Abstract
Natural killer (NK) cells are crucial effector cells of the innate immune response to viral infections, including HIV, through cytolytic activity and the production of cytokines with anti-HIV activities. We recruited 15 treatment naïve HIV patients and 16 healthy controls (HC) to assess NK cell subsets or expression of multiple markers by flow cytometry. The frequency of circulating CD56brightCD16−ve and CD56dimCD16bright NK cell subsets was significantly lower among the HIV group than in HC. The CD56−veCD16bright subset was higher in HIV patients, but this was only apparent when gated among total NK cells, not total lymphocytes. NK cells among HIV participants also showed a lower and higher frequency of CD8 and HLA-DR expressing cells, respectively. In addition, CD7 median fluorescent intensity and CD2+CD7− frequencies were significantly lower in HIV patients. A distinct population of KIR3DL1/S1 cells was unexpectedly higher among CD56brightCD16−ve NK cells in HIV patients. In conclusion, this study in the Ethiopian setting confirms many previous findings, but the down-regulation of CD7 and enhanced KIR3DL1/S1 within the CD56bright subsets have not been widely reported among HIV patients and merit further research.
Subject terms: Immunology, Microbiology, Infectious diseases
Introduction
An effective immune response towards invading microbes requires the combined action of innate and adaptive immunity. Natural killer (NK) cells are effector lymphocytes of the innate immune response, and lyse cells infected with the virus, including human immunodeficiency virus (HIV)1. They also participate in determining the fate of adaptive immune responses through secretion of cytokines, such as interferon γ (IFN- γ)2. NK cell function is primarily determined by germline-encoded activating and inhibitory receptors, which after integration results in NK cell activation or not3. NK cells are typically identified as CD3−CD56+ cells. They can be further classified based on the expression of CD164. The majority of peripheral blood NK cells are CD56dimCD16bright and usually cytotoxic5, whereas most of the remaining are CD56brightCD16−ve and produce more cytokines and chemokines than the former6. The CD56bright subsets are highly proliferating, and considered immature precursor subsets for CD56dim cells. The CD56dim has been proposed to undergo further irreversible differentiation, progressively losing immature inhibitory NK cell receptors, such as natural killer group 2 A (NKG2A), and begin the sequential acquisition of killer like immunoglobulin receptors (KIRs) and CD577,8.
Numerous studies have shown that HIV infection alters subsets distribution and the expression of receptors on NK cells. This includes the enhanced appearance of a functionally impaired subset, the normally infrequent CD56−veCD16bright subset, and the decrease in frequencies of the CD56brightCD16− and CD56dimCD16bright subsets6,9–11. HIV also upregulates inhibitory while downregulating activation receptors that could support disease progression12,13. Although highly active anti-retroviral therapy (HAART) restores most NK cell functions and normalizes subset distribution, incomplete recovery of subsets has been reported even after long-term HAART14. Incomplete recovery may, in turn, affect the recovery of adaptive immune cell functions, including the chronic alterations of T cells and associated immune dysfunction15.
The expression of certain markers, such as CD8 and KIR3DL1/S1 on NK cells is associated with slow HIV progression to AIDS16,17. A higher frequency of CD57+ NK cells has also been shown to be associated with better CD4 recovery and low HIV viral load18. Furthermore, activated NK cells expressing HLA-DR among HIV patients may be related to delayed recovery of CD4 count after HAART19. Adaptive or memory-like functions or phenotypes in human cytomegalovirus (HCMV) infections have been identified among NKG2C+ NK cells. Hence, NK cell phenotypes may reveal valuable information about disease prognosis, particularly among HAART naïve individuals.
While the global number of new HIV infections is declining, southern and eastern African regions comprise 47% of the global infections20. The prevalence of HIV infection among individuals aged 15–49 in Ethiopia is 0.9%21. Few comprehensive studies evaluating NK phenotype have been done in sub-Saharan Africa (SSA), and none to the best of our knowledge in Ethiopia. Given that genetic and environmental factors impacting disease may differ in these settings, our objective in the current study was to assess NK phenotypes of HIV patients and controls, and determine whether or not such findings were consistent or not with prior studies.
Results
Characteristics of the study participants
A total of 31 study participants (15 HAART naïve HIV patients and 16 healthy controls) were enrolled. The majority were female, comprising 9/15 (60%) in the HIV group and 10/16 (62.5%) in healthy controls (HC) groups. The median age of HIV and HC groups was 32 (range 20–46) and 28 (range 19–42), respectively (Table 1). The median viral load in HAART naïve HIV patients was 102,566.5 copies/ml (range 4039–257,615). All HIV patients had the disease for greater than 6 months.
Table 1.
Characteristics of HAART naïve HIV patients and healthy controls.
| Characteristics | HIV patients | Healthy controls |
|---|---|---|
| Case | 15 | 16 |
| Sex (male/female) | 6/9 | 6/10 |
| Age (median with range) | 32 (20–46) | 28 (19–43) |
| Residence (urban/rural) | 14/1 | 12/4 |
| HIV viral load level (median with range) | 102,566.5 copies/ml (4039–257,615) | N/A |
| Duration of infection at the time of diagnosis | > 6 months | N/A |
NK cells in HIV patients and healthy controls
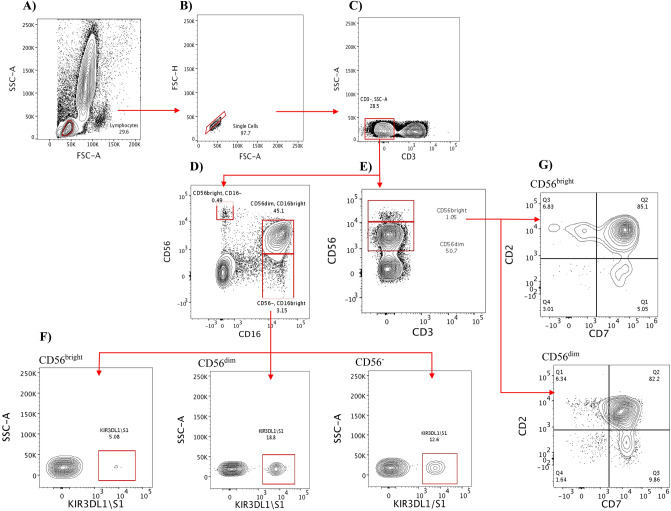
In this study, we defined NK cells by the expression of CD56 and/or CD16 among CD3 negative cells into three subsets: CD56brightCD16−ve, CD56dimCD16bright, and CD56−veCD16bright (Fig. 1). In some experiments, only CD56 was used to define CD56bright and CD56dim subsets, as described previously in the literature22. The frequency of total NK cells (Fig. 2) and the subsets (Fig. 3) were subsequently analyzed and compared across study groups.
Figure 1.
Gating strategy for NK cell subsets. (A) The lymphocytes were first identified based on forward (FSC) and side scatter (SSC) analysis; doublets were subsequently excluded with a FSC-H vs FSC-A plot (B). (C) CD3− cells were then gated, followed by the definition of the three NK cell subsets in the panels containing CD56 and CD16 (D), or two subsets in the panels without CD56 antibody alone (E). (F) The three subsets (CD56brightCD16−ve in the lower-left corner, CD56dimCD16bright in the middle, and CD56−veCD16bright in the lower right corner) were further examined for CD8 and KIR3DL1/S1 expression. (G) The expression of CD2 and CD7 among CD56brightCD16−ve (top right corner) and CD56dimCD16bright (lower right corner) is shown. The representative sample for gating illustration was taken from healthy controls.
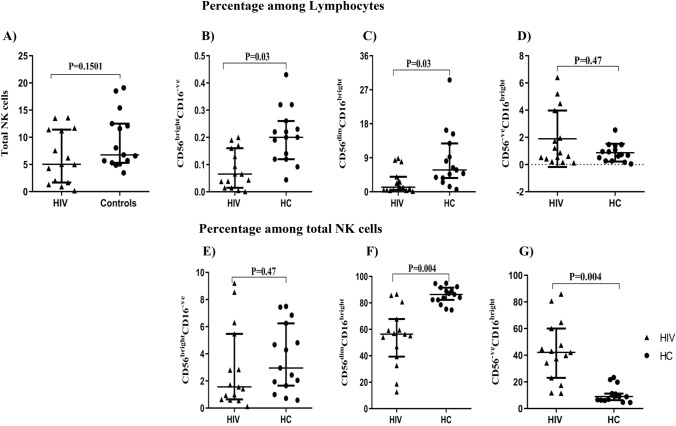
Figure 2.
Total NK cells and subset distribution among the study groups. The frequency of total NK cells is indicated in the upper left corner, and was calculated based on the sum of CD56brightCD16−ve, CD56dimCD16bright, and CD56−veCD16bright subsets (A). The percentage of NK subsets was gated among lymphocytes, the great grandparent node as defined in Fig. 1 (B–D), or total NK cells, the sum of all three NK subsets (E–G) from HIV (triangles, n = 15) or HC (circles, n = 15). Mann–Whitney tests were performed with a p-value < 0.05 considered statistically significant. The median and 25th and 75th percentile are indicated on each plot.
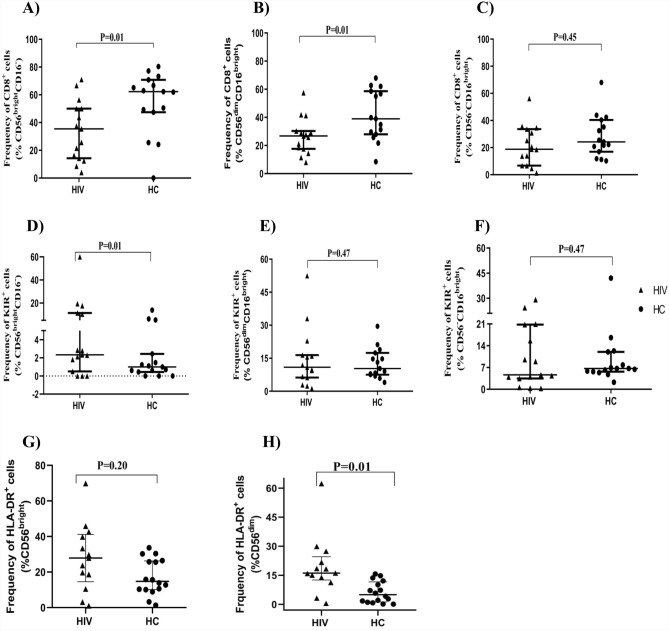
Figure 3.
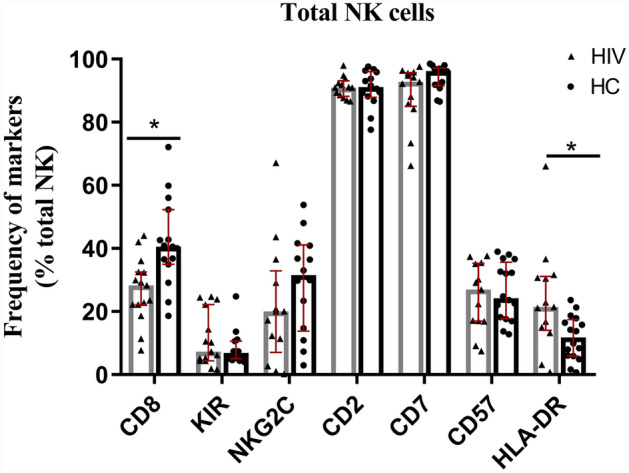
Expression of multiple markers on NK cells among the study groups. The analysis of CD8 and KIR3DL1/S1 expression was quantified on total NK cells comprising CD56brightCD16−, CD56dimCD16bright, and CD56−veCD16bright, whereas the expression of NKG2C, CD2, CD7, CD57, and HLA-DR were determined from NK cells comprising the CD56bright and CD56dim subsets. A total of 15 subjects were evaluated for HIV and 15 for HC subjects for CD8 and KIR markers; 13 (HIV) and 14 (HC) subjects for NKG2C, CD2, CD7; and 13 (HIV) and 16 (HC) subjects for CD57 and HLA-DR marker expression. Abbreviations on X-axis refer to the different NK cell markers. Mann–Whitney tests were performed with a p-value < 0.05 considered statistically significant. The median and 25th and 75th percentile are indicated on each plot.
NK cell subset distribution among the study groups
The frequency of total NK cells and subsets thereof were analyzed and compared between HC and HIV groups. We assessed NK cell subsets as a fraction of total NK cells (defined as the sum of the three subsets) or as a fraction of total lymphocytes (Fig. 2). Among lymphocytes, we observed a lower frequency of both CD56dimCD16bright and CD56brightCD16−ve in HIV compared to HC subjects (p = 0.03 and 0.03, respectively). There was no significant difference in the CD56−veCD16bright NK cell subset frequency between the groups. However, when frequencies were analyzed among total NK cells, we continued to note a significantly lower frequency in the CD56dimCD16bright subset (p = 0.004), but a significantly higher frequency in the CD56−veCD16bright subset in the HIV compared to HC groups (p = 0.004). Furthermore, among total NK cells, a strong inverse correlation was observed between CD56−veCD16bright and CD56dimCD16bright subsets (r = − 0.9296, p = 0.0001).
Expression of CD8, KIR, NKG2C, CD2, CD7, CD57, and HLA-DR on NK cells
We examined total NK cells for the expression of CD8, KIR3DL1/S1 (CD158e1/e2), CD159C (NKG2C), CD2, CD7, CD57, and HLA-DR between the healthy control (HC) and HIV groups. As shown in Fig. 3, NK cells from the HIV group had a significantly lower frequency of CD8+ cells (p = 0.001) and a higher frequency of HLA-DR+ cells (p = 0.045), relative to HC. In contrast, the expression of KIR3DL1/S1, NKG2C, CD7, and CD57 did not significantly differ between the cohorts.
The expression of NK cell receptors within the subsets
To determine the differences in the expression of different NK cell markers, we first examined the frequencies of CD8 and CD158e1/e2 (KIR3DL1/S1) expressing cells among the three subsets. We found a small percentage of KIR+ cells among the CD56brightCD16−ve subset, and this frequency was significantly higher (p = 0.01) in HIV than in HC subjects, shown in Fig. 4. Notably, KIR frequencies were higher in the other two NK subsets as well, though these differences did not reach statistical significance. The frequency of CD8+ cells was significantly lower in HIV compared with HC subjects within the CD56brightCD16−ve (p = 0.01), and CD56dimCD16bright (p = 0.01) subsets, whereas CD8 frequencies among the CD56−veCD16bright subsets were similar in the two subject cohorts (p = 0.47).
Figure 4.
Frequency of KIR3DL1/S1+, CD8+ and HLA-DR cells among NK cells subsets. The frequency of CD8+ and KIR3DL1/S1+ cells within the CD56brightCD16−ve (A,D), CD56dimCD16bright (B,E) and CD56−veCD16bright (C,F) subsets are depicted for both HIV (n = 15) and HC groups (n = 15). The frequency of HLA-DR positive cells is shown for CD56bright (G) and CD56dim (H) subsets (n = 13, HIV group, n = 16 HC group). Mann–Whitney tests were performed with a p-value < 0.05 considered statistically significant. The median and 25th and 75th percentile are indicated on each plot.
In addition, we evaluated the surface expression of CD57, HLA-DR, and NKG2C markers, focusing on the CD56bright and CD56dim NK subsets. As shown in Fig. 4, the frequency of HLA-DR+ cells on the CD56dim subset was significantly higher in the HIV group than in HC (p = 0.01). We did not detect significant differences between the HIV and control cohorts in the expression of CD57 and NKG2C among CD56bright and CD56dim (data not shown).
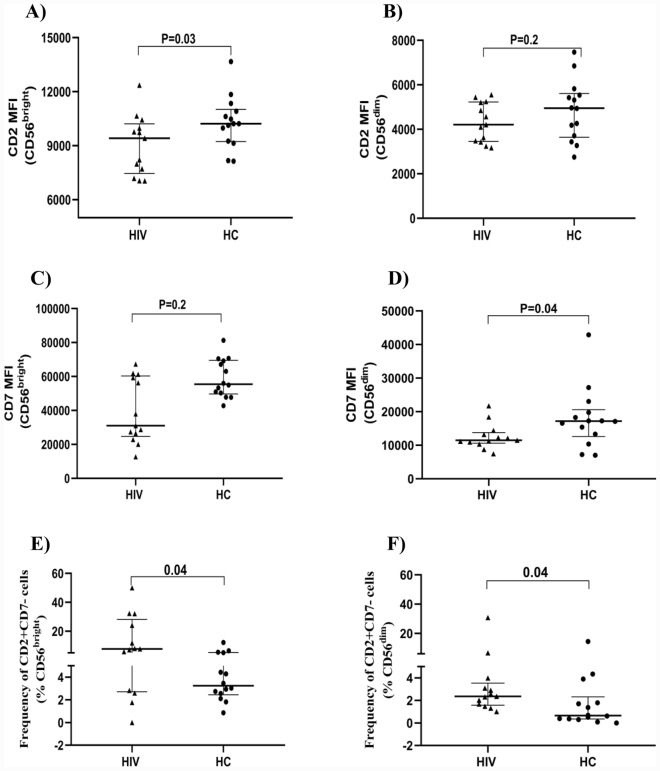
Expression of CD7 and CD2 by NK cell subsets
We further evaluated the two subsets for the expression of CD2 and CD7 (Fig. 5). CD2 is a major coactivating receptor expressed on NK that recognizes CD58, a ligand expressed on a wide variety of tissues. Most NK cells express CD7, the density of which may be related to maturation or activation11,23. We found no significant differences in the frequency of CD7 expressing NK cells between the HIV and HC study groups. However, the density of CD7, as indicated by the median fluorescent intensity, was lower in HIV patients. This was apparent in total NK cells (p = 0.02). and among the CD56dim subset the density was lower among HIV than HC (p = 0.04)). CD7 was also lower among HIV patients within the CD56bright subset, but this did not reach statistical significance (p = 0.2).
Figure 5.
Expression of CD2 and CD7 by the subsets. The dot plot graph depicts the median fluorescent intensity of CD2 and CD7 on CD56bright (A,C) and CD56dim (B,D) NK cell subsets (HIV = 13, HC, 14 subjects). The frequency of CD7−CD2+ cells among CD56bright (E) and CD56dim (F) subsets is depicted. Mann–Whitney tests were performed with a p-value < 0.05 considered statistically significant. The median and 25th and 75th percentile are indicated on each plot.
The frequency of CD2 positive cells did not differ between the two cohorts among either total NK cells, or CD56bright or CD56dim subsets. Furthermore, we did not observe differences in CD2 density between HIV or HC subjects (p = 0.3). However, the CD56brightCD16−ve subset did exhibit lower CD2 density among HIV relative to HC (p = 0.03). A significant difference between the two cohorts was not seen among CD56dim cells (p = 0.2).
Co-expression of CD7 and CD2 was evaluated on both the CD56bright and CD56dim subsets. There was a large proportion of CD7+CD2+ cells within both subsets, without differences in the two clinical cohorts. However, we observed a significantly higher frequency of CD7−CD2+ cells among both CD56bright (p = 0.04) and CD56dim (p = 0.04) subsets (Fig. 5E,F).
Correlation of NK cell subsets and molecules with viral load
We assessed whether any of the frequencies of NK subsets or molecules expressed on these subsets were associated with viral load. Neither NK cells, the three NK subsets, nor any of the molecules expressed on them showed statistically significant correlations with viral load. However, three populations did show borderline associations. NKG2C expressing NK cells were inversely correlated with viral load (r = − 0.47, p = 0.055). Similarly, CD57 NK cells were negatively associated with viral load (r = − 0.46, p = 0.08), whereas CD2+CD7− NK cells were positive correlated with viral load (r = 0.53, p = 0.06).
Discussion
NK cells represent an important component of the immune response and are associated with reduced HIV disease progression both in observational studies and in vaccine trials24–26. Since many of the studies on NK phenotype and function have been performed in developed countries and few in resource limited regions and given that local factors could impact disease, the goal of the current study was to characterize the phenotypic of NK subsets among HAART naïve HIV subjects compared with healthy controls in the Ethiopian setting. Our key findings are as follows: (1) The three previously identified NK subsets, CD56brightCD16−ve, CD56dim CD16bright and CD56−veCD16bright, were observed to significantly change according to patterns previously identified when evaluated as a fraction of total NK cells. However, the CD56−veCD16bright subset, hypothesized to expand in HIV infection, was not found to be significantly changed when expressed as a function of total lymphocytes, raising the possibility that in our cohort much of the observed effect may not be related to expansion. (2) We identified elevated levels of HLA-DR molecules on NK, particularly the CD56dimCD16bright subset confirming previous studies. Moreover, we observed down-regulation of CD7 molecules in these subsets, presumably a related effect of NK activation. (3) We confirmed other research illustrating a significant decrease in CD8 positive NK cells among HIV infected subjects. Finally, we observed a relatively high fraction of KIR3DL1 positive cells among the CD56brightCD16−ve NK cells in HIV patients, an unexpected finding given current views of the CD56brightCD16−ve as a relatively immature and KIR negative population.
The CD56−veCD16bright population has gained attention for its overexpression in HIV patients, with further studies illustrating reduced functional capacity in these cells. This has led to the conclusion that it represents an expanded population of NK cells. Initial evidence for this was obtained by demonstrating enhanced concentration of CD56−veCD16bright NK cells in the blood of HIV patients9,11,27. Many subsequent studies have assumed expansion based on the observed elevated percentage of this subset among total gated NK cells28,29. However, in this study, we observed significantly enhanced percentages of the CD56−veCD16bright cells only among gated NK cells, not among lymphocytes. This suggests an additional mechanism that may contribute to the apparent elevated frequency of CD56−veCD16bright cells, namely the depletion of other NK subsets. As in other recent studies, we did not assess NK subset number as a function of blood volume, but our findings suggest in the future this should be considered in parallel, rather than simply relying on the subset percentages of gated NK.
While it is clear that cell activation is crucial for protective immune cell function in general, and NK cell function in particular, it is well established that HIV pathogenesis is unusual in that widespread and inappropriate activation of the immune system occurs and can result in reduced function. In the context of NK cells, it is tempting to speculate that the elevated expression of HLA-DR molecules on NK cells in HIV patients reflects such a process. Indeed, some reports have indicated that frequencies of activated NK cells in HIV patients are predictive of a poor prognosis14,22,30,31. It is possible that such activation could be a prelude to the aforementioned differentiation into dysfunctional CD16+CD56−ve cells11, enhanced susceptibilities to apoptosis leading to diminished NK numbers or reduced function by HIV products such as nef32. Given the role of translocated intestinal lumen bacterial products in immune activation in HIV patients33, it is also possible that such microbial products may contribute either directly or indirectly to NK activation via monocyte/dendritic lineage cells.
CD7 has been shown to be down-regulated as a consequence of activation on both NK cells and T cells. It may be seen at depressed levels in chronic diseases, with most evidence demonstrated in cancer models, and little evidence in HIV disease34–36. In the present study we observed reduced intensity of CD7 on NK cells in HIV patients. In addition, we also observed a small population of CD7−CD2+ cells enriched in these subjects. While CD7 negative cells have been reported, others have suggested these may represent myeloid lineage cells. Further studies will be required to clarify this, but given the generalized decrease in CD7 in HIV patients, it is conceivable that these could represent NK cells with down-regulated CD7 molecules. With the demonstrated functional importance of CD737,38, our results suggest that further study of NK cells with reduced CD7 may shed further light on NK dysfunction in HIV disease.
We observed significantly depressed frequencies of CD8 expressing NK cells. This was observed equally among the CD56dimCD16bright and CD56brightCD16−ve subsets, confirming earlier studies that reported a selective loss of CD16+CD56+CD8+ or CD16+CD8+ NK cells in HIV infection17,28,39. Though not evaluated here, CD8 positive NK cells have been shown to exhibit polyfunctionality, with both high cytokine production and cytolytic capacities17, a property thought to be involved in protection by T cells against HIV progression.
KIRs are generally expressed by mature CD56dimCD16brightcells40. However, we observed a significant fraction of CD56brightCD16−ve cells which were positive for KIR3DL1/S1. To our knowledge, only one other study reported such a finding in HIV patients41. While CD56brightCD16−ve cells are relatively infrequent in blood they are more commonly expressed in tissues, and though typically KIR negative, increased KIR on decidual NK CD56brightCD16−ve cells have been reported42,43. The relationship with CD56brightCD16−ve cells in tissues with those in the blood is unclear43, but it is conceivable that under some conditions, a subset of CD56brightCD16−ve cells may undergo differentiation expressing KIR and retaining a high level of CD56 at least during the initial stages. Such a possibility would also be consistent with an in vitro study in which CD56bright NK cells activated in vitro retained the CD56bright phenotype but acquired KIR over several days in culture44. Further studies are needed to address the role of KIR on CD56brightCD16−ve cells in HIV disease.
Our study was limited by a relatively small sample size, reducing the power of the study. Several observations, in particular correlations of marker expression with HIV viral load, showed suggestive associations with borderline statistical significance. Future studies in this setting with larger sample sizes will be needed to confirm and extend these observations.
Conclusions
This is the first study to comprehensively examine the phenotypes of multiple NK subsets in Ethiopia among HIV patients. We confirmed many of the previously defined phenotypic characteristics of NK cells described in multiple studies elsewhere. Further, our results highlight the importance of gating for enumeration of the overexpressed CD56−veCD16bright NK subset, the potential importance of CD7 as activation and functional marker for NK cells, and finally the overexpression of KIR3DL1/S1 among the CD56brightCD16−ve subset among HIV patients, observations which have received little attention in previous studies.
Materials and methods
Study participants
A total of 31 study participants, 15 HAART naïve HIV patients (HIV group) and 16 healthy controls (HC) aged 18–49 years were enrolled in the study. In some experiments, a subset of these was utilized. The participants were recruited from health centers and hospitals in Addis Ababa, Ethiopia. All participants were screened for HIV by current national test algorithms and initial positives were further tested with the recency test (Asante recency test), with all patients exhibiting chronic disease > 6 months. HIV subjects were tested for viral load at it is a routinely done test, instead of CD4 T cell count. Healthy study participants were appointed to come back after a month, at which point the HIV test was repeated to confirm negativity. None of the participants had conditions such as tuberculosis, diabetes mellitus, pregnancy or were on treatment with immune suppressive medication (Supplementary Table S1).
Staining of cell surface antigens and flow cytometric acquisition
Whole blood samples were collected from each participant by venous puncture, and whole blood staining was performed. Briefly, 100 μl of blood was transferred to each tube containing a mixture of antibodies labeled as: panel-1 with CD3-APC-Cy7 (Becton-Dickinson (BD), USA), CD16-FITC (BD, USA), CD56-PE (BD, USA), KIR3DL1/S1-APC (Beckman coulter), and CD8-PerCP (BD, USA), panel-2 with CD3-APC-Cy7, CD56-PE and NKG2C-Alexa488 (R&D, England), panel-3 with CD3-APC-cy7, CD56-PE, CD2-FITIC (Becton-Dickinson, USA) and CD7-PerCP (BD, USA), panel-4 with CD3-APC-cy7, CD56-PE, CD57-FITC (BD, USA), HLA-DR-PERCP (BD, USA) and CD19-APC (BD, USA), panel-5 with no antibody (unstained tube). The tubes were then incubated for 20 min in the dark at 4 °C. Red blood cells (RBCs) were lysed with 1 ml of BD FACS lysing solution for 10 min at room temperature, and washed with addition of 3 ml of FACS buffer (phosphate buffer saline 0.1 mM EDTA and 0.1% fetal bovine serum), centrifugation at 400×g for 7 min, and resuspended with 400 µl of FACS buffer. Compensation bead containing tubes were prepared with each fluorochrome and used during flow cytometer set up. 100,000 events were acquired from all tubes except panel-5 (20,000 events only), using a FACS Canto II flow cytometer equipped with FACSDiva software. In some experiments for some samples staining was poor resulting in sample exclusion from analysis, resulting in sample numbers for a given marker set less than the maximum 15 HIV and 16 HC subjects. Sample numbers are indicated in the figure legends.
FlowJo analysis
FlowJo (BD) software was used for analysis with prepared templates, incorporating gates adjusted for each sample. Lymphocytes were gated based on forward scatter (FSC) and side scatter (SSC) and singlets were subsequently selected using an FSC-height (FSC-H) versus FSC-area (SSC-A) plot. After gating on CD3 negative cells, NK cells were then identified by the expression CD16 and/or CD56 as depicted in Fig. 1. Percentages of individual NK subsets (CD56brightCD16−ve, CD56dimCD16bright and CD56−veCD16bright) were defined among total NK cells defined as the sum of the three subsets. NK subsets were defined also as a percentage of lymphocytes as the great-grandparent node using FlowJo software. This figure is mathematically identical to 100 times the product of the fraction of singlets among gated lymphocytes, the fraction of CD3− cells among gated singlets, and the fraction of the NK subset of interest. The three NK subsets were further analyzed for expression of CD8 and KIR3DL1/S1 using panel-1. However, for the rest of the panels, only CD56dim and CD56bright subsets were identified as depicted in Fig. 1 and analyzed for NKG2C, CD8, CD7, CD2, CD57, and HLA-DR.
Data analysis
Data obtained from FlowJo Version 10.1 software and the viral load data from the Abbott machine were exported into JMP software (SAS, Cary, NC) after importation of an intermediate Excel (Microsoft) file for statistical analysis. Cell populations of interest were compared according to the study cohort as independent variables using the Mann–Whitney U test with p-values < 0.05 considered statistically significant. Spearman’s correlation was done to examine the correlation between NK subset frequencies and HIV viral load for HIV-infected participants.
Ethical approval and informed consent
Participation in this study was voluntary and the study was approved by both the ethical review board at the University of Gondar, School of Biomedical and Laboratory science and Armauer Hansen Research Institute All Africa Leprosy, Tuberculosis, and Rehabilitation Training Center Ethics Review Committee (AERC), protocol number P006/18. Written informed consent was obtained from the study participants before involvement in the study. The AERC approved the study of the patient material collected. All experiments were performed in accordance with relevant guidelines and regulations.
Supplementary Information
Acknowledgements
We thank Ms. Wegene for providing us HLA-DR antibodies whenever necessary and all TB research unity staff at the Armauer Hansen Research Institute (AHRI) for assisting us with the staining. Finally, we would like to forward our heartfelt thanks to University of Gondar for supporting HA, AHRI for sponsoring the project, and DTU for sponsoring the M.Sc. program at the University of Gondar.
Abbreviations
- CD
Cluster of differentiation
- CLR
C-type lecithin receptor
- DC
Dendritic cells
- FACS
Fluorescent activated cell sorter
- FSC
Forward scatter
- HAART
Highly active anti-retroviral treatment
- HIV
Human immune deficiency virus
- HLA
Human leukocyte antigens
- Ig
Immunoglobulin
- KIR
Killer like immunoglobulin receptor
- NK
Natural killer cells
- SSC
Side scatter
- AIDS
Acquired immune deficiency syndrome
- N/A
Not applicable
Author contributions
H.A. performed the flow cytometry experiment, analyzed the data, and wrote the manuscript. R.H. designed the experiment and assisted in the analysis and preparation of the manuscript. M.L., A.K., M.W., M.Z., and A.M. have been involved analysis of the data and manuscript preparation. B.A., Y.T., L.H. and A.G. are involved in the data collection, laboratory analysis, and writing of the manuscript. All authors read and approved the final manuscript. All authors have approved the manuscript for publication.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-18413-3.
References
- 1.Scully E, Alter G. NK cells in HIV disease. Curr. HIV/AIDS Rep. 2016;13(2):85–94. doi: 10.1007/s11904-016-0310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luetke-Eversloh M, Killig M, Romagnani C. Signatures of human NK cell development and terminal differentiation. Front. Immunol. 2013;4:499. doi: 10.3389/fimmu.2013.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405(6786):537. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 4.Michel T, Poli A, Cuapio A, et al. Human CD56bright NK cells: An update. J. Immunol. 2016;196(7):2923–2931. doi: 10.4049/jimmunol.1502570. [DOI] [PubMed] [Google Scholar]
- 5.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkström NK, Ljunggren H-G, Sandberg JK. CD56 negative NK cells: Origin, function, and role in chronic viral disease. Trends Immunol. 2010;31(11):401–406. doi: 10.1016/j.it.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: Correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS One. 2010;5(8):e11966. doi: 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Björkström NK, Riese P, Heuts F, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood J. Am. Soc. Hematol. 2010;116(19):3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 9.Hong HS, Eberhard JM, Keudel P, et al. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J. Virol. 2010;84(2):1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alter G, Teigen N, Davis BT, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106(10):3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 11.Milush JM, López-Vergès S, York VA, et al. CD56 neg CD16+ NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology. 2013;10(1):1–13. doi: 10.1186/1742-4690-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kottilil S, Shin K, Planta M, et al. Expression of chemokine and inhibitory receptors on natural killer cells: Effect of immune activation and HIV viremia. J. Infect. Dis. 2004;189(7):1193–1198. doi: 10.1086/382090. [DOI] [PubMed] [Google Scholar]
- 13.Mavilio D, Benjamin J, Daucher M, et al. Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl. Acad. Sci. 2003;100(25):15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nabatanzi R, Bayigga L, Cose S, et al. Aberrant natural killer (NK) cell activation and dysfunction among ART-treated HIV-infected adults in an African cohort. Clin. Immunol. 2019;201:55–60. doi: 10.1016/j.clim.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nabatanzi R, Bayigga L, Ssinabulya I, et al. Low antigen-specific CD4 T-cell immune responses despite normal absolute CD4 counts after long-term antiretroviral therapy an African cohort. Immunol. Lett. 2014;162(2):264–272. doi: 10.1016/j.imlet.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alter G, Altfeld M. NK cells in HIV-1 infection: Evidence for their role in the control of HIV-1 infection. J. Intern. Med. 2009;265(1):29–42. doi: 10.1111/j.1365-2796.2008.02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad F, Hong HS, Jäckel M, et al. High frequencies of polyfunctional CD8+ NK cells in chronic HIV-1 infection are associated with slower disease progression. J. Virol. 2014;88(21):12397–12408. doi: 10.1128/JVI.01420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gondois-Rey F, Chéret A, Mallet F, et al. A mature NK profile at the time of HIV primary infection is associated with an early response to cART. Front. Immunol. 2017;8:54. doi: 10.3389/fimmu.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erokhina SA, Streltsova MA, Kanevskiy LM, Telford WG, Sapozhnikov AM, Kovalenko EI. HLA-DR+ NK cells are mostly characterized by less mature phenotype and high functional activity. Immunol. Cell Biol. 2018;96(2):212–228. doi: 10.1111/imcb.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Joint United Nations Programme on HIV/AIDS (UNAIDS). UNAIDS data (2019). [PubMed]
- 21.Federal HIV/AIDS Prevention and Control Office. HIV Prevention in Ethiopia National Road Map 2018–2020 (2018).
- 22.Slyker JA, Lohman-Payne BL, John-Stewart G, et al. The impact of HIV-1 infection and exposure on natural killer (NK) cell phenotype in Kenyan infants during the first year of life. Front. Immunol. 2012;3:399. doi: 10.3389/fimmu.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milush JM, Long BR, Snyder-Cappione JE, et al. Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood J. Am. Soc. Hematol. 2009;114(23):4823–4831. doi: 10.1182/blood-2009-04-216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings J-S, Moreno-Nieves UY, Arnold V, et al. Natural killer cell responses to dendritic cells infected by the ANRS HIV-1 vaccine candidate, MVAHIV. Vaccine. 2014;32(43):5577–5584. doi: 10.1016/j.vaccine.2014.07.094. [DOI] [PubMed] [Google Scholar]
- 25.Martin MP, Gao X, Lee J-H, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 26.Taborda NA, Hernández JC, Lajoie J, et al. Low expression of activation and inhibitory molecules on NK cells and CD4+ T cells is associated with viral control. AIDS Res. Hum. Retroviruses. 2015;31(6):636–640. doi: 10.1089/aid.2014.0325. [DOI] [PubMed] [Google Scholar]
- 27.Mavilio D, Lombardo G, Benjamin J, et al. Characterization of CD56–/CD16+ natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc. Natl. Acad. Sci. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucia B, Jennings C, Cauda R, Ortona L, Landay AL. Evidence of a selective depletion of a CD16+ CD56+ CD8+ natural killer cell subset during HIV infection. Cytometry. 1995;22(1):10–15. doi: 10.1002/cyto.990220103. [DOI] [PubMed] [Google Scholar]
- 29.Hu P-F, Hultin LE, Hultin P, et al. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+ CD56+ cells and expansion of a population of CD16dimCD56-cells with low lytic activity. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;10(3):331–340. [PubMed] [Google Scholar]
- 30.Kuri-Cervantes L, de Oca GSM, Ávila-Ríos S, Hernández-Juan R, Reyes-Terán G. Activation of NK cells is associated with HIV-1 disease progression. J. Leukoc. Biol. 2014;96(1):7–16. doi: 10.1189/jlb.0913514. [DOI] [PubMed] [Google Scholar]
- 31.Naluyima P, Eller MA, Laeyendecker O, et al. Impaired natural killer cell responses are associated with loss of the highly activated NKG2A+ CD57+ CD56dim subset in HIV-1 subtype D infection in Uganda. AIDS (London, England). 2014;28(9):1273. doi: 10.1097/QAD.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quaranta MG, Napolitano A, Sanchez M, Giordani L, Mattioli B, Viora M. HIV-1 Nef impairs the dynamic of DC/NK crosstalk: Different outcome of CD56dim and CD56bright NK cell subsets. FASEB J. 2007;21(10):2323–2334. doi: 10.1096/fj.06-7883com. [DOI] [PubMed] [Google Scholar]
- 33.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima M, Almeida J, dos Anjos TM, Queirós ML, Justiça B, Orfao A. The, “ex vivo” patterns of CD2/CD7, CD57/CD11c, CD38/CD11b, CD45RA/CD45RO, and CD11a/HLA-DR expression identify acute/early and chronic/late NK-cell activation states. Blood Cells Mol. Dis. 2002;28(2):181–190. doi: 10.1006/bcmd.2002.0506. [DOI] [PubMed] [Google Scholar]
- 35.Yoo E-H, Kim H-J, Lee S-T, Kim W-S, Kim S-H. Frequent CD7 antigen loss in aggressive natural killer-cell leukemia: A useful diagnostic marker. Korean J. Lab. Med. 2009;29(6):491–496. doi: 10.3343/kjlm.2009.29.6.491. [DOI] [PubMed] [Google Scholar]
- 36.Ginaldi L, De Martinis M, D’Ostilio A, Di Gennaro A, Marini L, Quaglino D. Altered lymphocyte antigen expressions in HIV infection: A study by quantitative flow cytometry. Am. J. Clin. Pathol. 1997;108(5):585–592. doi: 10.1093/ajcp/108.5.585. [DOI] [PubMed] [Google Scholar]
- 37.Lee DM, Staats HF, Sundy JS, et al. Immunologic characterization of CD7-deficient mice. J. Immunol. 1998;160(12):5749–5756. [PubMed] [Google Scholar]
- 38.Rabinowich H, Pricop L, Herberman RB, Whiteside TL. Expression and function of CD7 molecule on human natural killer cells. J. Immunol. 1994;152(2):517–526. [PubMed] [Google Scholar]
- 39.Mansour I, Doinel C, Rouger P. CD16+ NK cells decrease in all stages of HIV infection through a selective depletion of the CD16+ CD8+ CD3-subset. AIDS Res. Hum. Retroviruses. 1990;6(12):1451–1457. doi: 10.1089/aid.1990.6.1451. [DOI] [PubMed] [Google Scholar]
- 40.Pende D, Falco M, Vitale M, et al. Killer Ig-like receptors (KIRs): Their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantegani P, Tambussi G, Galli L, Din CT, Lazzarin A, Fortis C. Perturbation of the natural killer cell compartment during primary human immunodeficiency virus 1 infection primarily involving the CD56bright subset. Immunology. 2010;129(2):220–233. doi: 10.1111/j.1365-2567.2009.03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma S, King A, Loke YW. Expression of killer cell inhibitory receptors on human uterine natural killer cells. Eur. J. Immunol. 1997;27(4):979–983. doi: 10.1002/eji.1830270426. [DOI] [PubMed] [Google Scholar]
- 43.Jabrane-Ferrat N. Features of human decidual NK cells in healthy pregnancy and during viral infection. Front. Immunol. 2019;10:1397. doi: 10.3389/fimmu.2019.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romagnani C, Juelke K, Falco M, et al. CD56brightCD16− killer Ig-like receptor− NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J. Immunol. 2007;178(8):4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.