Abstract
BACKGROUND:
Abnormalities in Ca2+ homeostasis are associated with cardiac arrhythmias and heart failure. Triadin plays an important role in Ca2+ homeostasis in cardiomyocytes (CMs). Alternative splicing of a single triadin gene produces multiple triadin isoforms. The cardiac predominant isoform, mouse MT-1 or human Trisk32 is encoded by triadin exons 1 to 8. In humans, mutations in the triadin gene that lead to a reduction in Trisk32 levels in the heart can cause cardiac dysfunction and arrhythmias. Decreased levels of Trisk32 in the heart are also common in patients with heart failure. However, mechanisms that maintain triadin isoform composition in the heart remain elusive.
METHODS:
We analyzed triadin expression in heart explants from patients with heart failure and cardiac arrhythmias and in hearts from mice carrying a knockout allele for Trdn-as, a CM-specific long noncoding RNA encoded by the antisense strand of the triadin gene, between exon 9 and 11. Catecholamine challenge with isoproterenol was performed on Trdn-as knockout mice to assess the role of Trdn-as in cardiac arrhythmogenesis, as assessed by ECG. Ca2+ transients in adult mouse cardiomyocytes were measured using the IonOptix platform or the GCaMP system. Biochemistry assays, single molecule fluorescence in situ hybridization, subcellular localization imaging, RNA-sequencing, and molecular rescue assays were used to investigate the mechanisms by which Trdn-as regulates cardiac function and triadin levels in the heart.
RESULTS:
We report that Trdn-as maintains cardiac function, at least in part, by regulating alternative splicing of the triadin gene. Knockout of Trdn-as in mice down-regulates cardiac triadin, impairs Ca2+ handling, and causes premature death. Trdn-as knockout mice are susceptible to cardiac arrhythmias in response to catecholamine challenge. Normalization of cardiac triadin levels in Trdn-as knockout CMs is sufficient to restore Ca2+ handling. Finally, Trdn-as colocalizes and interacts with serine/arginine splicing factors in CM nuclei and is essential for efficient recruitment of splicing factors to triadin pre-mRNA.
CONCLUSIONS:
These findings reveal regulation of alternative splicing as a novel mechanism by which a lncRNA controls cardiac function. This study indicates potential therapeutics for heart disease by targeting the lncRNA or pathways regulating alternative splicing.
Keywords: Long noncoding RNA, cardiomyocytes, gene splicing, cardiac arrhythmias, heart failure, Ca2+ handling
Introduction
Dysregulation of intracellular Ca2+ homeostasis in cardiomyocytes (CMs) is common in cardiac arrhythmias and heart failure, affecting millions of people 1-3. Intracellular Ca2+ in CMs is maintained by the function of the dihydropyridine receptor (DHPR), sarcoplasmic reticulum (SR) ryanodine receptor 2 (RYR2), and SR Ca2+ ATPase (SERCA2A). In the junctional SR, DHPR channels juxtaposed to RYR2 form Ca2+-release units, which transfer membrane depolarization to induce myocyte contraction through excitation-contraction coupling by releasing Ca2+ from the SR to increase the free intracellular Ca2+ concentration. The SR membrane protein Triadin, which is important to Ca2+ homeostasis, physically interacts with the RYR2 and the Ca2+-buffering protein calsequestrin 2 (Casq2) through a KEKE motif between amino acids 200-232 4-5. There are several triadin isoforms in muscle cells, with the ~32-kDa isoform, called MT-1 (mouse) or Trisk32 (human) predominant in the heart, whereas the 95-kDa isoform, called Trdn95, is dominant in skeletal muscle 6-8. All triadin isoforms are produced via alternative splicing of a single TRDN/Trdn gene, and share identical sequences between amino acids 1 and 264, which includes a protein-protein interaction motif, the KEKE motif 7, 9. Homozygous or compound heterozygous mutations in TRDN, the gene that encodes triadin, have been identified as an underlying cause for cardiac arrhythmias such as catecholaminergic polymorphic ventricular tachycardia (CPVT) and sudden death, called triadin knockout syndrome (TKOS) 10-13. TKOS is refractory to conventional therapy such as β-blocker therapy, highlighting the urgent and unmet need for novel therapeutics to treat this severe heart disease 12. The majority of TRDN homozygous variants were frameshift mutations, whereas the proportions of compound heterozygous variants were nonsense (~39%), splice-site altering (~28%), missense (~17%), and frameshift mutations (~17%) 12. These mutations in the TRDN gene can cause an early stop codon, nonsense-mediated decay, or protein instability 10-13. Trdn knockout mice are susceptible to cardiac arrhythmias caused by catecholamine challenge with isoproterenol (ISO) 14. In addition to cardiac arrhythmias caused by triadin perturbation, cardiac triadin is also significantly down-regulated in hearts from patients with heart failure 15. However, the mechanisms that regulate levels of cardiac triadin isoforms in the heart remain elusive.
The majority of the genome is transcribed as non-coding RNA. Among the non-coding RNA species, long non-coding RNAs (lncRNAs), defined as an RNA molecule containing more than 200 nucleotides which is not protein-coding, are the largest subclass. Studies over the past decade have demonstrated that lncRNAs play roles in cardiac development and disease through various mechanisms. For example, Bvht and Fendrr interact with polycomb repressor complex 2 (PRC2) to allow activation of the cardiac gene network 16-17. HBL1, a human-specific lncRNA, negatively regulates cardiac differentiation from human pluripotent stem cells by acting as a sponge for miR-1 18. Uph is required for expression of the cardiac transcription factor Hand2 by stabilizing its super-enhancer 19. In addition to crucial roles in development, lncRNAs also play important roles in cardiovascular disease, the leading cause of death. Mhrt, a conserved lncRNA, prevents pathological hypertrophy by antagonizing the function of Brg1, a catalytic subunit of the SWI/SNF chromatin-remodeling complex 20. Chaer and Chast are required for cardiac hypertrophy and remodeling 21-22. Chaer promotes cardiac hypertrophy by interfering with PRC2 binding to chromatin via physical interaction with the catalytic subunit of the complex. ZNF593-AS regulates Ca2+ handling by functioning as a guide RNA scaffold and recruiting heterogeneous nuclear ribonucleoprotein C (HNRNPC) to RYR2 mRNA 23. Whether lncRNAs can regulate cardiac homeostasis and pathogenesis by additional mechanisms remains an important question.
To discover specific lncRNAs that regulate cardiac gene expression, we performed transcriptome analysis of fibroblasts, fibroblasts trans-differentiating into induced CMs (iCMs) at various stages, and CMs. These analyses revealed a CM-specific lncRNA, Trdn-as, encoded by the antisense strand of the Trdn gene. This lncRNA is conserved in genomic position across species. Through loss- and gain-of-function approaches in vitro and in vivo, we show that Trdn-as is both necessary and sufficient to control levels of the cardiac isoform of triadin, MT-1. We show that both TRDN-AS and cardiac triadin are significantly down-regulated in human patients with heart failure and cardiac arrhythmias. We demonstrate that Trdn-as colocalizes and interacts with serine/arginine (SR) splicing factors in CMs. We also demonstrate that Trdn-as deletion impairs recruitment of SR splicing factors to Trdn mRNA. Finally, we demonstrate that Trdn-as KO mice are susceptible to cardiac arrhythmias in response to catecholamine challenge, which mimics the phenotype in Trdn KO mice 14. Via its role in regulating Triadin splicing, we present evidence that Trdn-as plays an important role in regulating cardiac calcium homeostasis, and when disrupted can lead to arrhythmogenesis.
Methods
Detailed methods are available in the Supplemental Methods of the Supplemental Material.
Data Availability Statement
Source and original data are available from the corresponding author upon reasonable request.
Animal Studies and Human Subjects
Animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Colorado Anschutz Medical Campus (CU). Human hearts from the tissue bank maintained by the Division of Cardiology at CU reviewed and approved by the Colorado Multiple Institutions Review Board.
Statistical Analysis
All data are presented as mean ± SE, mean + SE, mean ± SD, or mean + SD, as indicated in figure legends. Statistical analysis was conducted with GraphPad Prism.
Results
TRDN-AS/Trdn-as is cardiac-specific and enriched in nuclei of cardiomyocytes.
Mouse fibroblasts can be reprogrammed into functional iCMs, with low efficiency 24. To search for lncRNAs that are able to improve functionalities of iCMs, we performed microarray analysis in reprogramming fibroblasts at various time points and generated a list of uncharacterized genes based on their expression levels in neonatal mouse ventricular cardiomyocytes (NMCMs) (Figure S1A). As a proof of principle, on this list, was C130080G10Rik (also known as CPR), a cardiac lncRNA, that has been shown to control CM proliferation by recruiting DNMT3A to promote CpG methylation 25. A putative lncRNA, D830005E20Rik was selected for further studies because (1) this transcript was not expressed in fibroblasts but was highly expressed in CMs, (2) expression of the transcript was increased during reprogramming in a time-dependent manner, and (3) expression of the transcript was much lower than that in NMCMs (Figure S1A). D830005E20Rik is located on the antisense strand within the mouse Trdn gene between exons 9 and 11 (Figure S1B). Hereafter, D830005E20Rik will be referred to as Trdn-as. Rapid amplification of cDNA ends (RACE) analysis revealed the transcript of Trdn-as (Figure S1B). Over-expression of Trdn-as significantly increased proportions of spontaneously beating cells in culture, while knockdown of the transcript dramatically decreased fractions of spontaneously beating cells (Figure S1C). However, neither over-expression nor knockdown of Trdn-as affected expression of cardiomyocyte marker cTnT in reprogramming cells (Figure S1D-E). These data suggest that Trdn-as might play crucial roles in iCM contraction and maturation, and/or Ca2+ handling due to the close relationship between Ca2+ handling and CM contraction.
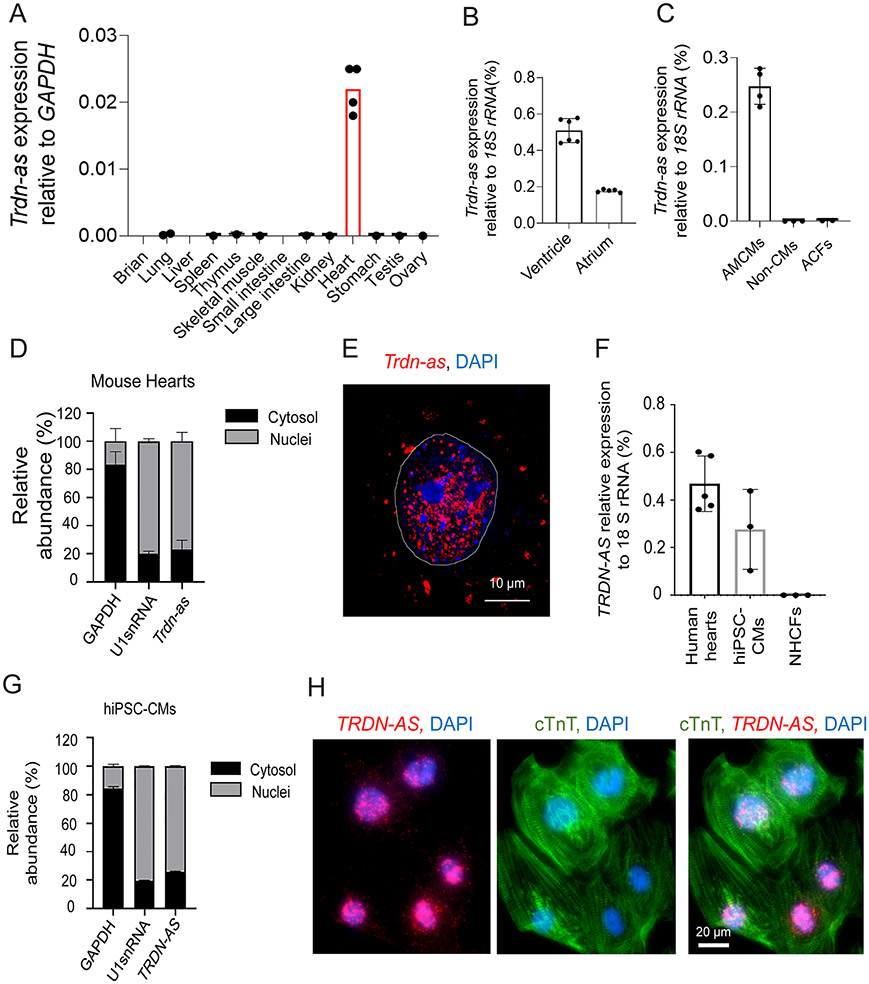
Trdn-as is exclusively expressed in mouse hearts with the highest expression localized to the ventricle (Figure 1A-B) by qPCR analysis using specific primers (Table S1). To further delineate which cardiac cell types express Trdn-as, we isolated ventricular CMs and non-CMs from wild type (WT) mice at the age of 5 months (hereafter referred to as adult mouse ventricular CMs (AMCMs)) as described previously 26-27. Trdn-as expression was only detectable in AMCMs, but not in non-CMs or adult cardiac fibroblasts (ACFs) (Figure 1C). Subcellular fractionation demonstrated that ~ 80% of Trdn-as localized within the nucleus of AMCMs (Figure 1D). In addition, we analyzed subcellular distribution of Trdn-as in NMCMs by single molecule fluorescence in situ hybridization (smFISH) 28. We designed tiled oligonucleotides targeting Trdn-as exons (Table S2). To confirm the specificity of the probes, we performed smFISH in Trdn-as overexpressing as well as in non-transfected HeLa cells. In non-transfected cells wherein Trdn-as is not expressed, no smFISH signal was detected. In contrast, the hybridization signal strongly appeared in HeLa cells transfected with Trdn-as expression vectors (Figure S2), confirming that the oligonucleotide probes in this study specifically recognize Trdn-as in smFISH assays. We then conducted smFISH assays in NMCMs following adenoviral overexpression of Trdn-as. The smFISH assay demonstrated that the majority of Trdn-as localized in CM nuclei (Figure 1E), confirming the results of subcellular fractionation (Figure 1D).
Figure 1. Human TRDN-AS and mouse Trdn-as are exclusively expressed in cardiomyocytes (CMs) and enriched in nuclei.
A-C) Expression of Trdn-as in mouse tissues (A), adult mouse heart ventricle and atria (B), CMs (AMCMs), non-CMs, and cardiac fibroblasts (ACFs) isolated from adult mouse hearts (C) by qPCR. Tissues or cells were harvested from at least 2 mice. Each filled circle represents one animal. Data are presented as mean in A or mean ± SD in B and C. D) qPCR analysis of nuclear and cytoplasmic RNAs in adult mouse hearts. GAPDH and U1 snRNA are used as markers of cytoplasmic and nuclear fractions, respectively. N = 3 mouse hearts. E) Cellular distributions of Trdn-as in neonatal mouse cardiomyocytes (NMCMs) by smFISH. Trdn-as was overexpressed in NMCMs by adenoviral delivery with a MOI of 5. smFISH assays were performed 72 hours post-transduction. The dotted line surrounds a CM nucleus. F) Expression of TRDN-AS in healthy human hearts, hiPSC-CMs, and normal human cardiac fibroblasts (NHCFs) by qPCR. Each filled circle represents an independent induction for hiPSC-CMs or an individual human heart explant. G) qPCR analysis of nuclear and cytoplasmic RNAs in hiPSC-CMs. GAPDH and U1 snRNA are used as markers of cytoplasmic and nuclear fractions, respectively. N = 3 independent inductions. H) Cellular distributions of human TRDN-AS in hiPSC-CMs by FISH. TRDN-AS was over-expressed in hiPSC-CMs by adenoviral delivery with a MOI of 5. FISH was performed 48 hours post-infection. hiPSC-CMs are marked by cardiac troponin T (cTnT).
In humans, a putative lncRNA, RP11-532N4.2, is located on the antisense strand within the human TRDN gene between exons 9 and 11 (Figure S3A). Hereafter, RP11-532N4.2 is referred to as TRDN-AS. Transcriptomic analysis of heart explants from 14 healthy donors indicates that TRDN-AS is the most highly expressed lncRNA in the human heart 29 (Figure S3B). The transcript of TRDN-AS was determined by RACE assays (Figure S3A). TRDN-AS is primarily expressed in the ventricle according to the NIH Genotype-Tissue Expression (GTEx) project 30 (Figure S3C). Like most lncRNAs, the primary nucleotide sequence is not well conserved between species, with only ~45% homology between human and mouse. However, the genomic position and expression pattern are well conserved across species (Figure S3D and Ref. 31). Similar to the mouse lncRNA, TRDN-AS is expressed in human CMs rather than cardiac fibroblasts (Figure 1F). Cellular fractionation assays revealed that the majority of TRDN-AS localizes to nuclei of CMs derived from human induced pluripotent stem cells (hiPSC-CMs) (Figure 1G), confirmed by FISH analysis in hiPSC-CMs following adenoviral overexpression of TRDN-AS (Figure 1H).
Taken together, this antisense lncRNA is conserved in genomic position, primarily expressed in CMs, and localizes to nuclei.
Knockout of Trdn-as in mice impairs cardiac function and causes premature death.
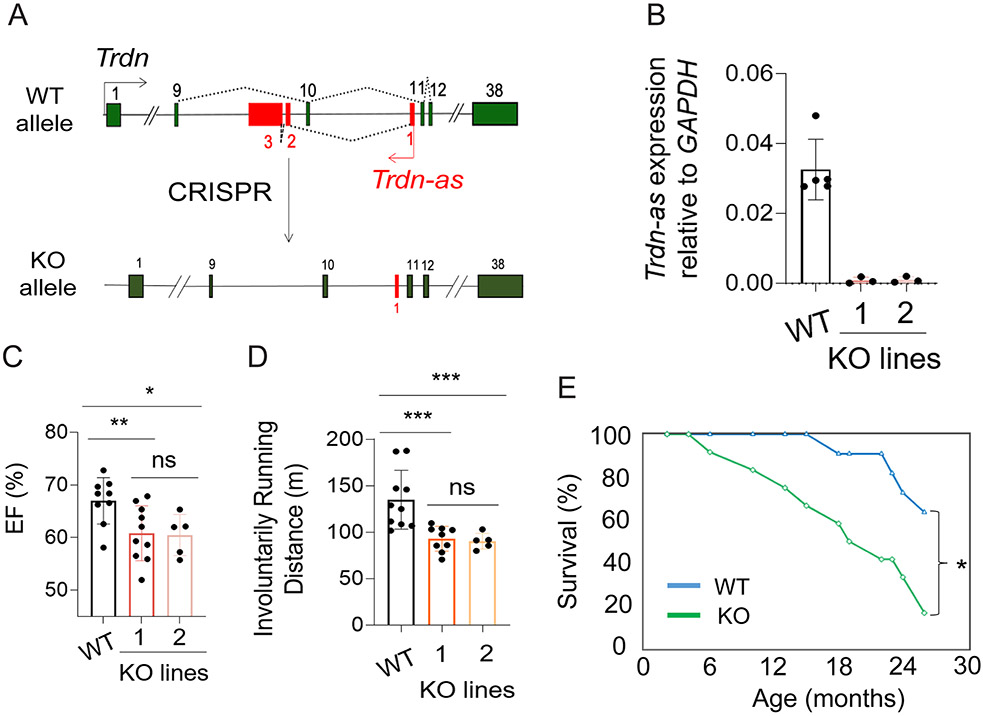
To examine the function of this lncRNA in vivo, we utilized CRISPR/Cas9 to delete exons 2 and 3 of mouse Trdn-as (Figure 2A). qPCR analysis confirmed that Trdn-as expression was undetectable in KO hearts (Figure 2B). To avoid CRISPR off-target effects, we generated two independent Trdn-as KO mouse lines. Heterozygous Trdn-as mice are normal in behavior and fertile. After we backcrossed heterozygous Trdn-as mice to WT C57BL6 mice for two generations, we crossed heterozygous Trdn-as mice with each other to generate mice with homozygous Trdn-as deletion. Homozygous Trdn-as KO mice were born at Mendelian ratios. We performed Sanger sequencing of the top 5 predicted off-target loci for each sgRNA but did not observe off-target editing by the sgRNA/Cas9 complex in either KO mouse line (Table S3). Deletion of Trdn-as did not dramatically change the anatomy or morphology of the heart (Figure S4). Echocardiographic measurement revealed a slight reduction in left ventricular ejection fraction (EF) in both Trdn-as KO mouse lines, relative to WT mice, at the age of 8 months, while these parameters were indistinguishable between these two KO lines (Figure 2C). Both KO mouse lines also displayed a significant reduction in involuntary exercise capacity compared to WT, while the exercise capacity of two KO lines was comparable (Figure 2D). The similarity of the two KO lines indicate that it is unlikely that the observed phenotype is due to CRISPR off-target effects. Therefore, we used the KO line 1 for all other studies unless otherwise specified. Strikingly, Trdn-as KO mice are susceptible to premature death compared to WT littermates, with fewer than 20% of KO mice surviving beyond 24 months of age, compared to over 60% of WT mice (Figure 2E). These data indicate that the lncRNA is required for normal cardiac function and longevity, and that loss of lncRNA expression is sufficient to induce abnormal cardiac function, reduced exercise capacity, and premature death.
Figure 2. Knockout of Trdn-as impaired cardiac function and caused premature death.
A) Knockout (KO) of Trdn-as by CRISPR/Cas9 genome editing. Trdn exons are shown in green and Trdn-as in red. B) Deletion of Trdn-as in hearts of two KO lines, line 1 and line 2 confirmed by qPCR. Each filled circle represents one mouse. Data are presented as mean ± SD. C-D) Left ventricular ejection fraction (EF) by echocardiography and running distance of indicated mice. Mice ran at speeds of 15, 25, 28, 31, 34, 37, and 40 m/min, each for 1 minute until mice were exhausted. Each filled circle represents one mouse. Data are presented as mean ± SD. One-way ANOVA with Tukey’s post test; *p = 0.05, **p < 0.05, ***p < 0.01, ns: not significant. E) Kaplan-Meier survival curve for WT (n = 11) and Trdn-as KO (n = 12) mice by OASIS2. Log-Rank test; *p<0.05. Both males and females were used for these assays.
Trdn-as mutant mice are susceptible to cardiac arrhythmias in response to catecholamine challenge.
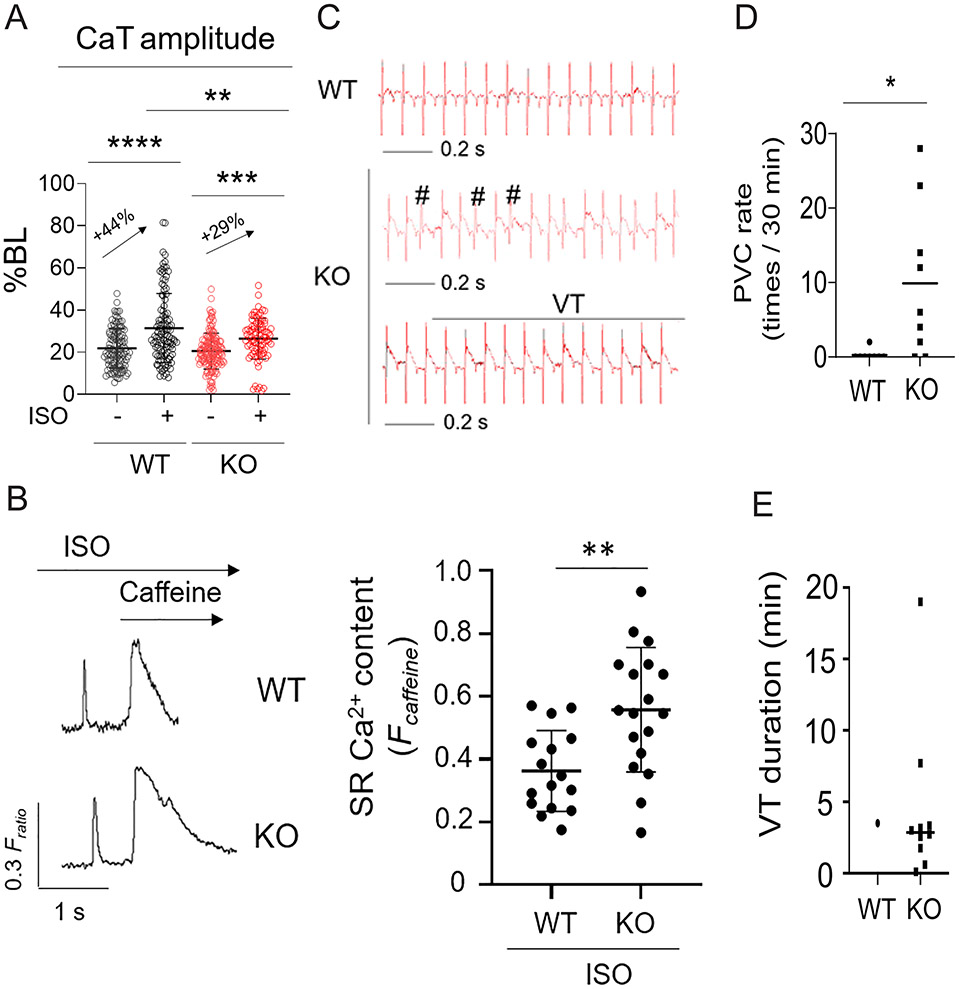
To better understand the mechanisms whereby loss of Trdn-as caused these phenotypes and thereby define the function of Trdn-as, we performed RNA-seq analysis. Deletion of Trdn-as in the mouse heart significantly dysregulated ~200 genes that are associated with GO terms such as “negative regulation of Ca2+ ion transmembrane transporter activity” and “positive regulation of ryanodine-sensitive Ca2+-release channel activity” (Figure S5A-B). These data suggest that Trdn-as influences Ca2+ handling. To test this hypothesis, we used the IonOptix platform to measure Ca2+ transients in Fura-2-loaded AMCMs isolated from WT or Trdn-as KO mice at the age of 5 months. AMCMs were field-stimulated at 1 Hz to induce cellular Ca2+ dynamics and contraction. At baseline, deletion of Trdn-as appeared to cause a slight, nonsignificant reduction in the amplitude of Ca2+ transients. Treatment with isoproterenol (ISO) was able to increase cytosolic Ca2+ levels in CMs during systole 32. However, this ISO-induced increase was significantly attenuated in Trdn-as KO AMCMs (Figure 3A), consistent with our observation that SR Ca2+ content was significantly increased in KO CMs compared to WT CMs under conditions of ISO stimulation (Figure 3B). To investigate effects of excess catecholamine signaling in the intact heart, we challenged WT and Trdn-as KO mice with ISO (0.1 mg/kg body weight) and monitored heart rhythm for 30 minutes by electrocardiography (ECG). This treatment barely induced abnormalities in heart rhythm in WT mice (Figure 3C-E). However, Trdn-as KO mice were much more susceptible to premature ventricular contractions (PVC) and ventricular tachycardia (VT) in response to catecholamine challenge with ISO (Figure 3C-E). Without ISO treatment, WT and Trdn-as KO mice displayed comparable ECG parameters (Table S4). Taken together, these data demonstrate that Trdn-as plays an essential role in regulation of CM Ca2+ homeostasis, and that disruption of this lncRNA can cause abnormalities in Ca2+ handling and cardiac arrhythmias under catecholamine stress.
Figure 3. Trdn-as KO mice are susceptible to cardiac arrhythmias in response to catecholamine challenge.
A) Ca2+ transients (CaT) of isolated AMCMs were field-stimulated at 1 Hz and recorded by the IonOptix system. AMCMs were treated with 0.1 μM ISO for 10 min prior to recording. 10 to 15 CMs per mouse, and 3 mice for each group were recorded. Each open circle represents an individual AMCM. Data are presented as mean ± SD. One-way ANOVA with Tukey’s post test, **p<0.01, ***p<0.001, ****p<0.0001. B) Modulation of SR Ca2+ load by Trdn-as under adrenergic stimulation. Representative IonOptix traces (left panel) of WT and Trdn-as KO AMCMS, which were isolated, loaded with Fura2-AM, pretreated with 500 nM Isoproterenol, and paced at 0.5 Hz. Where indicated, caffeine (10 mM) was added to the solution to induce Ca2+ release from SR, visible as high, extended peak. SR Ca2+ content was measured (right panel) by assessing the height of this peak 46, 47. 3 to 5 AMCMs per mouse, and 5 mice for each group were assayed. Each filled circle represents an individual AMCM. Data are presented as mean ± SD. P-values were calculated by Student’s t-test, **p<0.01. C) ECG records showing typical traces of WT mice and representative examples of PVC (#) and VT in KO mice after i.p. injection of ISO (0.1 mg/kg). D-E) Rate of PVC (D) and Duration of individual VT during a 30-min period after ISO injection (E). N = 8 WT mice and 9 KO mice. Each filled circle or square represents an individual WT or KO mouse. Data are presented as mean. P-values were calculated by Student’s t-test, *p<0.05.
Trdn-as is both sufficient and necessary for expression of cardiac triadin.
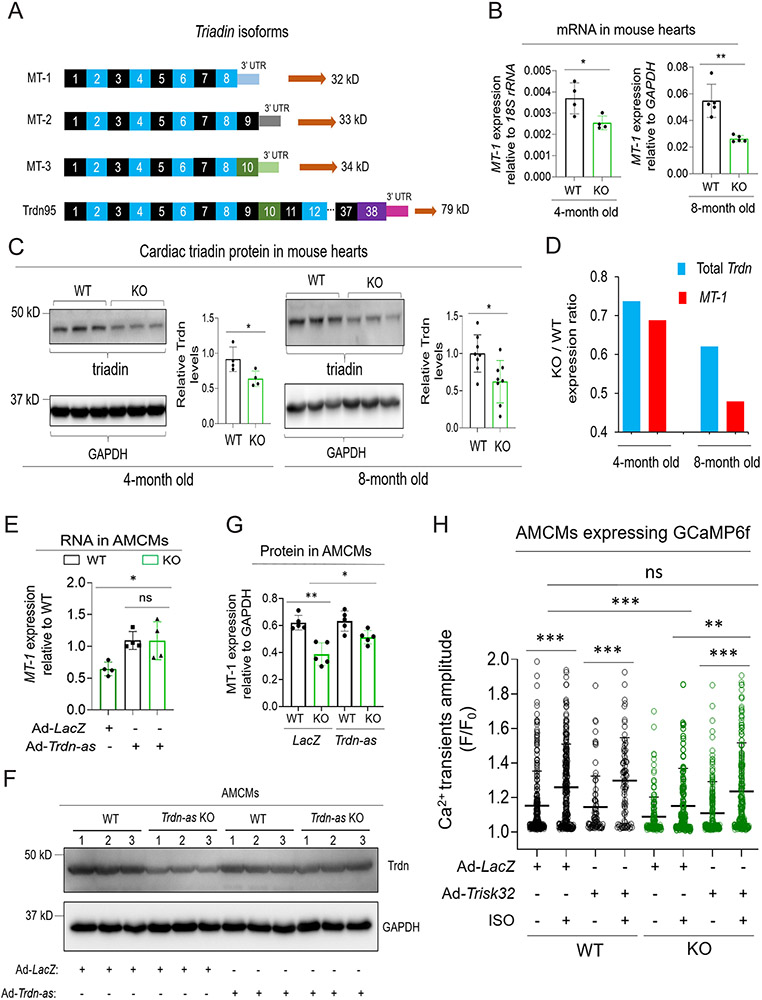
Next, we examined whether deletion of Trdn-as dysregulated Trdn expression. At least four triadin isoforms, produced by alternative splicing of the Trdn gene, have been identified in the mouse heart, with the predominant isoform (MT-1) encoded by exons 1 to 8 8. qPCR analysis using isoform-specific primers revealed that MT-1 expression was down-regulated by approximately 30% and 50% in Trdn-as KO hearts compared to WT counterparts at the age of 4 months and 8 months, respectively (Figure 4A-B). Triadin protein was also significantly down-regulated in Trdn-as KO mouse hearts (Figure 4C). Expression ratio of total Trdn in KO versus WT is greater than MT-1 expression ratio (Figure 4D); in other words, downregulation of MT-1 was more pronounced in KO mice than downregulation of other triadin isoforms. This observation may be due to increased expression of non-MT-1 isoforms in KO hearts which we observed with MT-3 (Figure S6A). These data indicate that Trdn-as positively regulates the expression of the predominant cardiac triadin isoform, MT-1, and is required for maintaining the normal level of triadin in the heart.
Figure 4. Trdn-as is essential and sufficient for maintenance of cardiac triadin levels in mouse hearts.
A) Triadin isoforms are alternative splicing products of a single gene, Trdn. Three short isoforms, MT-1 (ENSMUST00000219982.2), MT-2 (ENSMUST00000217779.2), and MT-3 (ENSMUST00000219931.2) are expressed in the heart, while full-length triadin, Trdn95 (ENSMUST00000095761.5), is mainly expressed in skeletal muscle. Trdn exons are numbered. 3’ UTR sequence for each isoform is unique. B) Expression of triadin MT-1 in WT or Trdn-as KO hearts by qPCR. Total RNA was extracted from adult mouse hearts at indicated ages. Expression of triadin isoforms were quantified by qPCR using paired primers amplifying specific isoforms of triadin. N = 4 or 5 animals per each group. Each filled circle represents a mouse. Data are presented as mean ± SD. C) Immunoblotting analysis of triadin in WT or Trdn-as KO hearts at indicated ages. Lysates of adult mouse hearts were subjected to western blot. A predominant triadin band of ~32 kD was quantified on the right. Trdn95 protein was not detected in heart lysates. N = 4 or 8 animals per each group. Each filled circle represents a mouse. Data are presented as mean ± SD. D) Mean expression of total Trdn and MT-1 in Trdn-as KO vs WT (KO / WT) hearts at indicated ages. Expression of Trdn exons 7-8 was quantified using qPCR to represent total Trdn mRNA levels. Mean expression of total Trdn and MT-1 in the heart was calculated from 4 mice per group at the age of 4 months or 8 mice per group at the age of 8 months. E-G) Re-expression of Trdn-as in Trdn-as KO AMCMs rescued MT-1 expression at mRNA and protein levels. LacZ or Trdn-as was delivered into AMCMs by adenovirus (Ad). Gene expression was analyzed by qPCR for MT-1 RNA in E 48 hours post-infection or by immunoblotting for triadin protein in F-G 72 hours post-infection. Levels of indicated protein in AMCMs isolated from three WT or KO hearts are shown in F. In E and G, each filled circle represents a mouse. Data are presented as mean ± SD. H) Overexpression of human cardiac triadin (Trisk32) rescued the defect in Ca2+ transients in Trdn-as KO AMCMs. LacZ or Trisk32, and GCaMP6f were delivered into AMCMs by adenovirus (Ad). 48 hours later, AMCMs were treated with 0.1 μM ISO for 15 mins. Ca2+ transients were imaged and analyzed using the GCaMP6-based system. AMCMs were isolated from 3 WT or 3 Trdn-as KO mice. Each open circle represents one AMCM. Data are presented as mean + SD. One-way ANOVA with Tukey’s post test (E, G, H) or Student’s t-test (B, C), *p<0.05, **p<0.01, ***p<0.001, ns, not significant.
We next determined whether forced expression of Trdn-as was sufficient to rescue MT-1 expression in KO AMCMs. We isolated AMCMs from WT and KO mice at the age of 5 months and overexpressed Trdn-as or LacZ (as a control) in KO AMCMs by adenoviral delivery. Re-expression of Trdn-as was sufficient to normalize MT-1 expression in KO AMCMs at both the mRNA and protein levels (Figures 4E-G, S6B), suggesting the essentiality of Trdn-as for maintenance of proper cardiac triadin levels. To further confirm whether Trdn-as is sufficient for MT-1 expression, we transiently expressed Trdn-as in C2C12 cells, skeletal muscle myoblasts. In skeletal muscle cells, Trdn-as is not expressed (Figure 1A) and MT-1 is not the predominant triadin isoform, making C2C12 cells a useful complementary system to Trdn-as KO cells. Overexpression of Trdn-as significantly increased expression of MT-1 in C2C12 cells (Figure S6C). In addition, in reprogrammed iCMs, over-expression of Trdn-as significantly increased MT-1 expression as well as the fraction of beating cells, while knockdown of Trdn-as had the opposite effect (Figures S1C and S6D-E). Taken together, these data indicate that Trdn-as is necessary and sufficient for the expression of cardiac triadin.
Next, we investigated whether forced expression of cardiac triadin in Trdn-as KO CMs was sufficient to reverse the defects caused by lncRNA deficiency. As shown in Figure 3A, the amplitude of Ca2+ transients was significantly lower in Trdn-as KO AMCMs compared to WT AMCMs under ISO treatment. Although we were able to use the IonOptix platform to measure Ca2+ transients in freshly isolated AMCMs, we encountered technical difficulties doing so in AMCMs cultured for multiple days. To measure Ca2+ transients in these cells, we introduced GCaMP6f, a fast Ca2+ sensor 33 into AMCMs by adenoviral delivery. GCaMP has been utilized to measure Ca2+ transients in mouse CMs in vitro and in vivo 34. Ca2+ transients were imaged and measured in AMCMs expressing GCaMP6f for 48 hours. At baseline, Trdn-as KO AMCMs displayed a nonsignificant downward trend in Ca2+ transient amplitude compared to WT AMCMs. While ISO treatment significantly increased the amplitude of Ca2+ transients in WT AMCMs, this ISO-induced increase was significantly attenuated in Trdn-as KO AMCMs (Figure 4H). These measurements, obtained using the GCaMP6f approach, were comparable to those measured by the IonOptix platform (Figure 3A versus 4H). Strikingly, over-expression of human cardiac triadin (Trisk32) in Trdn-as KO AMCMs restored the amplitude of Ca2+ transients to the level in WT AMCMs following ISO treatment, thereby resensitizing Trdn-as KO AMCMs to catecholamine signaling (Figure 4H). These data indicate that Trdn-as regulates Ca2+ handling in CMs, at least in part, through maintenance of proper levels of cardiac Triadin.
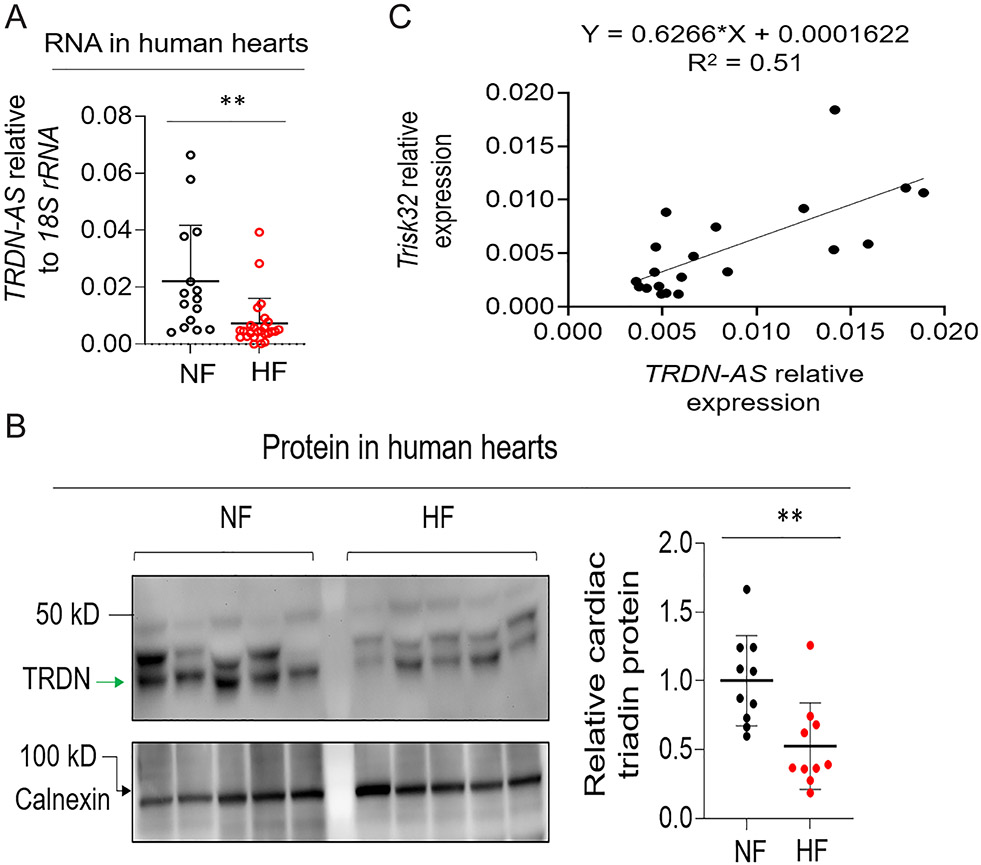
While TRDN-AS is highly expressed in healthy human hearts (Figure S3C), we speculated that it might be differentially expressed in heart failure. Both TRDN-AS and cardiac triadin, Trisk32, were significantly down-regulated in heart biopsies from patients with heart failure as well as ventricular arrhythmias, compared to control donors (Figure 5A-B). Moreover, TRDN-AS positively correlates with Trisk32 levels in human hearts (Figure 5C), suggesting a potential role of TRDN-AS in the regulation of cardiac triadin levels during cardiac pathogenesis in humans.
Figure 5. TRDN-AS was down-regulated in patients with heart failure.
A) Expression of TRDN-AS in hearts of healthy donors (Non-failing; NF) or patients with HF. Each open circle represents an individual human subject. Data are presented as mean + SD. B) Expression of cardiac triadin in NF or HF human hearts. Multiple bands with molecular weights of 30 to 50 kD appeared due to multiple isoforms and/or glycosylation of triadin in the heart, as shown in previous studies 5, 6. Trdn95 was not detectable in any samples by immunoblotting. Calnexin serves as a loading control. Non-glycosylated triadin, denoted by an arrow, was quantified. Each filled circle represents an individual human subject. Data are presented as mean ± SD. C) Expression correlation between TRDN-AS and cardiac triadin TRISK32 was analyzed using a simple linear regression model in GraphPad Prism 9.3. Each filled circle represents an individual human subject. Student’s t-test (A, B), **p<0.01.
Trdn-as colocalizes with serine/arginine splicing factors.
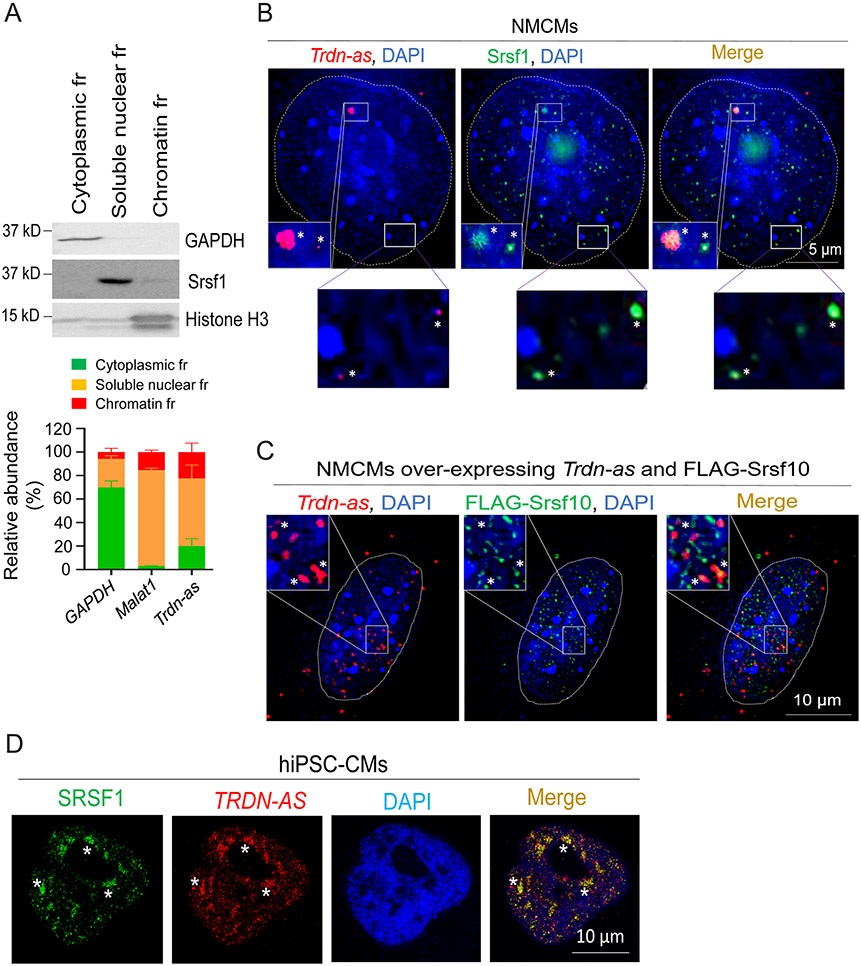
We next sought to determine the molecular mechanisms by which Trdn-as controls triadin levels in the heart. As shown in Figure 4A-D, deletion of Trdn-as altered the relative levels of alternative splicing Trdn products in the heart. We then asked whether Trdn-as controls cardiac triadin levels by interfering with splicing events. As Trdn-as is enriched in CM nuclei (Figure 1D-E), we carried out subcellular fractionation, which demonstrated that ~ 60% of Trdn-as was not associated with chromatin (Figure 6A), suggesting that Trdn-as could regulate biological processes such as gene splicing in chromatin-free regions. Serine/arginine splicing factors (SRSFs) are important components of spliceosomes and are essential for alternative splicing 35. We therefore asked whether Trdn-as colocalizes with SRSFs by smFISH and immunofluorescence imaging. We focused on Srsf1 and Srsf10 (also known as SRp38), which are essential for alternative splicing in the heart 36, 37. We initially characterized these potential colocalizations in non-CMs. We overexpressed either TRDN-AS or Trdn-as in HEK293 cells and performed FISH to label the lncRNA and co-immunostained for SRSF1. Both TRDN-AS and Trdn-as formed puncta and colocalized with SRSF1 in the nuclei of HEK293 cells (Figure S7A-B). Next, we examined lncRNA-SRSF colocalization in CMs. Using smFISH probes specific for Trdn-as and immunostaining for SRSFs in NMCMs, we demonstrated colocalization of both endogenous and over-expressed Trdn-as and Srsf1/Srsf10 in nuclei (Figure 6B-C). NMCMs overexpressing Trdn-as displayed increased levels of Trdn-as smFISH signals (Figure 6C vs 6B), confirming the specificity of the Trdn-as probes. FISH followed by co-immunostaining for SRSF1 also demonstrated that overexpressed TRDN-AS colocalized with SRSF1 in nuclei of hiPSC-CMs (Figure 6D). Results from non-CMs and CMs indicate that the lncRNA colocalizes with SR splicing factors in nuclei.
Figure 6. Trdn-as and TRDN-AS colocalize with SR splicing factors in CMs.
A) qPCR analysis of the relative abundance of transcripts from subcellular fractions (fr) of AMCMs. Immunoblotting for GAPDH as a marker for cytoplasm, Srsf1 for soluble nuclear, and histone H3 for chromatin fractions are shown in the top panel. GAPDH and Malat1 RNAs are used as markers for cytoplasmic and soluble nuclear fractions, respectively. N = 3 independent experiments. Data are presented as mean + SD. B-C) Confocal imaging analysis of Trdn-as and Srsf1 in B and over-expressed Trdn-as and FLAG-Srsf10 in C in NMCMs. Endogenous (in B) or over-expressed Trdn-as (in C) were detected by smFISH. In C, NMCMs were infected with adenovirus vectors carrying Trdn-as or FLAG-Srsf10 with a MOI of 5. 48 hours later, over-expressed Trdn-as and FLAG-Srsf10 were analyzed using smFISH and immunostaining, respectively. Nuclei are surrounded by white lines. Insets are enlarged on the right corner. Colocalizations are indicated by stars. D) Confocal imaging analysis of TRDN-AS and SRSF1 in a hiPSC-CM by FISH. TRDN-AS was over-expressed in hiPSC-CMs by adenovirus with a MOI of 5. 48 hours later, TRDN-AS and SRSF1 were analyzed using FISH and immunostaining, respectively. Colocalizations are indicated by stars.
Trdn-as interacts with serine/arginine splicing factors and Trdn pre-mRNA.
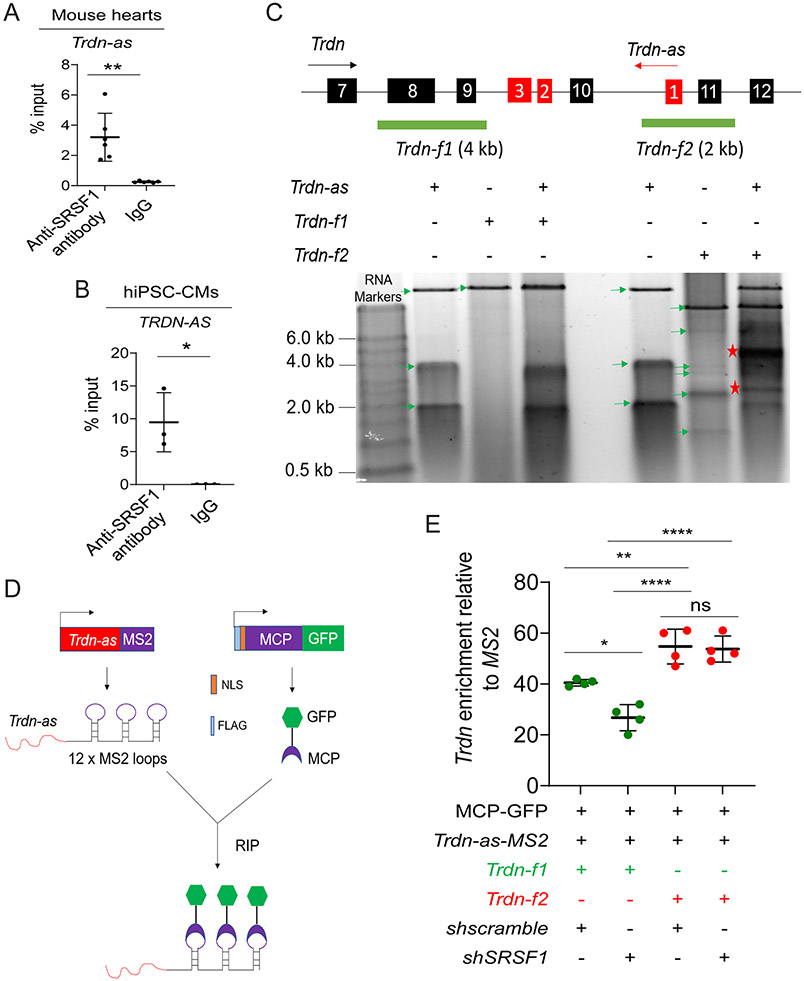
There are multiple predicted binding motifs for SRSF1 and SRSF10 within regions in the third exon of Trdn-as and TRDN-AS (Figure S8A-B), suggesting potential interactions between the lncRNA and SR splicing factors. To examine the specificity of the interaction between Trdn-as and SR splicing factors, we performed RNA coimmunoprecipitation (RIP) assays on nuclear extracts using an anti-SRSF1 antibody. First, we conducted RIP using nuclear extracts from HEK293 cells. Since commercial anti-SRSF10 antibodies were incapable of pulling down endogenous SRSF10 in our hands, we overexpressed Trdn-as and either FLAG-tagged SRSF10 or LacZ by transfection. qPCR analysis of RIP samples (RIP-qPCR) precipitated by anti-SRSF1 or anti-FLAG antibodies using Trdn-as-specific primers demonstrated a specific interaction between Trdn-as and SRSF1 or SRSF10 in non-myocytes (Figure S8C-D). Interestingly, co-overexpression of SRSF10 increased the interaction between Trdn-as and SRSF1 (Figure S8C), indicating that SR splicing factors synergistically interact with Trdn-as. Second, we performed RIP using an anti-SRSF1 antibody in nuclear extracts from WT mouse hearts. RIP-qPCR analysis also revealed a specific interaction between SRSF1 and Trdn-as in the mouse heart (Figure 7A). Finally, RIP-qPCR also revealed that TRDN-AS interacted with SRSF1 in hiPSC-CMs (Figure 7B). Taken together, these data indicate that Trdn-as and TRDN-AS physically interact with splicing factors.
Figure 7. Trdn-as interact with SR splicing factors and Trdn pre-mRNA.
A) RIP-qPCR analysis of the interaction of Trdn-as with Srsf1 in adult mouse hearts. n=6 independent experiments. Data are presented as mean ± SD. Student’s t-test, **p<0.01. B) RIP-qPCR analysis of the interaction of TRDN-AS with SRSF1 in hiPSC-CMs. n=3 independent experiments. Data are presented as mean ± SD. Student’s t-test, *p<0.05. C) Analysis of interactions between Trdn-as and Trdn pre-mRNA by native agarose gel electrophoresis. Trdn and Trdn-as genes schematics are shown on top. Exons of Trdn and Trdn-as are numbered in black and red boxes, respectively. The black and red arrows indicate the direction of Trdn or Trdn-as transcription, respectively. Full-length Trdn-as and two partial fragments of Trdn pre-mRNA, Trdn-f1 and Trdn-f2 were in vitro transcribed. RNA was purified after treatment with proteinase K and RNase-free DNase I. The mixture of Trdn-as and either Trdn-f1 or Trdn-f2 was resolved in native agarose gel electrophoresis. Green arrows indicate individual RNAs. Asterisks indicate RNA-RNA duplex. D) A schematic diagram shows pulldown of lncRNA-associated complexes using the MS2-MCP system. MS2 RNA stem loops strongly interact with the MS2 coat protein (MCP). Trdn-as was fused to 12 x MS2 stem loops (Trdn-as-MS2). Nuclear-localized, FLAG-tagged MCP was fused to GFP (FLAG-NLS-MCP-GFP). Trdn-as associated complexes are precipitated using a GFP antibody. E) qPCR analysis of Trdn pre-mRNA levels in complexes precipitated by the GFP antibody. HEK293 cells were transfected with various combinations of expression plasmids encoding MCP-GFP, shscramble, shSRSF1, MS2, or Trdn-as-MS2. 48 hours later, nuclear extracts of HEK293 cells were incubated with in vitro transcribed Trdn pre-mRNA Trdn-f1 or Trdn-f2. MS2 or Trdn-as-MS2 associated complexes were precipitated by anti-GFP antibody. Trdn pre-mRNA was analyzed using qPCR to amplify Trdn exon 8 for Trdn-f1 or Trdn exon 11 for Trdn-f2, respectively. N = 4 independent experiments. Data are presented as mean ± SD. One-way ANOVA with Tukey’s post hoc test. *p<0.05, **p<0.01, ****p<0.0001, ns, not significant.
Next, we analyzed the interaction between the lncRNA and triadin pre-mRNA. Full-length Trdn-as and Trdn pre-mRNA were in vitro transcribed, and the lncRNA/pre-mRNA mixture was resolved by native agarose gel electrophoresis. These assays demonstrated that Trdn-as directly interacts with Trdn pre-mRNA via complementary bases (Figure 7C). Next, we determined whether Trdn-as, SR splicing factors, and Trdn pre-mRNA form a complex in vitro using an MS2-MCP system 38 (Figure 7D). An MS2 RNA stem loop strongly interacts with the MS2 coat protein (MCP). We therefore generated a construct encoding full-length Trdn-as fused to 12 copies of the stem-loop of phage MS2 (Trdn-as-MS2) under the control of a CMV promoter. We co-expressed Trdn-as-MS2 and a nuclear localized MCP-GFP fusion protein (FLAG-NLS-MCP-GFP) in HEK293 cells by transfection, and incubated 48-hour transfected cell nuclear extracts with an anti-GFP antibody and in vitro transcribed partial Trdn pre-mRNA Trdn-f1 or Trdn-f2, as shown in Figure 7C. We examined association of in vitro transcribed pre-mRNA with Trdn-as by performing RIP with the anti-GFP antibody. SRSF1 was assayed by immunoblotting (Figure S9A). Trdn-as and Trdn pre-mRNA were analyzed by qPCR (Figures S9B and 7E). Trdn-f1 pre-mRNA that did not directly interact with Trdn-as (Figure 7C) was able to be detected in Trdn-as-associated complexes (Figure 7E) because SRSF1/SRSF10 can interact with both Trdn-as (Figures 6 and 7A-B) and pre-mRNA. However, more Trdn-f2 pre-mRNA that directly formed an RNA/RNA duplex with Trdn-as (Figure 7C) was precipitated by Trdn-as (Figure 7E). Importantly, knockdown of SRSF1 significantly decreased the Trdn-f1 amount in Trdn-as-associated complexes, but did not alter the association of Trdn-f2 with Trdn-as (Figure 7E), indicating that the interaction between Trdn-as and Trdn pre-mRNA is not reliant on SRSF1. These assays demonstrated that Trdn-as directly interacts with Trdn pre-mRNA and SR splicing factors in vitro.
Trdn-as is essential for recruitment of splicing factors to triadin transcripts in vivo.
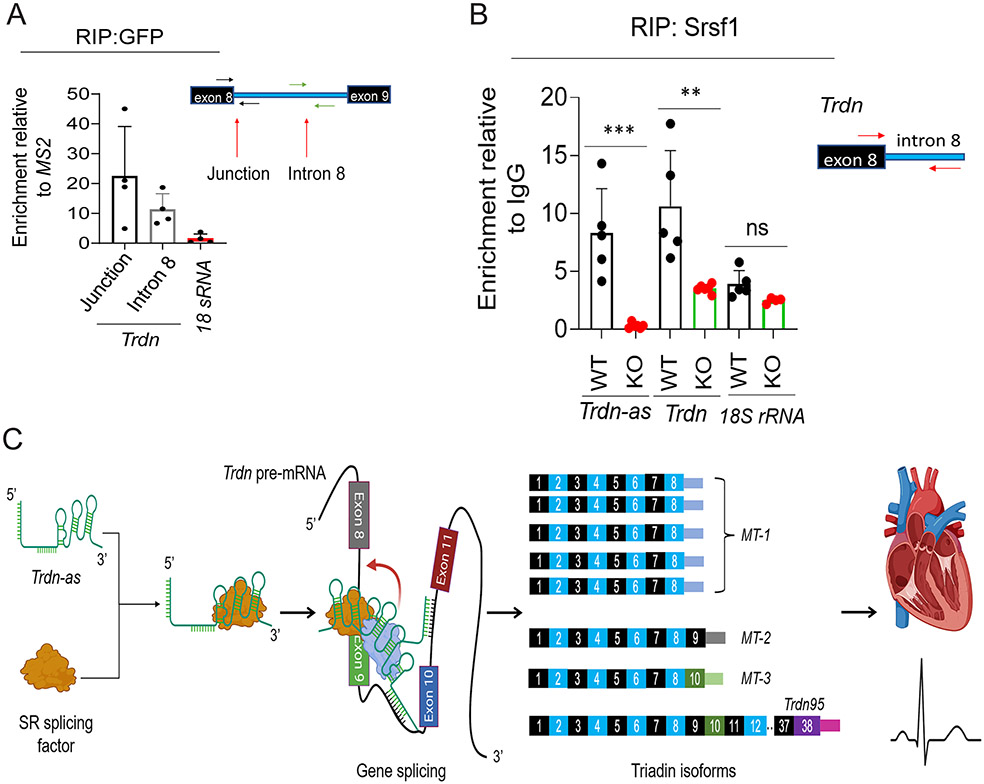
Knockout of Trdn-as altered alternative splicing products of the Trdn gene (Figure 4B-D). Trdn-as directly interacts with Trdn pre-mRNA in vitro (Figure 7C). To investigate whether Trdn-as could be recruited to and retained by Trdn pre-mRNA in vivo, we pulled down the lncRNA-associated complexes using the MS2-MCP system, as shown in Figure 7D. We used adenoviral vectors to overexpress MS2 or Trdn-as-MS2 and FLAG-NLS-MCP-GFP in AMCMs isolated from WT mice. We performed RIP using an anti-GFP antibody from nuclear extracts of AMCMs 48 hours post-transduction. qPCR analysis using specific primers to target intron 8 of the Trdn gene revealed an association of Trdn-as with Trdn pre-mRNA (Figure 8A). Next, we determined the effects of Trdn-as on the recruitment of splicing factors to Trdn mRNA. We performed RIP using an anti-SRSF1 antibody in nuclear extracts from WT and Trdn-as KO mouse hearts. qPCR analysis of RNA in the precipitated fraction revealed that deletion of Trdn-as significantly decreased association of Srsf1 with Trdn mRNA in the heart (Figure 8B). It is well established that alterations in phosphorylation of SR splicing factors modulate alternative splicing by interfering with their recruitment to pre-mRNA and/or the activity of spliceosomes 39-40. Immunoblotting assays demonstrated that Trdn-as deficiency did not alter phosphorylation of Srsf1 in the mouse heart (Figure S10A-B). Taken together, these data indicate that Trdn-as is required for recruitment of SR splicing factors to maintain the normal composition of triadin isoforms in the heart.
Figure 8. Trdn-as is required for recruitment of SR splicing factors to Trdn transcripts in the heart.
A) RIP of Trdn-as associated complexes in AMCMs using the MS2-MCP system in Figure 7D. AMCMs were infected by adenoviruses carrying either MS2 or Trdn-as-MS2 and MCP-GFP. RIP with an anti-GFP antibody was performed 48 hours post-infection. Junction of Trdn exon 8 and intron 8, Trdn intron 8 denoting Trdn pre-mRNA, and 18s rRNA were then quantified by qPCR. N = 4 under each condition. Relative enrichment was calculated by normalization of RNA levels in AMCMs expressing Trdn-as-MS2 to those in AMCMs expressing MS2 post-RIP with the antibody against GFP. Data are presented as mean + SD. B) RIP with anti-SRSF1 antibody followed by qPCR analysis of RNAs in Srsf1-associated complexes in WT or Trdn-as KO mouse hearts. Trdn was quantified by qPCR to amplify the exon 8/intron 8 junction. N = 4-6 mouse hearts under each condition. Data are presented as mean + SD. One-way ANOVA with Tukey’s post test, ***p<0.001, ns, not significant. C) Model of Trdn-as to modulate triadin isoform composition in the heart. Trdn-as colocalizes and interacts with SR splicing factors in nuclear speckles. Trdn-as forms an RNA/RNA duplex with Trdn pre-mRNA and facilitates the recruitment of SR splicing factors to Trdn pre-mRNA to produce the predominant triadin isoform MT-1 in the heart. Knockout of Trdn-as impairs the recruitment of splicing factors to Trdn pre-mRNA, leading to abnormal composition of triadin isoforms in the heart, Ca2+ mishandling, and susceptibility to cardiac arrhythmias.
Discussion
In the heart, lncRNAs regulate gene expression by diverse mechanisms, including involvement in chromosomal epigenetic modifications and serving as a sponge for microRNAs. Here, we have discovered an additional mechanism by which a cardiac-specific and conserved lncRNA controls composition of triadin isoforms, via lncRNA-mediated gene splicing (Figure 8C). Our findings reveal that Trdn-as/TRDN-AS is able to regulate alternative splicing of the triadin gene in the heart. Loss-of-function mutations in the TRDN gene have been associated with cardiac arrhythmias and sudden death in humans 12-13. A recent genome-wide association study (GWAS) of genetic effects on ECG morphology demonstrates that rs12209641, an intron variant within the TRDN and TRDN-AS locus is statistically associated with differences in the ECG’s Q wave (p = 4 × 10−10) (https://www.ebi.ac.uk/gwas/variants/rs12209641) 41. Knockout of either Trdn-as or Trdn in the mouse heart had no detectable effects on ECG parameters at baseline (Table S4 and Ref. 14). However, both Trdn-as−/− and Trdn−/− mice displayed a wide QRS complex under catecholamine stimulation (Figure 3C and Ref. 14). Our data, together with the above GWAS highlight crucial roles of TRDN-AS and/or triadin in cardiac rhythms. Therefore, our findings that TRDN-AS/Trdn-as regulates alternative splicing of the triadin gene to maintain the cardiac function could assist with designing of therapeutic strategies to treat cardiac arrhythmias.
A previous study demonstrated that the SR splicing factor Srsf10 regulates triadin MT-1 levels through modulating alternative splicing of Trdn pre-mRNA in the heart 36. However, Srsf10 also plays a crucial role in alternative splicing events in skeletal muscle cells 42. Why Srsf10 specifically controls alternative splicing of Trdn pre-mRNA in the heart remains elusive. The CM-specific lncRNA, Trdn-as, interacts with SR splicing factors including Srsf10 and is required for their recruitment to Trdn mRNA (Figures 6-8). Our findings support that a cell-specific lncRNA is able to determine the cell-specific role of SR splicing factors that are ubiquitously expressed.
Although the majority of Trdn-as is localized to the soluble fraction of CM nuclei, ~20% of the lncRNA localizes in the cytosol and an additional ~20% of the lncRNA associates with chromatin (Figures 1D-E, 1G-H, and 6A). These data suggest that this lncRNA could play additional cytosolic roles and/or be involved in chromatin-related biological processes in CMs, in addition to regulation of splicing events (Figure 8C). Given the striking phenotype caused by deletion of Trdn-as in mice (Figures 2-3), elucidating additional functions of this lncRNA and the mechanisms of its functions in the cytosol and on chromatin could facilitate discovery of therapeutics for cardiac arrhythmias and heart failure. Recently, it has been shown that some putative lncRNAs encode micropeptides to regulate physiological processes 43, 44. Van Heesch et al. detected a ~ 7 kDa peptide encoded by TRDN-AS in 2 out of 5 human hearts by mass spectrometry. However, the authors did not detect peptides encoded by Trdn-as in mouse hearts 44. A major fraction of TRDN-AS/Trdn-as localizes in CM nuclei (Figure 1D-H), suggesting that this lncRNA primarily functions at the RNA level in the heart. Nonetheless, we cannot completely rule out the possibility that a micropeptide encoded by TRDN-AS regulates cardiac physiology. It would be interesting to study the mechanism of the action of this potential micropeptide in the future.
Recently, Trdn-as was speculated to reduce the expression of the skeletal muscle triadin isoform in CMs 45. Zhang et al. used dCas9-VP64 to over-activate Trdn-as expression in the HL-1 CM cell line, which led to a mild reduction in expression of Trdn95, the predominant skeletal muscle triadin isoform, from relative expression of ~ 3.6 ± 1.5 at baseline to ~ 3.2 ± 1.2 (mean ± SD) by qPCR but did not change cardiac triadin levels. Although, this difference was significant, a change in Trdn95 levels of ~ 3.6 ± 1.5 and ~ 3.2 ± 1.2 is relative small in magnitude. Based on these in vitro data from an immortalized CM cell line, Zhang et al. proposed a hypothetical model that Trdn-as expression blocked transcription of skeletal muscle triadin, Trdn95, by RNA Polymerase II collision. In our studies, knockout of Trdn-as significantly decreased cardiac triadin levels (Figures 4B-D). In addition, TRDN-AS is significantly reduced in heart explants from patients with heart failure. However, we did not observe increased levels of skeletal muscle triadin in those failing hearts wherein cardiac triadin was significantly decreased (Figure 5B). Although the above data from human and mouse hearts strongly suggest that this lncRNA primarily controls cardiac triadin levels, we cannot completely exclude the possibility that it suppresses skeletal muscle triadin expression in the heart through the previously proposed RNA Polymerase II collision mechanism. It is therefore worth examining the role of lncRNA transcription in the suppression of skeletal muscle triadin expression and/or in the activation of cardiac triadin expression in future studies.
Down-regulation of cardiac triadin has been associated with heart failure and cardiac arrhythmias (TKOS) 12-15. Human TRDN-AS that is significantly down-regulated in hearts of patients with heart failure and ventricular arrhythmias shows a positive correlation with triadin expression in the heart (Figure 5). Here, we show that Trdn-as is a positive regulator of cardiac triadin expression in the mouse heart (Figures 4). Knockout of Trdn-as impairs cardiac function and exercise capacity, as well as causing premature death (Figure 2). Trdn-as mutant mice are predisposed to cardiac arrhythmias in response to catecholamine challenge (Figure 3), which was also observed in Trdn knockout mice 14. Overexpression of Trdn-as is sufficient to normalize cardiac triadin levels in Trdn-as KO AMCMs (Figure 4E-G). These studies indicate that this CM-specific and conserved lncRNA could be a potential target of gene therapy to treat heart failure and cardiac arrhythmias.
Supplementary Material
Clinical Perspective.
What is new?
The cardiomyocyte-specific lncRNA Trdn-as is essential for maintenance of cardiac function, exercise capacity, and normal life span.
Trdn-as knockout mice are predisposed to cardiac arrhythmias in response to catecholamine challenge.
Trdn-as controls levels of cardiac triadin isoforms by regulating splicing of the triadin gene.
What are the clinical implications?
Cardiac explants from human heart failure patients as well as patients with cardiac arrhythmias patients demonstrate reduced expression of TRDN-AS and TRDN.
The mechanism of the TRDN-AS/Trdn-as-mediated alternative splicing of the triadin gene to specifically control levels of cardiac isoforms of triadin in the heart provides a potential strategy for the treatment of cardiac arrhythmias and heart failure.
Acknowledgements:
The authors thanked Dr. McKinsey for critical reading of the manuscript. Figure 8C was created with BioRender.com.
Sources of Funding:
A.S.R. was supported by pre-doctoral fellowships from Colorado Clinical & Translational Sciences Institute (TL1 TR001081), and the American Heart Association (AHA) (18PRE34030030). W.E.K. was supported by NIH Cardiology training grant T32HL007822, AHA (19POST34380250), and an NIH/CCTSI CO-Pilot Mentored Faculty Award (TL1 TR002533). K.S. was supported by funds from the Boettcher Foundation, University of Colorado DOMOECSP, Gates Frontiers Fund, American Heart Association, and NIH R01HL133230. REDCap was supported by NIH/NCATS Colorado (UL1 TR002535). Echocardiographic analysis was supported by NIH grant 1S10OD018156-01.
Non-standard Abbreviations and Acronyms
- LncRNA
long non-coding ribonucleic acid
- TRDN
human triadin gene
- Trdn
mouse triadin gene
- MT-1
predominant triadin isoform in mouse heart
- Trisk32
predominant triadin isoform in human heart
- RYR2
Ryanodine receptor 2
- SR
Sarcoplasmic reticulum
- CM
cardiomyocyte
- cTnT
cardiac muscle troponin T
- CF
cardiac fibroblast
- Ca2+
Calcium ion
- SERCA2A
sarco/endoplasmic reticulum Ca2+-ATPase
- DHPR
dihydropyridine receptor
- Casq2
calsequestrin 2
- TKOS
triadin knockout syndrome
- ISO
isoproterenol
- PRC2
polycomb repressor complex 2
- SWI/SNF
SWItch/Sucrose Non Fermentable
- HNRNPC
heterogeneous nuclear ribonucleoprotein C
- iCM
induced CMs
- SRSF
serine/arginine (SR) splicing factor
- NMCM
neonatal mouse ventricular cardiomyocyte
- AMCM
adult mouse ventricular cardiomyocyte
- WT
wild type
- KO
knockout
- RACE
rapid amplification of cDNA ends
- FISH
fluorescence in situ hybridization
- smFISH
single molecule fluorescence in situ hybridization
- GTEx
Genotype-Tissue Expression
- CRISPR
clustered regularly interspaced short palindromic repeats
- sgRNA
single-guide RNA
- hiPSC-CM
human induced pluripotent stem cell
- ECG
electrocardiography/electrocardiogram
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- PVC
premature ventricular contraction
- VT
ventricular tachycardia
- EF
ejection fraction
- LacZ
β-galactosidase
- GFP
green fluorescent protein
- RIP
RNA coimmunoprecipitation
- qPCR
quantitative polymerase chain reaction
- GCaMP
genetically encoded calcium indicator that is a fusion of calmodulin, a peptide sequence from myosin light-chain kinase, and green fluorescent protein
- Pre-mRNA
precursor mRNA
- MS2
RNA stem-loop repeats derived from the MS2 bacteriophage
- MCP
MS2 coat protein
- NLS
nuclear localization signal
- GWAS
genome-wide association study
Footnotes
Disclosures
None.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Smith GL, Eisner DA. Calcium Buffering in the Heart in Health and Disease. Circulation. 2019;139:2358–2371. doi: 10.1161/CIRCULATIONAHA.118.039329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bers DM. Cardiac sarcoplasmic reticulum calcium leak: basis and roles in cardiac dysfunction. Annu Rev Physiol. 2014;76:107–127. doi: 10.1146/annurev-physiol-020911-153308 [DOI] [PubMed] [Google Scholar]
- 4.Guo W, Campbell KP. Association of triadin with the ryanodine receptor and calsequestrin in the lumen of the sarcoplasmic reticulum. J Biol Chem. 1995;270:9027–30. doi: 10.1074/jbc.270.16.9027. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi YM, Alseikhan BA, Jones LR. Localization and characterization of the calsequestrin-binding domain of triadin 1. Evidence for a charged beta-strand in mediating the protein-protein interaction. J Biol Chem. 2000;275:17639–46. doi: 10.1074/jbc.M002091200. [DOI] [PubMed] [Google Scholar]
- 6.Hong CS, Ji JH, Kim JP, Jung DH, Kim DH. Molecular cloning and characterization of mouse cardiac triadin isoforms. Gene. 2001;278:193–9. doi: 10.1016/s0378-1119(01)00718-1. [DOI] [PubMed] [Google Scholar]
- 7.Marty I, Fauré J, Fourest-Lieuvin A, Vassilopoulos S, Oddoux S, Brocard J. Triadin: what possible function 20 years later?. J Physiol. 2009;587:3117–3121. doi: 10.1113/jphysiol.2009.171892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez CF. On the footsteps of Triadin and its role in skeletal muscle. World J Biol Chem. 2011;2:177–83. doi: 10.4331/wjbc.v2.i8.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marty I. Triadin regulation of the ryanodine receptor complex. J Physiol. 2015;593:3261–6. doi: 10.1113/jphysiol.2014.281147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux-Buisson N, Cacheux M, Fourest-Lieuvin A, Fauconnier J, Brocard J, Denjoy I, Durand P, Guicheney P, Kyndt F, Leenhardt A, et al. Absence of triadin, a protein of the calcium release complex, is responsible for cardiac arrhythmia with sudden death in human. Hum Mol Genet. 2012;21:2759–67. doi: 10.1093/hmg/dds104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altmann HM, Tester DJ, Will ML, Middha S, Evans JM, Eckloff BW, Ackerman MJ. Homozygous/Compound Heterozygous Triadin Mutations Associated With Autosomal-Recessive Long-QT Syndrome and Pediatric Sudden Cardiac Arrest: Elucidation of the Triadin Knockout Syndrome. Circulation. 2015;131:2051–60. doi: 10.1161/CIRCULATIONAHA.115.015397. [DOI] [PubMed] [Google Scholar]
- 12.Clemens DJ, Tester DJ, Giudicessi JR, Bos JM, Rohatgi RK, Abrams DJ, Balaji S, Crotti L, Faure J, Napolitano C, et al. International Triadin Knockout Syndrome Registry. Circ Genom Precis Med. 2019;12:e002419. doi: 10.1161/CIRCGEN.118.002419. [DOI] [PubMed] [Google Scholar]
- 13.Wleklinski MJ, Kannankeril PJ, Knollmann BC. Molecular and tissue mechanisms of catecholaminergic polymorphic ventricular tachycardia. J Physiol. 2020;598:2817–2834. doi: 10.1113/JP276757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chopra N, Yang T, Asghari P, Moore ED, Huke S, Akin B, Cattolica RA, Perez CF, Hlaing T, Knollmann-Ritschel BE, et al. Ablation of triadin causes loss of cardiac Ca2+ release units, impaired excitation-contraction coupling, and cardiac arrhythmias. Proc Natl Acad Sci U S A. 2009;106:7636–41. doi: 10.1073/pnas.0902919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gergs U, Berndt T, Buskase J, Jones LR, Kirchhefer U, Müller FU, Schlüter KD, Schmitz W, Neumann J. On the role of junctin in cardiac Ca2+ handling, contractility, and heart failure. Am J Physiol Heart Circ Physiol. 2007;293:H728–34. doi: 10.1152/ajpheart.01187.2006. [DOI] [PubMed] [Google Scholar]
- 16.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev Cell. 2017;42:333–348.e5. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 23.Fan J, Li H, Xie R, Zhang X, Nie X, Shi X, Zhan J, Yin Z, Zhao Y, Dai B, et al. LncRNA ZNF593-AS Alleviates Contractile Dysfunction in Dilated Cardiomyopathy. Circ Res. 2021;128:1708–1723. doi: 10.1161/CIRCRESAHA.120.318437. [DOI] [PubMed] [Google Scholar]
- 24.Riching AS, Song K. Cardiac Regeneration: New Insights Into the Frontier of Ischemic Heart Failure Therapy. Front Bioeng Biotechnol. 2021;8:637538. doi: 10.3389/fbioe.2020.637538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponnusamy M, Liu F, Zhang YH, Li RB, Zhai M, Liu F, Zhou LY, Liu CY, Yan KW, Dong YH, et al. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation. 2019;139:2668–2684. doi: 10.1161/CIRCULATIONAHA.118.035832. [DOI] [PubMed] [Google Scholar]
- 26.Bruns DR, Tatman PD, Kalkur RS, Brown RD, Stenmark KR, Buttrick PM, Walker LA. The right ventricular fibroblast secretome drives cardiomyocyte dedifferentiation. PLoS One. 2019;14:e0220573. doi: 10.1371/journal.pone.0220573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight WE, Cao Y, Lin YH, Chi C, Bai B, Sparagna GC, Zhao Y, Du Y, Londono P, Reisz JA, et al. Maturation of Pluripotent Stem Cell-Derived Cardiomyocytes Enables Modeling of Human Hypertrophic Cardiomyopathy. Stem Cell Reports. 2021;16:519–533. doi: 10.1016/j.stemcr.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweet ME, Cocciolo A, Slavov D, Jones KL, Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L, et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics. 2018;19:812. doi: 10.1186/s12864-018-5213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Béguec C, Wucher V, Lagoutte L, Cadieu E, Botherel N, Hédan B, De Brito C, Guillory AS, André C, Derrien T, et al. Characterisation and functional predictions of canine long non-coding RNAs. Sci Rep. 2018;8:13444. doi: 10.1038/s41598-018-31770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adachi-Akahane S, Cleemann L, Morad M. Cross-signaling between L-type Ca2+ channels and ryanodine receptors in rat ventricular myocytes. J Gen Physiol. 1996;108:435–54. doi: 10.1085/jgp.108.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaestner L, Scholz A, Tian Q, Ruppenthal S, Tabellion W, Wiesen K, Katus HA, Müller OJ, Kotlikoff MI, Lipp P. Genetically encoded Ca2+ indicators in cardiac myocytes. Circ Res. 2014;114:1623–39. doi: 10.1161/CIRCRESAHA.114.303475. [DOI] [PubMed] [Google Scholar]
- 35.Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell. 2009;16:528–38. doi: 10.1016/j.devcel.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016;15:2025–37. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–44. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 40.Lipp JJ, Marvin MC, Shokat KM, Guthrie C. SR protein kinases promote splicing of nonconsensus introns. Nat Struct Mol Biol. 2015;22:611–7. doi: 10.1038/nsmb.3057. [DOI] [PubMed] [Google Scholar]
- 41.Verweij N, Benjamins JW, Morley MP, van de Vegte YJ, Teumer A, Trenkwalder T, Reinhard W, Cappola TP, van der Harst P. The Genetic Makeup of the Electrocardiogram. Cell Syst. 2020;11:229–238.e5. doi: 10.1016/j.cels.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei N, Cheng Y, Wang Z, Liu Y, Luo C, Liu L, Chen L, Xie Z, Lu Y, Feng Y. SRSF10 Plays a Role in Myoblast Differentiation and Glucose Production via Regulation of Alternative Splicing. Cell Rep. 2015;13:1647–57. doi: 10.1016/j.celrep.2015.10.038. [DOI] [PubMed] [Google Scholar]
- 43.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–5. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, et al. The Translational Landscape of the Human Heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Salgado-Somoza A, Vausort M, Leszek P, Devaux Y; Cardiolinc™ network. A heart-enriched antisense long non-coding RNA regulates the balance between cardiac and skeletal muscle triadin. Biochim Biophys Acta Mol Cell Res. 2018;1865:247–258. doi: 10.1016/j.bbamcr.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Knollmann BC, Chopra N, Hlaing T, Akin B, Yang T, Ettensohn K, Knollmann BE, Horton KD, Weissman NJ, Holinstat I, et al. Casq2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. J Clin Invest. 2006;116:2510–20. doi: 10.1172/JCI29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bers DM. Sarcoplasmic reticulum. Excitation and contraction coupling and cardiac contractile force. Kluwer Academic Publishers. Dordrecht, The Netherlands/Boston, Massachusetts, USA/London, United Kingdom. 2001:161–202. [Google Scholar]
- 48.Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O'Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vyboh K, Ajamian L, Mouland AJ. Detection of viral RNA by fluorescence in situ hybridization (FISH). J Vis Exp. 2012;63:e4002. doi: 10.3791/4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bak G, Han K, Kim KS, Lee Y. Electrophoretic mobility shift assay of RNA-RNA complexes. Methods Mol Biol. 2015;1240:153–63. doi: 10.1007/978-1-4939-1896-6_12. [DOI] [PubMed] [Google Scholar]
- 51.Gorski PA, Kho C, Oh JG. Measuring Cardiomyocyte Contractility and Calcium Handling In Vitro. Methods Mol Biol. 2018;1816:93–104. doi: 10.1007/978-1-4939-8597-5_7. [DOI] [PubMed] [Google Scholar]
- 52.Chi C, Leonard A, Knight WE, Beussman KM, Zhao Y, Cao Y, Londono P, Aune E, Trembley MA, Small EM, et al. LAMP-2B regulates human cardiomyocyte function by mediating autophagosome-lysosome fusion. Proc Natl Acad Sci U S A. 2019;116:556–565. doi: 10.1073/pnas.1808618116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiattarella GG, Altamirano F, Tong D, French KM, Villalobos E, Kim SY, Luo X, Jiang N, May HI, Wang ZV, et al. Nitrosative stress drives heart failure with preserved ejection fraction. Nature. 2019;568:351–356. doi: 10.1038/s41586-019-1100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roussel J, Champeroux P, Roy J, Richard S, Fauconnier J, Le Guennec JY, Thireau J. The Complex QT/RR Relationship in Mice. Sci Rep. 2016;6:25388. doi: 10.1038/srep25388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell GF, Jeron A, Koren G. Measurement of heart rate and Q-T interval in the conscious mouse. Am J Physiol. 1998;274:H747–51. doi: 10.1152/ajpheart.1998.274.3.H747. [DOI] [PubMed] [Google Scholar]
- 56.Larson ED, St Clair JR, Sumner WA, Bannister RA, Proenza C. Depressed pacemaker activity of sinoatrial node myocytes contributes to the age-dependent decline in maximum heart rate. Proc Natl Acad Sci U S A. 2013;110:18011–6. doi: 10.1073/pnas.1308477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shkreta L, Delannoy A, Salvetti A, Chabot B. SRSF10: an atypical splicing regulator with critical roles in stress response, organ development, and viral replication. RNA. 2021;27:1302–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source and original data are available from the corresponding author upon reasonable request.