Abstract
In Eubacteria, expression of genes transcribed by an RNA polymerase holoenzyme containing the alternate sigma factor ς54 is positively regulated by proteins belonging to the family of enhancer-binding proteins (EBPs). These proteins bind to upstream activation sequences and are required for the initiation of transcription at the ς54-dependent promoters. They are typically inactive until modified in their N-terminal regulatory domain either by specific phosphorylation or by the binding of a small effector molecule. EBPs lacking this domain, such as the PspF activator of the ς54-dependent pspA promoter, are constitutively active. We describe here the in vivo and in vitro properties of the PspA protein of Escherichia coli, which negatively regulates expression of the pspA promoter without binding DNA directly.
Infection of Escherichia coli with filamentous bacteriophage f1 results in the strong and specific induction of the pspABCDE operon. This operon contains five open reading frames, of which at least four (pspA, -B, -C, and -E) code for expressed proteins. Following cloning of the pspABCDE operon, deletion analysis suggested that the pspA gene encoded a protein which negatively regulated expression of the operon (7). Later experiments demonstrated that the PspA protein, when transcribed from a heterologous promoter, was sufficient to negatively regulate expression of the operon in trans and that the absence of full-length PspA in vivo resulted in constitutive high-level expression of a mutant, truncated PspA protein, independent of the presence of any other psp proteins (44). Further, the inability of a frameshifted mutant PspA to inhibit psp expression indicates that it is the protein that is responsible for inhibition.
Transcription of the psp operon is dependent on an RNA polymerase (RNAP) holoenzyme containing the alternate sigma factor ς54 (13, 44). Like that of other ς54-dependent genes, transcription of pspA requires activation by a protein, in this case PspF, which belongs to the family of enhancer-binding proteins (EBPs) (13, 22). Through an ATP hydrolysis-dependent mechanism, these proteins convert the closed complex formed by ς54 and RNAP at the promoter into an open complex permissive for initiation (28). This conversion is the result of DNA loop-mediated, protein-protein contacts between the EBP and the β and ς54 subunits of the RNAP holoenzyme (31). Typically, EBPs are inactive until they are modified in their N-terminal domain either by binding an effector molecule (e.g., xylene for XylR [15]) or through a specific phosphorylation event (e.g., phosphorylation of Asp54 of NRI by NRII [37]). By contrast, PspF lacks this entire domain (as do the HrpR and HrpS proteins of Pseudomonas syringae [48]) and is constitutively active both in vivo and in vitro (22). In addition, PspF autoregulates its own expression by binding to sites overlapping its promoter, and its levels are constant in the presence or absence of inducing stimuli (19). Thus, regulation of pspA transcription cannot occur through the EBP modification pathway used by other ς54-dependent systems. Since the in vivo analysis of PspA demonstrated that it is required for this negative regulation, we studied the action of PspA in vitro.
The pspA gene encodes the 25.5-kDa PspA protein that, according to Chou-Fasman analysis (10), contains four long α-helices. Analysis of the protein sequence with the Macstripe program (26), based on the Lupas algorithm (34), strongly predicts that these helices will form a coiled coil comprising nearly the entire length of the protein. Proteins with coiled-coiled regions as extensive as that of PspA are relatively unusual in prokaryotes, with the TlpA protein of Salmonella enterica serovar Typhimurium providing one notable counterexample (27). While PspA does not contain sequences characteristic of integral inner membrane proteins (12), approximately 50% of the total cellular PspA is associated with the inner membrane of E. coli, and PspA is thus considered a peripheral membrane protein (6). The lack of any obvious DNA-binding motif and its acidic pI (5.56) suggests that PspA is not likely to bind DNA.
Three homologs of PspA have been identified: the SCYCSLRD protein from the cyanobacterium Synechocystis sp. strain PCC6803 (23), the cold-shock-induced PspB protein from Bacillus subtilis (16), and the IM30 protein of pea chloroplasts (33). The IM30 protein is localized to both of the envelope membranes and the thylakoid membrane of the chloroplast; however, little else is known about it or any of the other PspA homologs.
In addition to its roles in psp regulation, PspA appears to participate in several aspects of cellular physiology. PspA is a major component of the limited protein synthesis that occurs in late stationary phase, and cells which lack pspA have reduced viability under alkaline conditions as well as in late stationary phase (45). PspA also appears to aid in the maintenance of the proton motive force under stress conditions (25) and can stimulate the export of secreted proteins (24). Whether these seemingly disparate phenotypes reflect a common role for PspA remains unclear. We chose, however, to focus on the mechanism of PspA autoregulation, and we describe here experiments directed at elucidating this mechanism.
MATERIALS AND METHODS
Plasmid and bacterial strain construction.
Bacterial strains and plasmids used are listed in Table 1. Plasmid pJD40 was constructed with primers JD88 (5′-GGCTCTGCAGAGATCTGATTGAAGAATCAACA) and JD90 (5′-GGCTGAATTCACCTTAACTTAATGATTTTTAC) in a PCR with Pfu polymerase (Stratagene) and pMJ3 (22) as the template. The PCR product was digested with PstI and EcoRI and ligated into the PstI and EcoRI sites of pBR322. pJD42 was constructed with primers JD54 (5′-GGCTGGTACCTAGCGAGTTCATCAAGAAATA) and JD87 (5′-GGCTAAGCTTCGGAATAGCCAGAAATAGCG) in a PCR with Pfu polymerase and pBRPS-1 (7) as the template. The PCR product was digested with BglII and HindIII and ligated into the BamHI and HindIII sites of pGZ119EH (32). pJD23 was generated with primers JD91 (5′-GGCTGTCGACCGGTATTTTTTCTCGCTTTGC) and JD87 in a PCR with Pfu polymerase and pJD42 as the template. The PCR product was digested with SalI and ScaI and ligated to the SalI and EcoRV sites of plasmid pJH391 (18). pJD25 was generated by ligating the 1.3-kb PstI fragment containing the cat gene from pSKS114 into the PstI site of pJD23. pJD26 was constructed by ligating the same 1.3-kb PstI fragment into the PstI site of pJLB24. pJD43 was constructed by using primers JD76 (5′-GGCTACGCATATGGGTATTTTTTCTCGCTTTGCC) and JD78 (5′-GGCTGGATCCTTATTGATTGTCTTGCTTCATTTT) in a PCR with Pfu polymerase and pPS-1 (7) as the template. The PCR product was digested with NdeI and BamHI and ligated to pET15b (Novagen) digested with NdeI and BamHI. pJD31 was constructed with primers JD50 (5′-GGCTGAATTCTAGCGAGTTCATCAAGAAATA) and JD51 (5′-GGCTGGATCCAATGTTGTCCTCTTGATTTCT) in a PCR with Taq polymerase and pPS-1 as the template. The PCR product was digested with BamHI and EcoRI and ligated to plasmid pRS415 (43) digested with BamHI and EcoRI. pJD42 was constructed with primers JD54 and JD87 in a PCR with Pfu polymerase and pBRPS-1 (7) as the template. The PCR product was digested with BglII and HindIII and ligated to pGZ119EH digested with BamHI and HindIII. pJD45 was constructed by digesting pBRPS-1 with EcoRV and SphI and ligating the blunt ends, which resulted in the loss of the BamHI site. The plasmid was then digested with SnaBI, which removed a fragment containing the pspF, -A, -B, and -C genes, and a BamHI linker was inserted. The BamHI fragment of pSKS101 (9) containing the kan gene was then inserted into the BamHI site created by the linker.
TABLE 1.
Bacterial strains, plasmids, and phages
| Strain, plasmid, or phage | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| JD50 | K1342 Tets | This study |
| JD54 | JD50 ΔpspABC::kan | This study |
| JD59 | JD50 pspF877::tet | This study |
| JD61 | MC4100F+ pspFABC::kan λJH202 | This study |
| JH372 | MC1061 F′ 128 lacIqlacZ::Tn5 | 18 |
| K1342 | fadL701 phoM510 mcrB rrnB | Lab collection |
| tonA22 ompF relA1 pit-10 spoT1 lacIqlacZΔM15 zah::Tn10 | ||
| K1462 | C600 λDE3 | R. Webster |
| MC4100F+ | araD lacΔU169 relA thi rpsL (Strpr) | Lab collection |
| Plasmids | ||
| pFC50 | glnH promoter | 11 |
| pJD10 | pspA wt promoter | 13 |
| pJD12 | pspA ΔUASI and II promoters | 13 |
| pJD23 | pspA-λ repressor (1–102) in pJH391 | This study |
| pJD25 | pJD23 Cmr | This study |
| pJD26 | pJLB24 Cmr | This study |
| pJD31 | pspA-lacZ gene fusion | This study |
| pJD40 | pspF877 under the control of pspFp | This study |
| pJD42 | lac-pspA Cmr | This study |
| pJD43 | his6-pspA fusion, Ampr | This study |
| pJD45 | pspFABC::kan Ampr | This study |
| pJH137 | wt λ repressor Ampr | 18 |
| pJH370 | λ repressor-GCN4 Ampr | 18 |
| pJH391 | λ repressor fusion vector | 18 |
| pJLB24 | lac-pspA Ampr | 44 |
| pMJ3 | pspF::miniTn10-tet | 22 |
| pBRPS-1 | psp operon in pBR322 | 7 |
| Phages | ||
| λJH202 | λ pR-lacZ | 18 |
| λpsp3 | pspA-lacZ gene fusion | This study |
wt, wild type. pspA-λ repressor (1–102) refers to the fusion of the N-terminal domain of the λ repressor to the entire pspA sequence.
The λpsp3 phage that carries a pspA-lacZ fusion was generated by growing the λB305 phage (3) that contains the carboxy-terminal coding sequences of the lacZ and bla genes on a strain carrying the Ampr plasmid pJD31. Transfer of pspA-lacZ to the phage was signaled by the reconstitution of the functional lacZ and bla genes. Strains were made lysogenic for λpsp3 by conventional methods (42).
Strain JD50 was selected from K1342 (lacIq lacZΔM15 zah::Tn10 Tets) by use of the fusaric acid technique (5). P1 transduction from strain J134 (44) (pspABC::kan) into JD50 yielded JD54. JD59 was generated by P1 transduction from strain K1527 (22) (pspF877::tet). The ΔpspFABC mutation was introduced by homologous recombination (46) into strain MC4100F+, yielding strain JD61. Plasmid pJD45 was linearized with NcoI and transformed into strain JC7623, a recB recC sbcB mutant. DNA made from a Kanr transformant was examined by Taq polymerase-based PCR with the primers JABR6 and IR1070Bam (22), which are complementary to sequences in the pspA and pspF genes, respectively. Additionally, transduction of this mutation into a strain containing the λpsp3 lysogen resulted in a severe reduction in pspA-lacZ expression (data not shown). Plasmid pJD41 containing the pspF gene was transformed into this strain, which resulted in β-galactosidase levels higher than that of a wild-type strain, suggesting that PspA was absent (data not shown). Transformation of a second plasmid, pJD42, containing pspA under lac control reduced this increased level of pspA-lacZ expression, thus confirming that the strain lacked both the pspA and pspF genes (data not shown).
β-Galactosidase assay.
All measurements of β-galactosidase activity were conducted according to the established protocol (35).
In vitro transcription assay.
The protocol used for in vitro transcription was as described previously (13), unless explicitly noted in the figure legends. Purification of PspF and PspF with a deletion of the helix-turn-helix (PspFΔHTH) was as described previously (21).
Purification of PspA.
Strain K1462 (λDE3)/pJD43 was grown at 37°C with aeration in 250 ml of FB (per liter, 25 g of tryptone [Difco], 7.5 g of yeast extract [Difco], 6 g of NaCl, 1 g of glucose, 50 ml of 1 M Tris-Cl [pH 7.6]) from a dilution of 5 ml of a culture grown overnight in FB until an optical density at 660 nm of 0.4 was reached. Then, 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added and the cells were allowed to grow 5 h more at 37°C with high aeration. The cells were chilled and centrifuged at 5,000 × g for 10 min at 4°C. The supernatant was removed, and the pellet was either resuspended as described below or stored at −20°C.
All subsequent steps were done at 0 to 4°C. The pellet was resuspended in 2.5 ml of cold sonication buffer (100 mM Tris-Cl [pH 7.5], 50 mM NaCl). The suspension was sonicated (three times, 20 s at setting 60, Sonicator cell disruptor) and was then centrifuged for 8 min at 15,000 × g. The supernatant was removed, and the pellet was homogenized and resuspended in 3 ml of buffer A (100 mM Tris-Cl [pH 7.5], 300 mM NaCl), to which was added 0.2 ml of a freshly made 20% solution of CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (in glass-distilled water; Sigma) and 0.2 ml of 5 M NaCl, giving final concentrations of 1.1% CHAPS and 0.6 M NaCl. The suspension was rocked on a Nutator platform for 120 min and then centrifuged at 15,000 × g for 6 min. The supernatant was removed and added to 0.5 ml of Talon resin (Clontech) which had been washed twice with 5 ml of buffer A. The mixture was nutated for 120 min and then placed into a 5-ml gravity column (Clontech) and allowed to settle for 30 min. The supernatant was allowed to flow through, and the column was washed with 5 ml of a solution containing 100 mM Tris-Cl [pH 7.5], 300 mM NaCl, and 10 mM imidazole. Protein was eluted with fractions of 100 mM Tris-Cl (pH 7.5)–60 mM NaCl–100 mM imidazole. Individual fractions were dialyzed against three changes (350 ml each) of 20 mM Tris-Cl (pH 7.5)–60 mM NaCl, glycerol was added to 20%, and the fractions were stored at −70°C.
Purification was assessed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (29), followed by staining with Coomassie brilliant blue as described previously (41). Protein concentrations were determined with a DC protein assay kit (Bio-Rad).
Gel mobility shift assay.
The 260 fragment (260 bp) contains the entire psp promoter region, including sequences from −188 to +72 relative to the start site of pspA transcription (20). The gel mobility shift assay using either cell extract from a strain overproducing PspF or purified PspA protein at specified concentrations was performed with 2 ng of the 260 fragment as described previously (20).
RESULTS
PspA acts in vivo as a negative regulator of pspA transcription.
Induction of the psp operon is accompanied by an increase in the amount of pspABCDE-specific mRNA (7), but neither that study nor the subsequent in vivo study of PspA (44) demonstrated that PspA acts directly on transcription. All previous assays of PspA function had relied on measurement of PspA protein levels and thus could not exclude posttranscriptional regulation. We fused the pspA promoter with the lacZ gene (with the lac ribosome-binding site) carried on a lambda lysogen to assay pspA promoter activity. In a wild-type psp+ strain, lacZ expression was quite modest from the pspA promoter, but in a ΔpspABC deletion mutant, there was a 50-fold increase in lacZ expression (Table 2). Production of PspA from a plasmid containing pspA under lac control repressed expression of the fusion (Table 2) but had no effect on expression of lacZ under the control of its own promoter (data not shown). This experiment shows that PspA is sufficient to inhibit pspA transcription. Further, since the fusion contains psp-specific sequences only up to +30 relative to the start site of transcription (and thus lacks the pspA ribosome-binding site and the pspA gene), the negative regulation does not require downstream sequences.
TABLE 2.
In vivo analysis of PspA functiona
| Strain/plasmid | Chromosome | Protein(s) of plasmidborne gene | β-Galactosidase activity (Miller units) |
|---|---|---|---|
| JD50λpsp3 | wt | None | 45 |
| JD54λpsp3 | ΔpspABC | None | 2,011 |
| JD50λpsp3/pJD26 | ΔpspABC | PspA | 12 |
| JD59λpsp3 | pspF877 | None | 8 |
| MC4100λpsp3 | wt | None | 80 |
| JD61λpsp3 | ΔpspFABC | None | 11 |
| JD61λpsp3/pJD40 | ΔpspFABC | PspFΔHTH | 231 |
| JD61λpsp3/pJD42 | ΔpspFABC | PspA | 12 |
| JD61λpsp3/pJD40/pJD42 | ΔpspFABC | PspFΔHTH, PspA | 32 |
The results of in vivo experiments showing PspA to be a negative regulator of pspA transcription. β-Galactosidase assays were performed as described in Materials and Methods. Values are means of results from at least three experiments, with a standard deviation of <15% in all cases. PspA expression from plasmid pJD26 was under Plac control; induction was for 90 min with 1 mM IPTG. Expression of PspFΔHTH was under the control of its own, constitutive promoter. wt, wild type.
We then addressed the issue of whether PspA affects the ability of PspF to bind DNA or whether PspA might act by affecting the DNA geometry of the pspA promoter region as does the Nac protein of Klebsiella aerogenes at the nac promoter (14). To test this possibility, the effect of PspA on PspFΔHTH was examined. This protein, encoded by pspF877, lacks the C-terminal 31 amino acids of PspF that comprise nearly the entire helix-turn-helix motif thought to constitute the DNA-binding domain. While it does not bind DNA (21), PspFΔHTH can still activate pspA transcription when present at high concentrations (13). pspA-lacZ expression in a pspF877 strain was reduced to almost background levels (Table 2); this reduction may simply have been the result of the low level of activation by the PspFΔHTH protein in single copy or may reflect both the weakened transcriptional activation by PspFΔHTH and repression by PspA.
To distinguish between these two possibilities, a plasmid containing the pspF877(PspFΔHTH) gene (under the control of its native promoter) was introduced into a strain containing a pspFABC deletion but lacking such a plasmid. In this strain, lacZ expression from the pspA-lacZ construct was very low (Table 2) and was not further reduced by introduction of a plasmid expressing PspA protein under lac control. Introduction of a plasmid bearing pspF877 into the ΔpspFABC strain increased lacZ expression 20-fold (Table 2), and this increase was repressed by the presence and induction of a compatible plasmid containing pspA under lac control (Table 2). Thus, PspA negatively regulates transcription even when the activator does not bind DNA.
PspA acts as a dimer in vivo.
Given that coiled-coiled proteins form dimers or higher-order oligomers (1), we asked whether this was true of PspA. A system developed to assay the sequence requirements for the leucine zipper of the Saccharomyces cerevisiae transcriptional activator GCN4 offers a useful method to assay protein dimerization in vivo (18). The system takes advantage of the observation that the N-terminal DNA-binding domain of the λ repressor dimerizes inefficiently and requires a separate C-terminal dimerization domain in order to bind to its operator and effect repression. We constructed a fusion of this N-terminal domain to PspA and tested its ability to repress a λ pR-lacZ fusion carried on a lambda lysogen.
The background expression of the fusion was repressed by either the wild-type λ repressor or the λ repressor-GCN4 fusion (Table 3). While PspA alone had no effect on expression of the fusion, the λ repressor-PspA fusion repressed strongly (Table 3). This result suggests that PspA is able to mediate dimerization of two λ repressor N-terminal domains through the formation of a PspA homodimer. Additionally, analysis of purified PspA on nondenaturing gels suggests that it forms dimers (and perhaps higher-order multimers [G. Jovanovic, unpublished data]). We also asked whether the λ repressor-PspA fusion could negatively regulate expression from the pspA (as distinct from the λ pR) promoter. Use of a strain carrying the pspA-lacZ promoter fusion together with the ΔpspABC deletion showed that the λ repressor-PspA fusion negatively regulates pspA transcription, albeit slightly less effectively than wild-type PspA (Table 3). Thus, even though PspA is fused to this heterologous DNA-binding domain, it remains active.
TABLE 3.
Analysis of dimerization of PspAa
| Strain/plasmid | Reporter | Protein | β-Galactosidase activity (Miller units) | % Repression |
|---|---|---|---|---|
| JH372λJH202 | λ pR-lacZ | None | 2,286 | 0 |
| JH372λJH202/pJH137 | λ pR-lacZ | λ repressor | 67 | 97 |
| JH372λJH202/pJH370 | λ pR-lacZ | λ repressor- GCN4 | 163 | 93 |
| JH372λJH202/pJLB24 | λ pR-lacZ | PspA | 2,299 | 0 |
| JH372λJH202/pJD23 | λ pR-lacZ | λrepressor- PspA | 172 | 92 |
| JD54λpsp3 | pspA-lacZ | None | 2,430 | 0 |
| JD54λpsp3/pJD26 | pspA-lacZ | PspA wt | 8 | 99.7 |
| JD54λpsp3/pJD25 | pspA-lacZ | λ repressor- PspA | 164 | 93 |
Results of in vivo experiments of PspA dimerization are shown. β-Galactosidase assays were performed as described in Materials and Methods. Values are means of results from at least three experiments, with a standard deviation of <15% in all cases. Expression of all proteins was under Plac control; induction was for 120 min with 1 mM IPTG. The percent repression was calculated as follows: 1 − [(β-galactosidase with repressor)/β-galactosidase with no repressor)]. λ repressor refers to the wild-type λ repressor. λ repressor-GCN4 refers to fusion of the N-terminal domain of the λ repressor with the GCN4 leucine zipper domain. λ repressor-PspA refers to fusion of the N-terminal domain of the λ repressor with the entire PspA sequence. wt, wild type.
Purification of PspA.
We purified PspA by His6 tag-Ni2+ affinity chromatography (39). The pspA gene was cloned downstream and in frame with a DNA sequence coding for His6 in the pET15B expression vector. Before purification was initiated, the activity of His6-PspA as a negative regulator of pspA transcription was tested by expressing the plasmid in a strain containing a pspA-lacZ reporter and demonstrating that it was as effective as a plasmid expressing wild-type PspA (data not shown).
Previous characterization of PspA demonstrated that it was recovered approximately equally in the cytoplasmic and membrane fractions (6). Initial efforts aimed at purification starting with the soluble fraction obtained following cell lysis, sonication, centrifugation, and elution from the Talon resin-containing column yielded small quantities of PspA relative to the total cellular content. Given the original observations of the subcellular localization of PspA, it seemed possible that PspA would partition with the insoluble fraction because of its affinity for the hydrophobic components in the initial postsonication pellet. Several detergents were assayed for their ability to release PspA from the insoluble fraction. While the nonionic detergents like Triton X-100 were not particularly effective, the zwitterionic detergent CHAPS and, to a somewhat greater extent, the ionic detergents deoxycholate and sodium Sarkosyl released most of the PspA (data not shown). CHAPS was chosen because up to 2% CHAPS did not interfere with binding of His6-PspA to the Talon resin.
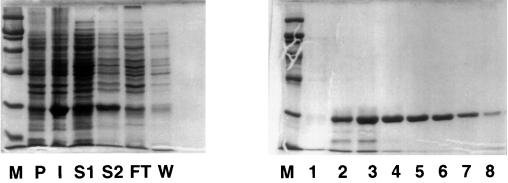
The overexpressed protein was largely in the detergent supernatant fraction (S2) rather than in the soluble fraction (S1) (Fig. 1). The S2 fraction contained approximately 25% of the total PspA. Following incubation of S2 with the Talon resin, most of the His6-PspA was bound to the resin, since there was little in the flowthrough from the gravity column. It was specifically bound, because a wash with a low concentration of imidazole (10 mM) released little His6-PspA protein but substantial amounts of other proteins. Results of elution with successive fractions of 0.1 M imidazole are presented in the right panel of Fig. 1. Fractions 4, 5, 6, and 7 show His6-PspA in comparatively pure form. The majority of the His6-PspA which was bound to the resin was released by elution with imidazole. These fractions were dialyzed to remove the imidazole (which inhibits transcription), pooled, and stored at −70°C in storage buffer (20 mM Tris-Cl [pH 7.5], 60 nM NaCl, 20% glycerol), where the protein appeared to be stable for more than 2 weeks.
FIG. 1.
Purification of PspA. The left-hand panel illustrates the steps in the purification of His6-PspA as described in Materials and Methods. Portions of each step in the purification were analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Lanes (percentages in parentheses are the portions of the step that were loaded): M, markers (from the bottom, 21, 30, 46, 66, and 97.4 kDa); P, preinduction K1462.pJD43; I, postinduction K1462.pJD43 (0.06%); S1, soluble fraction (0.1%); S2, soluble fraction following solubilization of the pellet with CHAPS-NaCl (0.1%); FT, flowthrough from the gravity column (0.1%); W, 10 mM imidazole wash (0.2%). The right-hand panel shows fractions from the elution with 100 mM imidazole in order of elution (2% of each fraction was loaded).
In vitro activity of PspA.
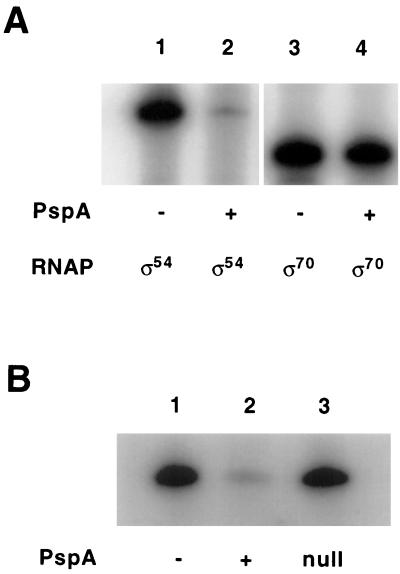
In vitro transcription assays containing purified components, including RNAP holoenzyme and a specific DNA template, demonstrated that His6-PspA has a strong negative effect on transcription from the ς54-dependent pspA promoter (pspA) but no effect on the ς70-dependent tac promoter (Fig. 2A). A sample of His6-PspA was boiled for 10 min before being added to the pspA in vitro transcription reaction mixture. Surprisingly, this treatment reduced the inhibitory activity only approximately twofold; by contrast, incubating the protein on ice for 3 days resulted in a near total loss of function (data not shown). A mock purification from the same strain used to purify His6-PspA, but containing a plasmid lacking the his6-pspA clone, had no effect on pspA transcription, whereas a purification conducted in parallel with the plasmid that overproduces His6-PspA yielded an activity with the inhibitory effect (Fig. 2B).
FIG. 2.
Controls for PspA inhibition of pspA transcription. (A) In vitro transcription reaction mixtures contained either supercoiled pJD10 (wild-type pspA promoter) or supercoiled pGZ119EH (tac promoter). PspF was included at 4 nM in lanes 1 and 2. ς54 and core RNAP were omitted from the reaction mixture, and ς70-RNAP holoenzyme was included instead at 13 nM. PspA (300 nM) was included in lanes 2 and 4. (B) In vitro transcription reaction mixtures contained supercoiled pJD10 (wild-type pspA promoter) and PspF at 4 nM. PspA (300 nM) was included in lane 2. Lane 3 contains a fraction from a purification of a strain carrying only the expression vector (see the text for more details).
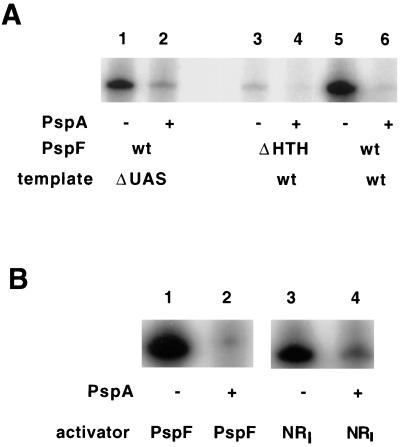
Since in vivo experiments demonstrated that PspA inhibits PspFΔHTH-dependent activation of pspA (Table 2), His6-PspA was assayed in in vitro pspA transcription reaction mixtures containing purified PspFΔHTH. Although PspFΔHTH was much less effective in stimulating transcription from the pspA promoter than PspF, it was inhibited by the same concentration of PspA (8-fold) (Fig. 3A, lanes 3 and 4) to the same extent as PspF (13-fold) (Fig. 3A, lanes 5 and 6).
FIG. 3.
PspA activity in vitro determined after inhibition of PspFΔHTH- and NRI-dependent activation of transcription. (A) In vitro transcription reactions were performed as described in Materials and Methods except that all components were incubated together at 37°C for 10 min without the template and then the template (15 nM) was added with [α-32P]CTP. The reaction then proceeded at 37°C for 10 min before addition of cold CTP. Templates were either the wild type (wt) (lanes 3 to 6, supercoiled pJD10, wt pspA promoter) or a ΔUAS mutant (lanes 1 and 2, supercoiled pJD12, pspA promoter without the UAS I and II sites). PspA (300 nM) was in lanes 2, 4, and 6; PspF (4 nM) was in lanes 1, 2, 5, and 6; and PspFΔHTH (70 nM) was in lanes 3 and 4. The arbitrary units of quantified pspA RNA transcripts were as follows: 3,110 (lane 1), 410 (lane 2), 105 (lane 3), 13 (lane 4), 5,784 (lane 5), and 434 (lane 6). (B) Reactions were performed as described for panel A with either supercoiled pJD10 (lanes 1 and 2; wt pspA promoter) or supercoiled pFC50 (lanes 3 and 4; glnH promoter). PspA (300 nM) was in lanes 2 and 4; PspF (4 nM) was in lanes 1 and 2; and NRI (30 nM, also 30 nM NRII) was in lanes 3 and 4. The arbitrary units of quantified pspA RNA transcripts were as follows: 7,785 (lane 1), 613 (lane 2), (glnH RNA transcript) 5,431 (lane 3), and (glnH RNA transcript) 486 (lane 4). HTH, helix-turn-helix.
PspA does not bind DNA.
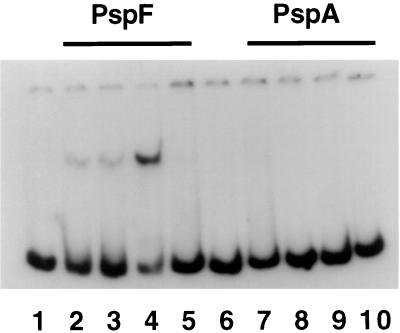
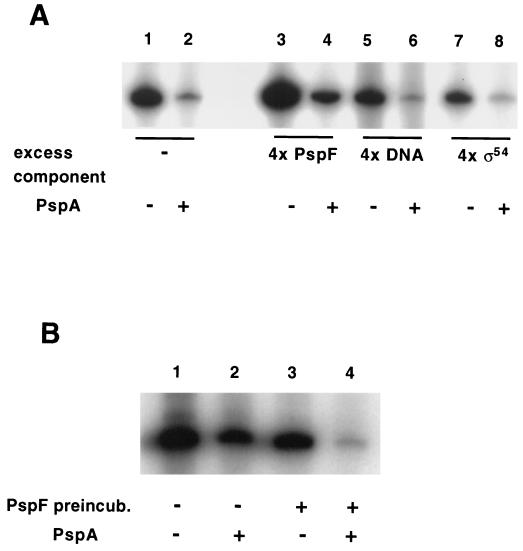
No property of PspA, including its sequence, suggests that it is a DNA-binding protein. When tested explicitly by a gel shift assay, PspA did not affect the mobility of a linear DNA fragment containing sequences (−188 to +72) spanning the pspA promoter (Fig. 4, lanes 6 to 10). By contrast, extract from cells overexpressing PspF shifted this fragment (Fig. 4, lanes 2 to 4) as has been reported previously (20). Additionally, since PspFΔHTH does not bind DNA in vitro (21), it is therefore unlikely that PspA-dependent inhibition of pspA is mediated by binding of PspA to DNA in the promoter region containing the upstream activation sequences (UASs). Further evidence for this interpretation comes from the observation that His6-PspA inhibition of pspA transcription was nearly as effective when the template lacked the UASs (Fig. 3A, lanes 1 and 2) as when the template was the wild type (lanes 5 and 6). Also consistent is the observation (see below) that PspA inhibits transcription at another ς54-dependent promoter (glnA) containing UASs completely different from those of pspA.
FIG. 4.
PspA does not bind DNA in the pspA promoter region. The 260 fragment containing sequences from −188 to +72 relative to the start site of pspA transcription (20) was used in a gel mobility shift assay using either crude cell extract from a strain overproducing PspF (lanes 2 to 4), purified PspA protein (lanes 6 to 9), or no added protein (lanes 1 and 5). In addition to 2 ng of 32P-labeled 260 fragment, each lane contained the following. Lane 2, 1 μl of crude cell extract; lane 3, 200 ng of nonspecific competitor [poly(dI-dC)] and 1 μl of crude cell extract; lane 4, 400 ng of nonspecific competitor and 2 μl of crude cell extract; lane 5, 400 ng of nonspecific competitor, 2 μl of crude cell extract, and 300 ng of specific competitor (unlabeled 260 fragment); lane 6, 10 nM purified PspA; lane 7, 20 nM purified PspA; lane 8, 50 nM purified PspA; lane 9, 100 nM purified PspA.
Specificity of PspA inhibition.
Since the promoter specificity of activation by EBPs is thought to reside in the sequences of their DNA-binding domains (36), the inhibition of PspΔHTH-dependent pspA transcription suggests that PspA may be active against other EBPs. That is, given that the PspFΔHTH protein is composed of only the central domain which contains the residues involved in ATP hydrolysis as well as the catalysis of open-complex formation, a protein able to inhibit PspFΔHTH activation must interact either with this domain or with its target, the RNAP holoenzyme. Since this central domain is highly conserved, a protein which interacts with PspFΔHTH might interact with the central domains of other EBPs and inhibit activation of transcription. In fact, NRI-dependent (11-fold) (Fig. 3B, lanes 3 and 4) activation of the glnH promoter is inhibited by His6-PspA to an extent similar to that of PspF-dependent activation of the pspA promoter (13-fold) (Fig. 3B, lanes 1 and 2). The in vivo significance of this inhibition of a heterologous EBP by PspA should be the subject of future investigations.
Titration of PspA inhibition of pspA transcription.
Titration of His6-PspA activity in inhibiting pspA (Fig. 5) demonstrated that, at a concentration of 300 nM, His6-PspA almost entirely eliminates PspF-dependent pspA-specific transcription but that this concentration has little effect on ς70-dependent tac transcription (Fig. 2A, lanes 3 and 4). The midpoint of this titration (∼100 nM) suggests that the interaction between PspA and its target protein is relatively weak. Also, this concentration is relatively high when compared to the concentrations of PspF (4 nM) and ς54 (45 nM). If His6-PspA targets some aspect of the interaction between PspF and ς54, then as a non-DNA-binding protein, it is at a disadvantage because the concentrations of PspF and ς54 relative to each other when both are bound to the DNA are higher than their simple solution concentrations.
FIG. 5.
Titration of PspA inhibition of pspA transcription. (A) In vitro transcription reactions were performed as described in Materials and Methods with supercoiled pJD10 (wild-type pspA promoter) except that all components were incubated together at 37°C for 10 min without the template, and then the template (15 nM) was added with [α-32P]CTP. The reaction then proceeded at 37°C for 10 min before addition of cold CTP. PspA was added in the concentration shown under each lane. PspF was added at 4 nM. (B) Quantification was carried out as described in Materials and Methods and was plotted as the quantity of the pspA-specific transcript (in arbitrary transcription units) against the concentration of PspA.
Mechanism of PspA inhibition.
In view of the hypothesis that PspA acts by binding either ς54 or PspF, and by preventing their interaction, we modified the in vitro transcription protocol. Since the interaction of DNA-binding proteins is facilitated by DNA, we incubated all protein components in the absence of DNA. This change in experimental protocol had little effect on the ability of PspA to inhibit pspA-specific transcription (Fig. 6A, lanes 1 and 2). We then increased the concentrations of various components of the reaction. Increasing the concentration of either the DNA template (Fig. 6A, lane 5) or ς54 (lane 7) fourfold (compared to that in Fig. 6A, lanes 1 and 2) was not stimulatory for pspA transcription; thus, neither factor is limiting. Addition of an excess of PspF (lane 3) stimulated transcription both in the presence and in the absence of PspA. A decrease in ς54 levels to below saturation had no effect on PspA inhibition of pspA transcription (data not shown).
FIG. 6.
Analysis of the PspA target in inhibition of pspA transcription. (A) In vitro transcription reactions used supercoiled pJD10 (wild-type pspA promoter) except that all components were incubated together at 37°C for 10 min without the template, and then the template (15 nM) was added with [α-32P]CTP. The reaction then proceeded at 37°C for 10 min before addition of cold CTP. PspA (300 nM) was present in lanes 2, 4, 6, and 8. PspF was present at 4 nM in all lanes except for lanes 3 and 4, where it was present at 16 nM. Also, in lanes 5 and 6, the template was present at 60 nM; in lanes 7 and 8, ς54 was present at 172 nM. The pspA RNA transcripts were quantified (in arbitrary units) as follows: 6,810 (lane 1), 731 (lane 2), 15,160 (lane 3), 2,320 (lane 4), 3,140 (lane 5), 419 (lane 6), 2,958 (lane 7), and 418 (lane 8). (B) In vitro transcription reactions were performed as described for panel A with pJD10 (wild-type pspA promoter) except that in lanes 1 and 2, PspF was added at the same time as the template and the [α-32P]CTP. PspA (300 nM) was present in lanes 2 and 4; PspF was present at 4 nM. The pspA RNA transcripts were quantified (in arbitrary units) as follows: 4,601 (lane 1), 2,025 (lane 2), 3,322 (lane 3), and 406 (lane 4). preincub., preincubation.
Since the ratio of pspA-specific transcripts in the presence or absence of PspA was unaffected by the increased concentrations of the reaction components, it seems unlikely that a simple sequestration mechanism involving tight binding and inactivation of any of these components is responsible for PspA's inhibitory function. The simplest explanation for these results is that PspA binds to PspF with relatively low affinity. A second explanation, that most of the added PspA is inactive, is rendered unlikely by the observation that the ratio of pspA transcription in the absence or presence of PspA did not change appreciably after the concentration of PspF was increased fourfold. If the apparent association constant of ∼100 nM determined from the results shown in Fig. 5 is correct, then under the conditions used here, where the concentration of PspA was 300 nM, an increase in the concentration of PspF from 4 to 16 nM would be expected to leave the extent of inhibition essentially unchanged.
We compared the effect on PspA inhibition of addition of PspF prior to the addition of DNA (the same protocol as described above) with the effect of the addition of PspF simultaneously with the DNA. In this case, incubation of PspA with PspF before addition of DNA (Fig. 6B, lanes 3 and 4) showed a greater inhibition of pspA-specific transcripts than addition of PspF at the same time as the DNA (Fig. 6B, lanes 1 and 2). Thus, PspF bound to DNA and presumably able to interact with the ς54-RNAP holoenzyme complex is more resistant to inhibition than PspF free in solution.
DISCUSSION
The dependence of transcriptional initiation by ς54-RNAP on activation by an EBP suggests several potential mechanisms of negative regulation. Typically, EBPs are activated by modification of their N-terminal domains, so a mechanism that prevents this modification from occurring would inhibit activation. Alternatively, a protein might interact with the central domain of the EBP that contains the residues essential for ATP hydrolysis and for activation and directly inhibit one of these catalytic activities. Finally, EBPs bound to the UAS must interact with ς54-RNAP bound at the promoter, so a protein might prevent this interaction either by binding to and/or modifying the domains of the proteins that mediate this contact(s) or by blocking the binding of the EBP to the UAS sequences.
Most ς54-dependent promoters studied to date employ some variation of the first mechanism. In contrast, the inhibitory NifL protein of Klebsiella pneumoniae, appears to act by stoichiometric interaction with EBP NifA, as suggested by in vivo experiments (17) and confirmed in vitro with NifL purified from K. pneumoniae (4) or from Azotobacter vinelandii (2). Interestingly, NifL acts to inhibit activity of a NifA mutant lacking both the N-terminal and C-terminal domains (4). This mutant retains NTPase activity, which is not affected by NifL, suggesting that NifL acts to prevent interaction between NifA and ς54-RNAP. Since NifL does not inhibit activation by other EBPs, including NtrC (2, 4, 30) and AnfA (2), the target of this interaction presumably is NifA rather than ς54-RNAP.
PspF lacks an endogenous N-terminal domain and is constitutively active in vivo and in vitro, suggesting that it is subject to an inhibitory regulatory mechanism dependent on a second protein. Here we have demonstrated that in vivo (Table 2) and in vitro (Fig. 2), pspA transcription in the presence of PspF and ς54-RNAP is subject to inhibition by PspA. This inhibition is independent of the ability of PspF to bind DNA because PspA inhibits PspFΔHTH-dependent activation in vivo (Table 2) and in vitro (Fig. 3). Thus, similarly to NifL, PspA must interfere with some aspect of PspF function involving the central domain. However, unlike NifL, PspA inhibits activation by another EBP, NRI (Fig. 3); thus, PspA must initially recognize and consequently inhibit a part of the ς54-RNAP activation pathway common to all EBPs. An interesting question raised by this inhibition is whether induction of PspA expression has any effect on other ς54-dependent promoters in vivo.
Also, unlike NifL, PspA is not active in near-unity, stoichiometric concentrations with its target EBP (Fig. 5). The observation (Fig. 6A) that a higher concentration of individual reaction components (e.g., PspF, ς54, and DNA) failed to reduce the magnitude of PspA inhibition is consistent with the interpretation that the high-concentration requirement of PspA is real. Further, this observation is not consistent with a model of PspA inhibition where PspA binds to proteins with high affinity and thereby acts to sequester them from participation in transcriptional activation, as is the case with anti-sigma factors (8).
PspA inhibition is enhanced when reaction conditions are such that PspA and PspF can interact in the absence of DNA (Fig. 6B, lanes 3 and 4) compared to in its presence (Fig. 6B, lanes 1 and 2). This result suggests that incubation of PspA and PspF when PspF is not able to bind DNA allows for an interaction essential for PspA inhibition. Since PspA inhibits PspFΔHTH-dependent transcription equally as well as PspF-dependent transcription, DNA binding can not per se be the target of PspA. EBPs form tetramers (40) and higher-order oligomers on DNA (47), and it is thought that these ATP-dependent structures (38) are necessary intermediates in the reaction whereby the EBPs convert the ς54-RNAP closed complex to an open complex. This oligomerization is characteristic not only of PspF but, interestingly, also of PspFΔHTH, despite its inability to bind DNA (21). Thus, PspA may target PspF monomers or dimers and so a fourfold increase in PspF concentration in the presence of DNA would yield mostly an increase in oligomer. Given the apparent importance (and as yet incompletely understood role) of these oligomers for the mechanism of transcriptional activation at ς54-dependent promoters, this possibility should be explored more fully in future experiments.
ACKNOWLEDGMENTS
We thank Alex Ninfa for NRI, NRII, and ς54 proteins; Jim Hu for the N-terminal λ repressor fusion kit; and members of our laboratory for helpful discussions.
This work was supported by NSF grant MCB 93-16625. J.D. held an NSF graduate fellowship and was supported by NIH training grant CA09673-19 and by a Norman and Rosita Winston Foundation fellowship.
REFERENCES
- 1.Adamson J G, Zhou N E, Hodges R S. Structure, function and application of the coiled-coil protein folding motif. Curr Opin Biotechnol. 1993;4:428–437. doi: 10.1016/0958-1669(93)90008-k. [DOI] [PubMed] [Google Scholar]
- 2.Austin S, Buck M, Cannon W, Eydmann T, Dixon R. Purification and in vitro activities of the native nitrogen fixation control proteins NifA and NifL. J Bacteriol. 1994;176:3460–3465. doi: 10.1128/jb.176.12.3460-3465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron J, Weisberg R A. Mutations of the phage lambda nutL region that prevent the action of Nun, a site-specific transcription termination factor. J Bacteriol. 1992;174:1983–1989. doi: 10.1128/jb.174.6.1983-1989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger D K, Narberhaus F, Kustu S. The isolated catalytic domain of NIFA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NIFL. Proc Natl Acad Sci USA. 1994;91:103–107. doi: 10.1073/pnas.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner B R, Huang H C, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brissette J L, Russel M, Weiner L, Model P. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brissette J L, Weiner L, Ripmaster T L, Model P. Characterization and sequence of the Escherichia coli stress-induced psp operon. J Mol Biol. 1991;220:35–48. doi: 10.1016/0022-2836(91)90379-k. [DOI] [PubMed] [Google Scholar]
- 8.Brown K L, Hughes K T. The role of anti-sigma factors in gene regulation. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou P Y, Fasman G D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 11.Claverie-Martin F, Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 13.Dworkin J, Jovanovic G, Model P. Role of upstream activation sequences and integration host factor in transcriptional activation by the constitutively active prokaryotic enhancer-binding protein PspF. J Mol Biol. 1997;273:377–388. doi: 10.1006/jmbi.1997.1317. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Goss T J, Bender R A, Ninfa A J. Repression of the Klebsiella aerogenes nac promoter. J Bacteriol. 1995;177:5535–5538. doi: 10.1128/jb.177.19.5535-5538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez S, de Lorenzo V, Perez-Martin J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 16.Graumann P, Schroder K, Schmid R, Marahiel M A. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol. 1996;178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson N, Austin S A, Dixon R A. Role of metal ions in negative regulation of nitrogen fixation by the nifL product from Klebsiella pneumoniae. Mol Gen Genet. 1989;216:484–491. [Google Scholar]
- 18.Hu J C, O'Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 19.Jovanovic G, Dworkin J, Model P. Autogenous control of PspF, the constitutively active enhancer-binding protein of Escherichia coli. J Bacteriol. 1997;179:5232–5237. doi: 10.1128/jb.179.16.5232-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jovanovic G, Model P. PspF and IHF bind cooperatively in the psp promoter-regulatory region of Escherichia coli. Mol Microbiol. 1997;25:473–481. doi: 10.1046/j.1365-2958.1997.4791844.x. [DOI] [PubMed] [Google Scholar]
- 21.Jovanovic G, Rakonjac J, Model P. In vivo and in vitro activities of the Escherichia coli ς54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J Mol Biol. 1999;285:469–483. doi: 10.1006/jmbi.1998.2263. [DOI] [PubMed] [Google Scholar]
- 22.Jovanovic G, Weiner L, Model P. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J Bacteriol. 1996;178:1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome (supplement) DNA Res. 1995;2:191–198. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 24.Kleerebezem M, Crielaard W, Tommassen J. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 1996;15:162–171. [PMC free article] [PubMed] [Google Scholar]
- 25.Kleerebezem M, Tommassen J. Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol Microbiol. 1993;7:947–956. doi: 10.1111/j.1365-2958.1993.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 26.Knight A E. Ph.D. thesis. Cambridge, United Kingdom: Cambridge University; 1994. [Google Scholar]
- 27.Koski P, Saarilahti H, Sukupolvi S, Taira S, Riikonen P, Osterlund K, Hurme R, Rhen M. A new alpha-helical coiled coil protein encoded by the Salmonella typhimurium virulence plasmid. J Biol Chem. 1992;267:12258–12265. [PubMed] [Google Scholar]
- 28.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee H S, Narberhaus F, Kustu S. In vitro activity of NifL, a signal transduction protein for biological nitrogen fixation. J Bacteriol. 1993;175:7683–7688. doi: 10.1128/jb.175.23.7683-7688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J H, Hoover T R. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a sigma 54-dependent transcriptional activator, interacts with sigma 54 and the beta subunit of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessl M, Balzer D, Lurz R, Waters V L, Guiney D G, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H M, Kaneko Y, Keegstra K. Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol Biol. 1994;25:619–632. doi: 10.1007/BF00029601. [DOI] [PubMed] [Google Scholar]
- 34.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 35.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 36.Morett E, Cannon W, Buck M. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nif promoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 1988;16:11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ninfa A J, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Martin J, de Lorenzo V. ATP binding to the sigma 54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 39.Porath J. Immobilized metal ion affinity chromatography. Protein Expr Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 40.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 43.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 44.Weiner L, Brissette J L, Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991;5:1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- 45.Weiner L, Model P. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc Natl Acad Sci USA. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 48.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]