumors harbor a similar rate of TP53 and RB1 alterations as other SCLC35. Here we performed an analysis of genomic characteristics of 12 SCLC-P using targeted broad NGS in comparison to 155 other SCLC. We confirmed that the prevalence of TP53 and RB1 was comparable in SCLC-P vs other SCLC. Notably, we identified that SCLC-P harbored a significant enrichment in PTEN and MYC alterations. The high rate of PTEN inactivation in SCLC-P (42% vs 8% in other SCLC) may be of interest therapeutically given that targeting of PTEN-deficient cancers is an area of active investigation36, 37. MYC amplification and overexpression has been long noted to be associated with NE-low SCLC (so called “variant” SCLC) in cell lines38, 39 and more recently in SCLC mouse models6, 40. Association of high MYC mRNA specifically with SCLC-P has also been documented4. To our knowledge, this is the first demonstration of enriched MYC amplification and protein expression by IHC in SCLC-P patient samples. The presence of high MYC protein expression in some MYC-unamplified SCLC-P suggests an alternative mechanism for MYC upregulation in some of these tumors. The high rate (42%) of 20q13 locus amplification in SCLC-P is also a novel finding. Although this locus houses several oncogenes, AURKA may be of particular interest given the demonstrated activity of Aurora kinase inhibitors in SCLC41–43, which in some studies was selective for tumors with high MYC expression10, 40, 44–46. These data provide initial insight that SCLC-P exhibits enrichment in several potentially therapeutically exploitable genomic alterations; however, expanded analysis of a larger set of these tumors is warranted in future studies.
Current understanding of subtype-dependent clinical outcomes in patients with SCLC is limited, although it has been suggested that SCLC-P subtype is associated with poor prognosis5. In our series, patients with SCLC-P had similar overall survival to those with other SCLC. However, studies in larger cohorts will be needed to further evaluate the prognostic and treatment implications of SCLC-P relative to other SCLC subtypes.
The presence of tuft cells (also known as brush or caveolated cells) has been described by electron microscopy in human and animal lung tissue decades ago3. Recently, these cells were visualized by immunofluorescence utilizing POU2F3 antibody in mouse lung4. However, to our knowledge, this is the first immunohistochemical illustration of POU2F3-positive native cells in normal human lung tissue. As predicted for tuft cells, POU2F3-positive cells were present as rare single cells in the lung airways. Interestingly, POU2F3-positive cells localized primarily to smaller, more distal airways, whereas these cells were largely absent in the larger proximal airways. This contrasts with the observations in mice, where it was shown by immunofluorescence that the density of POU2F3-positive cells was higher in the proximal than in the distal murine airways4. This could reflect functional differences of tuft cells in human vs murine lung. Furthermore, by performing triple IHC, we confirm that these cells are distinct from basal and NE cells. However, whether tuft cells represent the cell of origin of SCLC-P or whether expression of POU2F3 and other tuft cell genes is attributable to aberrant transdifferentiation is not clear. The rare examples of cases illustrated in our current and prior12 studies with co-existence of distinct POU2F3-, ASCL1- or NEUROD1-expressing components within a single tumor imply that at least in some cases POU2F3 expression reflects transdifferentiation rather than tuft cell origin.
In summary, we demonstrate the diagnostic utility of POU2F3 IHC in NE-low SCLC and identify distinct genomic characteristics of SCLC-P. We also demonstrate that POU2F3 expression in lung tumors extends beyond SCLC to include a fraction of LCNEC and basaloid SCC, while various other lung cancer types and potential histologic mimickers of SCLC are consistently POU2F3-negative. Given our limited understanding of the distribution and oncogenic function of POU2F3 in tumors other than SCLC, further examination of its expression in tumors of different histotypes and organ sites is needed. Understanding whether the distinct characteristics of SCLC-P extend to other tumors with POU2F3 expression may allow for common therapeutic strategies for POU2F3-positive malignancies beyond SCLC.
Supplementary Material
Funding:
This work was supported by the Fiona and Stanley Druckenmiller Center for Lung Cancer Research, the Ning Zhao and Ge Li Family Initiative for Lung Cancer Research and New Therapies, and by National Cancer Institute U24-CA213274 and R35-CA263816. Infrastructural support was provided by the National Cancer Institute Cancer Center Core Grant P30-CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that there are no competing interests.
Author contributions
Marina K Baine: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing - original draft
Christopher A. Febres-Aldana: Formal analysis, Methodology, Writing - review & editing
Jason C. Chang: Formal analysis, Writing - review & editing
Achim A. Jungbluth: Validation, Resources
Shenon Sethi: Resources, Writing - review & editing
Cristina R. Antonescu: Resources, Writing - review & editing
William D. Travis: Writing - review & editing
Min-Shu Hsieh: Data acquisition, Writing - review & editing
Mee Sook Roh: Resources, Writing - review & editing
Robert J. Homer: Resources, Writing - review & editing
Marc Ladanyi: Validation, Resources, Writing - review & editing
Jacklynn V. Egger: Data curation, Writing - review & editing
W. Victoria Lai: Writing - review & editing
Charles M. Rudin: Funding acquisition, Writing - review & editing
Natasha Rekhtman: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Writing - original draft
CRediT Roles: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Software; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing - review & editing.
References
- 1.Nevo S, Kadouri N, Abramson J. Tuft cells: From the mucosa to the thymus. Immunol Lett 2019;210:1–9. [DOI] [PubMed] [Google Scholar]
- 2.O’Leary CE, Schneider C, Locksley RM. Tuft Cells-Systemically Dispersed Sensory Epithelia Integrating Immune and Neural Circuitry. Annu Rev Immunol 2019;37:47–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid L, Meyrick B, Antony VB, et al. The mysterious pulmonary brush cell: a cell in search of a function. Am J Respir Crit Care Med 2005;172:136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YH, Klingbeil O, He XY, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 2018;32:915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346–360.e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ireland AS, Micinski AM, Kastner DW, et al. MYC Drives Temporal Evolution of Small Cell Lung Cancer Subtypes by Reprogramming Neuroendocrine Fate. Cancer Cell 2020;38:60–78.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearsall SM, Humphrey S, Revill M, et al. The Rare YAP1 Subtype of SCLC Revisited in a Biobank of 39 Circulating Tumor Cell Patient Derived Explant Models: A Brief Report. Journal of Thoracic Oncology 2020;15:1836–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson KL, Stoney R, Frese KK, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nature Cancer 2020;1:437–451. [DOI] [PubMed] [Google Scholar]
- 9.Augert A, Eastwood E, Ibrahim AH, et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardnell RJ, Li L, Sen T, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017;8:73419–73432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudin CM, Poirier JT, Senzer NN, et al. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res 2011;17:888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baine MK, Hsieh M-S, Lai WV, et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. Journal of Thoracic Oncology 2020;15:1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beasley MB, Brambilla E, MacMahon H, et al. Chapter 1. Small cell lung carcinoma. WHO Classification of Tumours Editorial Board Thoracic Tumours. Lyon: International Agency for Research of Cancer; 2021. [Google Scholar]
- 14.Rekhtman N Lung neuroendocrine neoplasms: recent progress and persistent challenges. Mod Pathol 2022;35:36–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nature Reviews Cancer 2019;19:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirier JT, George J, Owonikoko TK, et al. New Approaches to SCLC Therapy: From the Laboratory to the Clinic. Journal of Thoracic Oncology 2020;15:520–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldfarbmuren KC, Jackson ND, Sajuthi SP, et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun 2020;11:2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoro DT, Haber AL, Biton M, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018;560:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plasschaert LW, Zilionis R, Choo-Wing R, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018;560:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders CJ, Reynolds SD, Finger TE. Chemosensory Brush Cells of the Trachea. A Stable Population in a Dynamic Epithelium. American Journal of Respiratory Cell and Molecular Biology 2013;49:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laddha SV, da Silva EM, Robzyk K, et al. Integrative Genomic Characterization Identifies Molecular Subtypes of Lung Carcinoids. Cancer Res 2019;79:4339–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wohlhieter CA, Richards AL, Uddin F, et al. Concurrent Mutations in STK11 and KEAP1 Promote Ferroptosis Protection and SCD1 Dependence in Lung Cancer. Cell Rep 2020;33:108444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baine MK, Sinard JH, Cai G, et al. A Semiquantitative Scoring System May Allow Biopsy Diagnosis of Pulmonary Large Cell Neuroendocrine Carcinoma. Am J Clin Pathol 2020;153:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lezcano C, Muller AM, Frosina D, et al. Immunohistochemical Detection of Cancer-Testis Antigen PRAME. Int J Surg Pathol 2021;29:826–835. [DOI] [PubMed] [Google Scholar]
- 25.Busam KJ, Jungbluth AA, Rekthman N, et al. Merkel Cell Polyomavirus Expression in Merkel Cell Carcinomas and Its Absence in Combined Tumors and Pulmonary Neuroendocrine Carcinomas. American Journal of Surgical Pathology 2009;33:1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middha S, Zhang L, Nafa K, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precision Oncology 2017:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baine MK, Rekhtman N. Multiple faces of pulmonary large cell neuroendocrine carcinoma: update with a focus on practical approach to diagnosis. Translational Lung Cancer Research; Publish Ahead of Print 2020. [DOI] [PMC free article] [PubMed]
- 29.Thunnissen E, Borczuk AC, Flieder DB, et al. The Use of Immunohistochemistry Improves the Diagnosis of Small Cell Lung Cancer and Its Differential Diagnosis. An International Reproducibility Study in a Demanding Set of Cases. J Thorac Oncol 2017;12:334–346. [DOI] [PubMed] [Google Scholar]
- 30.George J, Walter V, Peifer M, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nature Communications 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rekhtman N, Pietanza MC, Hellmann MD, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin Cancer Res 2016;22:3618–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015;34:5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keyhanian K, Phillips WJ, Yeung BS, et al. Neuroendocrine differentiation distinguishes basaloid variant of lung squamous cell carcinoma. Diagn Pathol 2022;17:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Simon-Keller K, Belharazem-Vitacolonnna D, et al. A Tuft Cell-Like Signature Is Highly Prevalent in Thymic Squamous Cell Carcinoma and Delineates New Molecular Subsets Among the Major Lung Cancer Histotypes. J Thorac Oncol 2021. [DOI] [PubMed] [Google Scholar]
- 35.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dillon L, Miller T. Therapeutic Targeting of Cancers with Loss of PTEN Function. Current Drug Targets 2014;15:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLoughlin NM, Mueller C, Grossmann TN. The Therapeutic Potential of PTEN Modulation: Targeting Strategies from Gene to Protein. Cell Chemical Biology 2018;25:19–29. [DOI] [PubMed] [Google Scholar]
- 38.Gazdar AF, Carney DN, Nau MM, et al. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924–2930. [PubMed] [Google Scholar]
- 39.Little CD, Nau MM, Carney DN, et al. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983;306:194–196. [DOI] [PubMed] [Google Scholar]
- 40.Mollaoglu G, Guthrie MR, Böhm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong X, Du J, Parsons SH, et al. Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov 2019;9:248–263. [DOI] [PubMed] [Google Scholar]
- 42.Niu H, Manfredi M, Ecsedy JA. Scientific Rationale Supporting the Clinical Development Strategy for the Investigational Aurora A Kinase Inhibitor Alisertib in Cancer. Front Oncol 2015;5:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyu J, Yang EJ, Zhang B, et al. Synthetic lethality of RB1 and aurora A is driven by stathmin-mediated disruption of microtubule dynamics. Nat Commun 2020;11:5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sos ML, Dietlein F, Peifer M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proceedings of the National Academy of Sciences 2012;109:17034–17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dammert MA, Brägelmann J, Olsen RR, et al. MYC paralog-dependent apoptotic priming orchestrates a spectrum of vulnerabilities in small cell lung cancer. Nature Communications 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owonikoko TK, Niu H, Nackaerts K, et al. Randomized Phase II Study of Paclitaxel plus Alisertib versus Paclitaxel plus Placebo as Second-Line Therapy for SCLC: Primary and Correlative Biomarker Analyses. Journal of Thoracic Oncology 2020;15:274–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


