Abstract
The Drosophila Ntan1 gene encodes an N-terminal asparagine amidohydrolase that we show is highly conserved throughout evolution. Protein isoforms share more than 72% of similarity with their human counterparts. At the cellular level, this gene regulates the type of glial cell growth in Drosophila larvae by its different expression levels. The Drosophila Ntan1 gene has 4 transcripts that encode 2 protein isoforms. Here we describe that although this gene is expressed at all developmental stages and adult organs tested (eye, antennae and brain) there are some transcript-dependent specificities. Therefore, both quantitative and qualitative cues could account for gene function. However, widespread developmental stage and organ-dependent expression could be masking cell-specific constraints that can be explored in Drosophila by using Gal4 drivers. We report a new genetic driver within this gene, Mz317-Gal4, that recapitulates the Ntan1 gene expression pattern in adults. It shows specific expression for perineural glia in the olfactory organs but mixed expression with some neurons in the adult brain. Memory and social behavior disturbances in mice and cancer and schizophrenia in humans have been linked to the Ntan1 gene. Therefore, these new tools in Drosophila may contribute to our understanding of the cellular basis of these alterations.
Subject terms: Cell biology, Genetics, Neuroscience
Introduction
The Ntan1 gene encodes the Drosophila N-terminal asparagine amidohydrolase homolog (NTAN1). It regulates endoreplication rates through the N-end rule pathway1. It has been proposed that in Drosophila, similarly to mammalian NTAN12, this enzyme recognizes proteins with an N-terminal asparagine (Asn) and converts it to aspartate (Asp)1. After the action of other enzymes, such as ATE1, this site becomes then a potential substrate for ubiquitination and proteasomal degradation1,3,4.
At the cellular level, development of an organism consist on a myriad number of events that lead to the formation of an adult individual. Among these events, genetic regulation of cell number and size, controlled by growth and division5, is of extreme importance. In the nervous system, developmental processes need to strictly control and regulate glial and neuronal cell growth in order to achieve a functional organism.
In Drosophila, the surface of the entire nervous system is covered by two thin glial layers: the perineural glia (PNG) and the subperineural glia (SPG)6. Both types of glial cells are involved in the formation of the blood–brain barrier7,8. During larval growth, while the PNG grows exponentially in number by extensive cell division9,10 the SPG retains its small number and massively increases the size of its cells individually by both endoreplication and endomitosis becoming then polyploid11,12. It has been shown that in the blood–brain barrier of Drosophila the endoreplication of SPG cells is regulated by the Ntan1 gene1, previously named Öbek.
In fact, in Drosophila larvae, glial cells present asparagine amidohydrolase activity of Ntan11. The mechanism proposed in these cells for regulating their growth involves different expression levels of the Ntan1 gene. During larval development, in SPG cells high expression levels of Ntan1 limit cell cycle and replication rates controlling ploidy and number of nuclei1. On the contrary, PNG cells express moderate levels of Ntan1, allowing then efficient PNG proliferation1.
Although this gene has been thoroughly studied in larvae, a detailed description of gene expression that pays attention to its different transcripts both during development and in the adult phase is still lacking. In this article, we tackle this question by analyzing mRNA expression of the different transcripts of the Ntan1 gene during different developmental stages and several adult organs such as the brain, eyes and antennae as representatives of central or peripheral nervous systems. Moreover, we show tissue-specific expression of the Ntan1 gene in adults by using available Gal4 lines.
In the peripheral nervous system of Drosophila, such as that involved in olfactory reception, antennal glial cells begin to originate about 16 h after puparium formation (APF) and are closely associated with the development of sensory axons13–15. In the adult antennae, there are mainly two different types of glia, GH146-type and Mz317-type glia. GH146 glial cells correspond to ~ 30% of the glial antennal cells15. This glia originates in the brain and migrates to the third antennal segment where it ensheathes the axons of the ORNs which are projected from the antenna to the brain15. Mz317 glial cells corresponds to ~ 70% of antennal glial cells. They constitute a peripheral layer to GH146 glia and seem to surround the somas of olfactory sensory neurons15. Although Mz317::GFP is considered a marker of perineural and cell body glia labelling most of peripheral glia16, nothing is known about the molecular basis of this type of glia. In this report, we demonstrate that the Mz317 driver recapitulates the expression of the Ntan1 gene as well as the other Gal4 drivers located within this gene.
Results
Ntan1 gene is expressed in all developmental stages from embryo to adult Drosophila
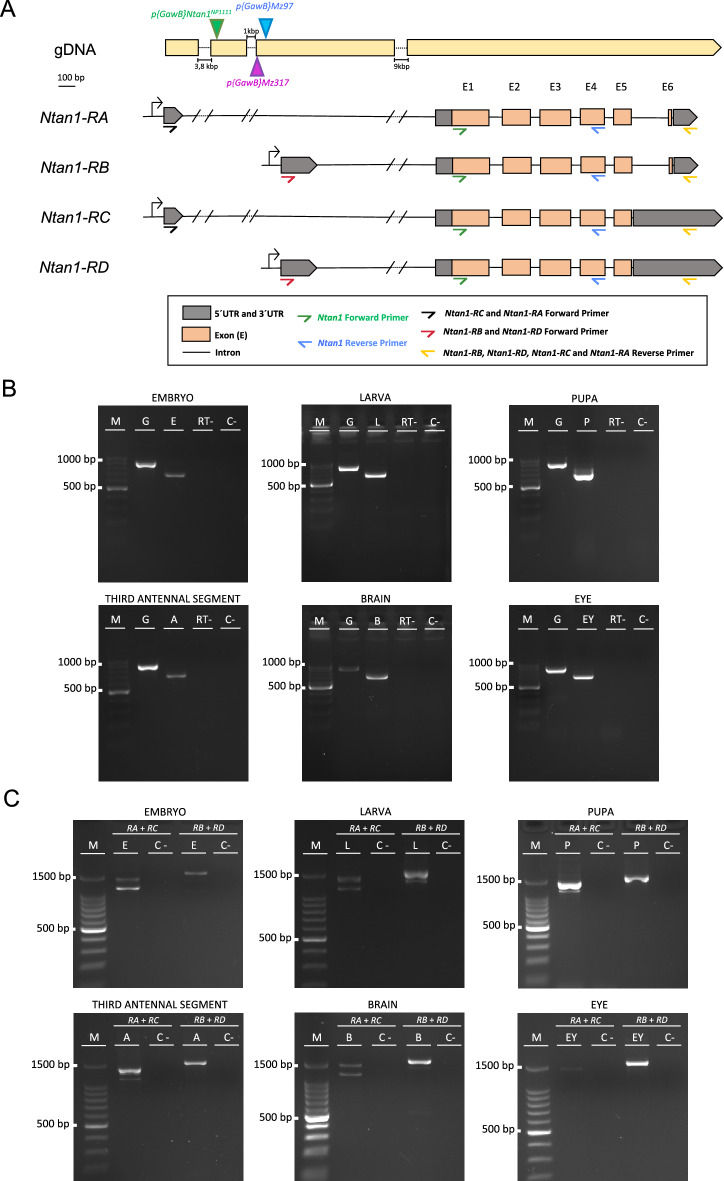
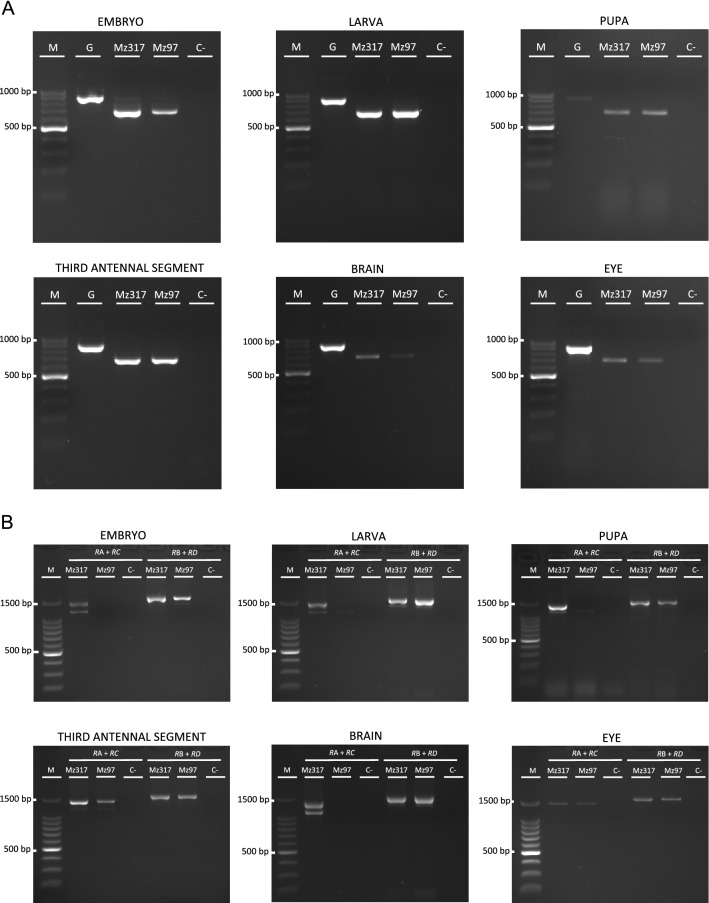
Although it was previously shown that NTAN1 was strongly expressed in embryo and larvae of Drosophila in a subset of glial cells in both the PNS and the CNS along with a weakly expression in other tissues1, little is known about the Ntan1 gene expression in other stages and adult organs. In order to test the native expression of the Ntan1 gene in different pre-adult developmental stages (embryo, larva and pupa) and some adult organs (third antennal segments, brain and eyes) of Drosophila melanogaster we performed RT-PCR. As described in the material and methods section, primers were designed in order to amplify different size products from gDNA and cDNA (Fig. 1A, Table 4). The expected size for the cDNA of the Ntan1 gene is 685 base pairs (bp), while for the genomic DNA is 901 bp. All products obtained in the wildtype genotype were of the expected sizes (Fig. 1B). These results confirm the expression of the Ntan1 gene in all the developmental stages and adult organs studied.
Figure 1.
Gene expression analysis of the Ntan1 gene in Drosophila melanogaster. (A) Schematic representation of the insertion sites of the Gal4 elements of Mz317-Gal4; Mz97-Gal4,UAS-Stinger and Ntan1NP1111-Gal4 fly lines in the Ntan1 gene. The P{GawB}Mz317 insertion and the P{GawB}Mz97 are located ∼ 150 bp and ∼ 100 bp upstream of the second transcriptional start site within the first intron, respectively. The P{GawB}Ntan1NP1111 insertion is located ∼ 1300 bp upstream of the second transcriptional start site within the first intron. 5′ UTR and 3′ UTR regions, exons and introns of each transcript and the location of the different primers used in RT-PCR are shown. (B) Gene expression of the Ntan1 gene by RT-PCR in different developmental stages and adult organs of wildtype flies (Canton-S). (C) Gene expression of Ntan1 transcripts by RT-PCR in different developmental stages and adult organs of wildtype flies (Canton-S). M: Molecular marker, G: Genomic DNA, E: Embryonic cDNA, L: Larval cDNA, P: Pupal cDNA, A: Third antennal segment cDNA, B: Brain cDNA, EY: Eye cDNA, RT−: RT-PCR without retrotranscriptase, C−: Negative control.
Table 4.
Primers designed for the RT-PCR gene expression analysis of G6PD, Ntan1 and Ntan1 transcripts.
| Gene/transcript | Primers (5′→ 3′) | Amplicon size (bp) |
|---|---|---|
| G6PD |
F: AGTCGCCTACAATGGTCTGC R:GTTCGAATCGTTGCTAACGG |
gDNA: 1106 cDNA: 959 |
| Ntan1 |
F: GGTGTGCTGCTGCAGGATGACT R: CGTAGTTAAAGGGTCCAATGC |
gDNA: 901 cDNA: 685 |
| Ntan1-RA |
F: GTGCACGAATCGCTGAACT R: CGTTCCCAGTTCAGTTCAGATCCAT |
cDNA:1136 |
| Ntan1-RB |
F: GTCTCTCTTTCCGGTTTGCGTC R: CGTTCCCAGTTCAGTTCAGATCCAT |
cDNA: 1336 |
| Ntan1-RC |
F: GTGCACGAATCGCTGAACT R: CGTTCCCAGTTCAGTTCAGATCCAT |
cDNA: 1405 |
| Ntan1-RD |
F: GTCTCTCTTTCCGGTTTGCGTC R: CGTTCCCAGTTCAGTTCAGATCCAT |
cDNA: 1601 |
The corresponding amplicon size for each primer pair is also indicated.
Gene expression of Ntan1 transcripts at different developmental stages and adult organs
According to Flybase, the Ntan1 gene has 4 transcripts: Ntan1-RA, Ntan1-RB, Ntan1-RC and Ntan1-RD, identical in their coding sequence two-by-two (Ntan1-RA and Ntan1-RB; Ntan1-RC and Ntan1-RD), but differentiable by their 5' and 3' UTR (UnTranslated Region) regions (Fig. 1A). The specific gene expression of each transcript of the Ntan1 gene was analysed in the same developmental stages and organs previously mentioned. Thus being able to determine if there is one or several transcripts specific to a particular stage or organ tested. Amplification of genomic DNA was used as a PCR positive control. The expected cDNA sizes of Ntan1-RA, Ntan1-RB, Ntan1-RC and Ntan1-RD transcripts are 1136 bp, 1336 bp, 1405 bp and 1601 bp, respectively (Fig. 1C, Table 1). Table 1 summarizes the expression data for all transcripts in the different samples studied. Ntan1-RA transcript is expressed in embryo, larva, pupa, brain and third antennal segments but not in the adult eye whereas Ntan1-RB transcript is only expressed in larva and brain. Finally, both Ntan1-RC and Ntan1-RD transcripts are expressed in all developmental stages and organs studied (Table 1, Fig. 1C, Fig. S4).
Table 1.
Summary of the results obtained in the gene expression analysis by RT-PCR of the Ntan1 transcripts in different developmental stages and adult organs of wildtype genotype Canton-S.
| Transcript | Embryo | Larva | Pupa | Adult | ||
|---|---|---|---|---|---|---|
| Brain | Eye | Third antennal segment | ||||
| Ntan1-RA | + | + | + | + | − | + |
| Ntan1-RB | − | + | − | + | − | − |
| Ntan1-RC | + | + | + | + | + | + |
| Ntan1-RD | + | + | + | + | + | + |
Negative symbol (−) indicates total absence of gene expression and positive symbol (+) indicates gene expression of the Ntan1 transcripts.
The Ntan1 gene is highly conserved throughout evolution
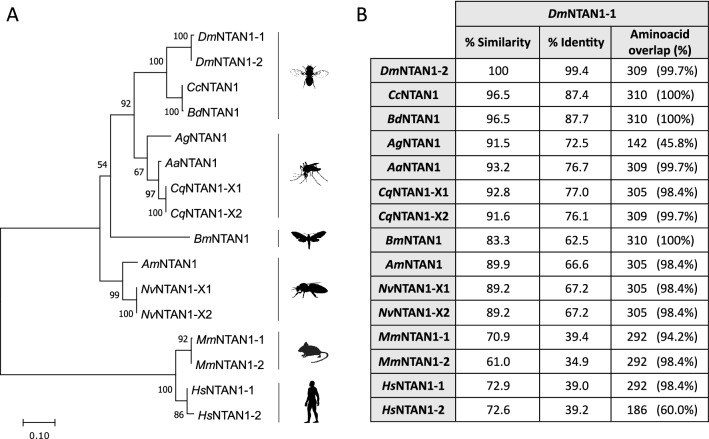
Although in Drosophila the Ntan1 gene has four transcripts that differ mainly in their UTR regions, they are translated into two different isoforms. As N-terminal asparagine amidohydrolases have been found in other species, we performed a comparative analysis of the aminoacid sequences of the NTAN1 proteins isoforms of various insects belonging to the Diptera, Hymenoptera and Lepidoptera Orders as well as their corresponding mouse and human orthologs. We infered their evolutionary relationships by constructing a phylogenetic tree that clearly illustrates the evolutionary conservation of the products of the Ntan1 gene (Fig. 2A). Although the percentage of identity between the Drosophila melanogaster NTAN1-1 isoform and the rest of ortholog isoforms from the species tested ranged from 34 to 87%, the percentage of similarity increases to the range between 61% to 96.5%. In fact, Drosophila and its human ortholog proteins share more than 72% of aminoacid similarity (Fig. 2B). In conclusion, we show that the Ntan1 gene is highly conserved through evolution, not only among insects but also in mammals (Fig. 2). It is generally accepted that high conservation of genes along evolution indicates an essential functional role.
Figure 2.
Comparative analysis of the aminoacid sequences of the NTAN1 isoforms of various insect and non-insect species. (A) Neighbor Joining tree of insects and other mammalian species NTAN1 orthologs. Dm (Drosophila melanogaster), Cc (Ceratitis capitata), Bd (Bactrocera dorsalis) Ag (Anopheles gambiae), Aa (Aedes aegypti), Cq (Culex quinquefasciatus), Bm (Bombyx mori), Am (Apis mellifera), Nv (Nasonia vitripennis), Mm (Mus musculus) and Hm (Homo sapiens). The tree was built with MEGA11. Bootstrap support (1000 replicates) is indicated. The scale bar units show aminoacid substitutions per site. (B) Comparison of sequence similarity and identity between the DmNTAN1-1 isoform and the other isoforms of the different species studied with the LALIGN/PLALIGN software.
Mz317-Gal4 insert is located into the Ntan1 gene and drives gene expression in adult brain and antennal glia
As it was previously shown that the Ntan1 gene was involved in controlling the levels of ploidy in subperineural and perineural glial cells during larval development1 we wondered if this gene was also being expressed in adult perineural glia. The Mz317-Gal4 line combined with a UAS-GFP reporter line drives the expression of GFP labeling the majority of peripheral glia including most antennal glia15 but the gene driving this especific mark was unknown. In order to determine the genomic location of the Gal4 insert of the Mz317-Gal4 line we performed the Splinkerette technique17. This method allows to determine P-element insertion sites with unknown genomic location. By PCR amplification and posterior sequencing of the 3′ P-element insertion site (Figs. S1 and S2) we were able to conclude that the Mz317-Gal4 line has its Gal4 element inserted into the Ntan1 gene, specifically around 150 bp upstream the second 5′ UTR region.
It was known from previous studies that the Mz317-Gal4 line drives gene expression in antennal perineural glia15 but in order to further investigate the cellular expression pattern of the Mz317-Gal4 line in different cell types we analyzed GFP expression in the genotype Mz317-Gal4;UAS-mCD8::GFP both in antennal cryosections and brain dissections of Drosophila melanogaster. For this purpose, immunohistochemistry was performed with anti-GFP antibody on antennal cryosections and with anti-GFP and anti-nc82 antibodies on brain samples. The anti-nc82 antibody recognizes the neuropil-specific BRUCHPILOT protein, which allows us to visualize the general structure of the brain18,19. Also, to distinguish the specific cell types observed in the Mz317-Gal;UAS-mCD8::GFP line, we restricted GFP expression in either neurons or glia by adding gene expression restriction elements such as elav-Gal80 and repo-Gal80, respectively. Genetic crosses were performed to obtain the following genotypes: Mz317-Gal;UAS-mCD8::GFP (GFP expression in all Mz317-type cells), Mz317-Gal4/repo-Gal80;UAS-mCD8::GFP (GFP expression absent in glia due to GAL80 expression driven by repo, a panglial marker), Mz317-Gal4/elav-Gal80;UAS-mCD8::GFP (GFP expression absent in neurons due to GAL80 expression driven by elav, a panneuronal marker). Immunostaining of the UAS-mCD8::GFP line by itself was used as a negative control.
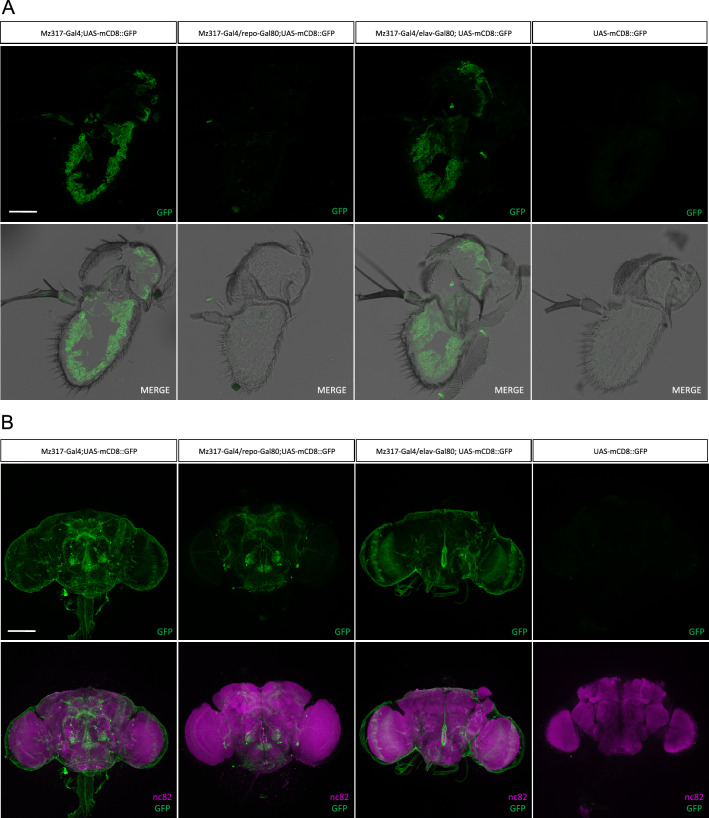
In the antenna, the Mz317-Gal4 line directs GFP expression exclusively in glial cells, as we did not observe any GFP expression in antennal neuronal cells (Fig. 3A). Furthermore, the antennal GFP expression pattern in Mz317-Gal4/elav-Gal80;UAS-mCD8::GFP, that restricts Ntan1-drived expression to non-neuronal cells, matches exactly the GFP expression pattern of the Mz317-Gal;UAS-mCD8::GFP individuals previously described15 (Fig. 3A). Antennal GFP expression is observed at the base of the sensilla, most likely surrounding the neuronal somas of most olfactory sensory neurons. Likewise, GFP expression is observed around the olfactory nerve. Thus, the expression pattern of the Mz317-Gal4 line corresponds exclusively to perineural antennal glial cells.
Figure 3.
Analysis of GFP expression in the Mz317-Gal4 line. (A) Immunohistochemistry on antennal cryosections (14 µm) with anti-GFP antibody (green) on the following genotypes: Mz317-Gal4;UAS-mCD8::GFP, Mz317-Gal4/repo-Gal80;UAS-mCD8::GFP, Mz317-Gal4/elav-Gal80;UAS-mCD8::GFP and UAS-mCD8::GFP. Scale bar represents 40 µm. (B) Immunohistochemistry on brain dissections with anti-GFP (green) and anti-nc82 (magenta) antibodies on Mz317-Gal4;UAS-mCD8::GFP, Mz317-Gal4/repo-Gal80;UAS-mCD8::GFP, Mz317-Gal4/elav-Gal80;UAS-mCD8::GFP and UAS-mCD8::GFP. Scale bar represents 100 µm.
As expected, GFP expression is clearly observed as a wrap around the whole brain structure (Fig. 3B). Additionally, we also observe staining in a selected number of non-glial cells in the brain (Fig. 3B). These cells correspond to neuronal cells as inferred from the restriction of GAL4 expression in elav-Gal80 expressing cells that suppressed this staining. These neurons are present in limited numbers in the subesophageal ganglion (SNG) (15 ± 2 cells), the mushroom bodies (MB) area and the upper part of the protocerebrum (SP) (12 ± 2 cells), the antero-ventrolateral part (AVLP) and projecting neurons of the optic lobes (OL) (32 ± 4 cells), and to an area close to the antennal lobes (AL) (6 ± 0 cells) where olfactory interneurons are usually located.
In summary, the Mz317-Gal4 line directs gene expression in the antenna exclusively in perineural glia and in the brain in perineural glial cells and in a limited number of neuronal cells.
Ntan1 gene as a marker for antennal and brain perineural glia
Once it was clear that the Gal4 insert of the Mz317-Gal4 line was located into the Ntan1 gene we wondered if there were other genetic driver lines with inserts in the Ntan1 gene that showed a similar expression pattern. Two other lines have driver insertions in the Ntan1 gene, Mz97-Gal4 and Ntan1NP1111-Gal4. In both cases, the Gal4 elements are inserted in the first intron of the Ntan1 gene (1 and https://flybase.org/ respectively). Note that the non-transcribed genomic DNA that is located in the middle of both 5’UTRs is considered the first intron according to the Ensembl database. While the Mz97-Gal4 insert is located ~ 100 bp upstream the second transcriptional start site the insertion of the Ntan1NP1111-Gal4 is located ~ 1300 bp upstream this region (Fig. 1A).
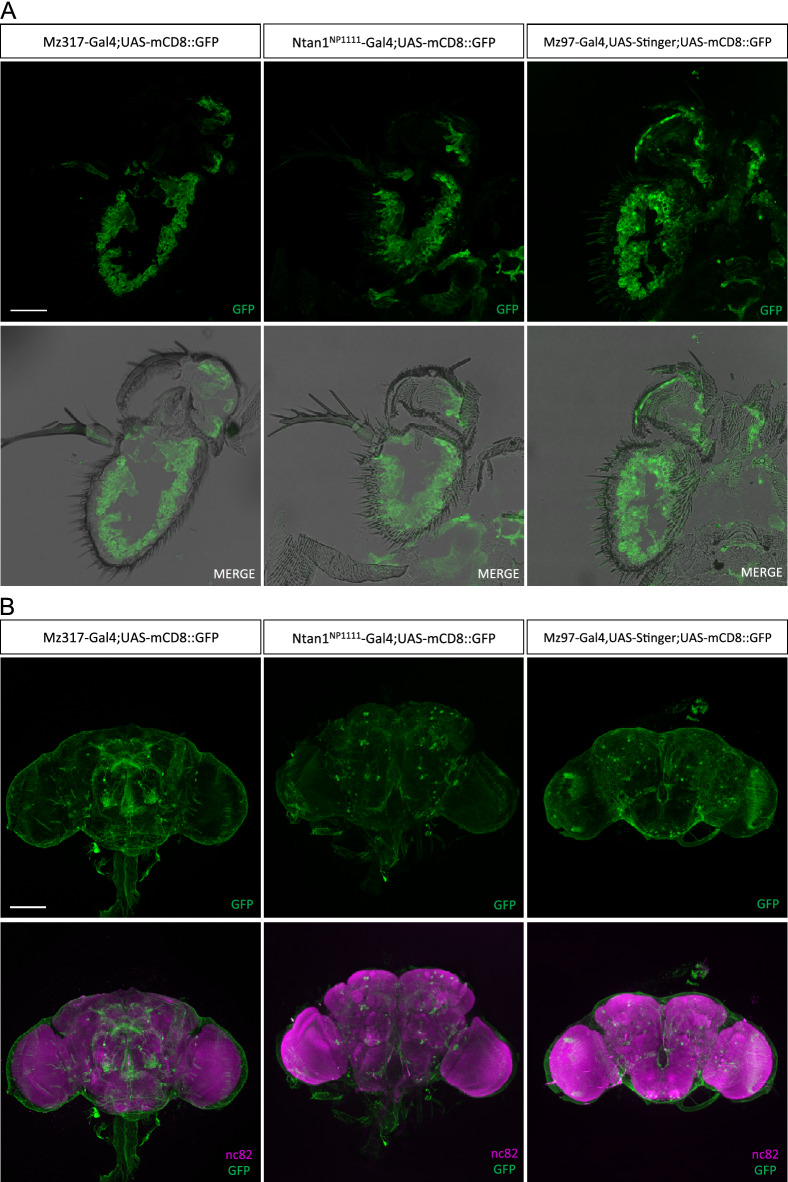
In order to compare the expression patterns of these Gal4 lines with Mz317-Gal4 we performed immunohistochemistry with anti-GFP antibody in the antenna and anti-GFP and anti-nc82 in the brain in the following genotypes: Mz317-Gal4;UAS-mCD8::GFP, Ntan1NP1111-Gal4;UAS-mCD8::GFP and Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP. In the last genotype, cell nuclei will also be GFP-stained together with membrane-bound GFP (mCD8) due to the additional UAS-Stinger element that is also present in the original MZ97-Gal4 line.
The antennal GFP expression observed was very similar in the three lines tested (Fig. 4A). Both in Mz97-Gal4 and Ntan1NP1111-Gal4 we observed glial GFP staining located in the base of the sensilla of the third antennal segment, most likely surrounding the neuronal somas of the olfactory sensory neurons as in Mz317-Gal4 (Figs. 3A and 4A). In the brain, there are slight differences in GFP expression among the different fly lines (Fig. 4B). All brains appear to have a glial wrap around their structure. However, Ntan1NP1111-Gal4;UAS-mCD8::GFP and Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP brains show a lower number of neuronal cells in the superior protocerebrum area, the mushroom body area and in the vicinity of the antennal lobes compared to Mz317-Gal4;UAS-mCD8::GFP (Fig. 4B). In the case of the Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP line there seems to be a higher expression of GFP, however, this is due to Stinger expression which also marks the cell nuclei. Taken together, these results indicate that the Ntan1 gene is mainly expressed in both antennal and brain perineural glia.
Figure 4.
Analysis of GFP expression in different Ntan1-related Gal4 lines in Drosophila melanogaster. (A) Immunohistochemistry on antennal cryosections (14 µm) with anti-GFP antibody (green) on the following genotypes: Mz317-Gal;UAS-mCD8::GFP, Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP and Ntan1NP1111-Gal4;UAS-mCD8::GFP. Scale bar represents 40 µm. (B) Immunohistochemistry on brain dissections with anti-GFP (green) and anti-nc82 (magenta) antibodies on Mz317-Gal;UAS-mCD8::GFP, Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP and Ntan1NP1111-Gal4;UAS-mCD8::GFP. Scale bar represents 100 µm.
Gene expression of Ntan1 transcripts in different Gal4 lines
After determining the developmental stages and organs in which the different transcripts of Ntan1 were expressed (Fig. 1C, Table 1) we wondered if the slight differences observed in brain staining among the different driver lines tested could be due to qualitative differences in transcript expression in the fly lines tested. For this purpose, we studied the expression of Ntan1 gene in different developmental stages and adult organs of Mz317-Gal4 and Mz97-Gal4,UAS-Stinger lines. In addition, we have determined whether or not there are one or more transcripts specific to a particular stage or adult organ in each fly line. As the Gal4 element of Mz97-Gal4,UAS-Stinger and Ntan1NP1111-Gal4 lines is inserted in the same area (first intron), the gene expression analysis of Ntan1 gene has been carried out only in the Mz97-Gal4,UAS-Stinger line. As before, an RT-PCR experiment was performed to check the native expression of the Ntan1 gene in Mz317-Gal4 and Mz97-Gal4,UAS-Stinger lines. Genomic DNA amplification was used as a positive control. The expected size of cDNA of the Ntan1 gene was 685 bp using the primers Ntan1-F GGTGTGCTGCTGCAGGATGACT and Ntan1-R CGTAGTTAAAGGGTCCAATGC. In both lines tested, the PCR products obtained had the expected sizes (Fig. 5A). These results confirm the expression of the Ntan1 gene in the different stages and organs studied in Mz317-Gal4 and Mz97-Gal4,UAS-Stinger lines. The expression of each transcript was also evaluated in both lines (Fig. 5B). Although in the wildtype genotype Ntan1-RB transcript is not expressed in embryo and pupa our results show that all transcripts are expressed in embryo, larva and pupa of Mz317-Gal4 line (Table 2). Also, some differences are observed in relation to Mz97-Gal4,UAS-Stinger line and the wildtype genotype. Specifically, there is no expression of Ntan1-RA transcript in embryo of Mz97-Gal4,UAS-Stinger line and additionally, there is no expression of Ntan1-RC transcript in embryo, larva or pupa of Mz97-Gal4,UAS-Stinger line (Table 2). On the other hand, in adult Drosophila, Ntan1-RA transcript is expressed in the brain and third antennal segments of the Mz317-Gal4 line, while it is only expressed in third antennal segments of Mz97-Gal4,UAS-Stinger. Ntan1-RB transcript is expressed only in the brain of both lines, whereas Ntan1-RC transcript is expressed in the brain, third antennal segments and eyes of the Mz317-Gal4 line and third antennal segments and eyes of the Mz97-Gal4,UAS-Stinger line. Finally, Ntan1-RD transcript is expressed in all three organs studied of both lines similarly to the wildtype genotype (Table 3). To sum up, the main gene expression differences in adult Drosophila between the wildtype genotype and the Gal4 lines studied are seen in the expression of Ntan1-RA and Ntan1-RC transcript of Mz97-Gal4,UAS-Stinger in the brain (Table 3). We observed no expression of the transcript Ntan1-RB in the embryo and in the pupa of the wildtype genotype (Fig. 1C, Table 1), whereas there is expression of this transcript in the Gal4 lines studied (Fig. 5B, Table 2). We performed an additional PCR using the G6PD housekeeping gene for the embryo and brain of the Mz97-Gal4 fly line, where we did not observe any expression of Ntan1-RA and Ntan1-RC (Fig. S3). With this data we can infer that even slight differences in the insertion sites of P(GawB) driver elements change gene expression of the Ntan1 transcripts that might account for slight differences also in GFP expression drived by the lines tested.
Figure 5.
Gene expression analysis of Ntan1 in Mz97-Gal4 and Mz317-Gal4 fly lines. Expression of the Ntan1 gene (A) and Ntan1 transcripts (B) by RT-PCR in Mz97-Gal4 and Mz317-Gal4 fly lines. M: Molecular marker, G: Genomic DNA of wildtype genotype, Mz317: Mz317-Gal4 cDNA, Mz97: Mz97-Gal4,UAS-Stinger cDNA and C−: Negative control.
Table 2.
Summary of the results obtained in the gene expression analysis by RT-PCR of the Ntan1 transcripts in different development stages (embryo, larva and pupa) in different genotypes.
| Transcript | Embryo | Larva | Pupa | ||||||
|---|---|---|---|---|---|---|---|---|---|
| wt | Mz317 | Mz97 | wt | Mz317 | Mz97 | wt | Mz317 | Mz97 | |
| Ntan1-RA | + | + | − | + | + | + | + | + | + |
| Ntan1-RB | − | + | + | + | + | + | − | + | + |
| Ntan1-RC | + | + | − | + | + | − | + | + | − |
| Ntan1-RD | + | + | + | + | + | + | + | + | + |
The genotypes tested are: Canton-S (wt), Mz317-Gal4 (Mz317) and Mz97-Gal4,UAS-Stinger (Mz97). Negative symbol (−) indicates total absence of gene expression and positive symbol (+) indicates gene expression of the Ntan1 transcripts.
Table 3.
Summary of the results obtained in the gene expression analysis by RT-PCR of the Ntan1 transcripts in different adult organs in different genotypes.
| Transcript | Adult | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Brain | Eye | Third antennal segment | |||||||
| wt | Mz317 | Mz97 | wt | Mz317 | Mz97 | wt | Mz317 | Mz97 | |
| Ntan1-RA | + | + | − | − | − | − | + | + | + |
| Ntan1-RB | + | + | + | − | − | − | − | − | − |
| Ntan1-RC | + | + | − | + | + | + | + | + | + |
| Ntan1-RD | + | + | + | + | + | + | + | + | + |
The genotypes tested are: Canton-S (wt), Mz317-Gal4 (Mz317) and Mz97-Gal4,UAS-Stinger (Mz97). Negative symbol (−) indicates total absence of gene expression and positive symbol (+) indicates gene expression of the Ntan1 transcripts.
Discussion
In this article, we offer a detailed gene expression study of the Ntan1 gene, previously characterized as encoding an N-terminal asparagine amidase in Drosophila1 as well as in other species of invertebrates and vertebrates2. At the cellular level its role has been related to the control of cell cycle and replication rates controlling ploidy and number of nuclei in some cellular types, especially in glia. In this cell type moderate expression levels associate to perineural glial cells (PNG) while high expression levels appear in subperineural glial cells during larval development in Drosophila1. In summary, the quantitative expression of the gene rather than just its presence or absence appears to be at the basis of its function deduced from studies of the entire gene. However, the gene encodes several transcripts and the importance of qualitative differences in expression depending on the developmental stage and cell type cannot be excluded beforehand.
In this report we study the expression of the Ntan1 gene and each of its 4 transcripts − RA to − RD during different developmental stages and in some adult organs to see if there is a specific expression pattern for any of them. It should be reminded that the differences in the sequence of the transcripts mainly affect their regulatory regions rather than the coding sequence that is identical between − RA and − RB (isoform 1) and between -RC and -RD (isoform 2), respectively. The results indicate that although there is expression of the Ntan1 gene in all stages and organs tested there are some specificities in the transcript expression (Fig. 1B,C, Table 1). Looking at the different developmental stages (embryo, larva and pupa), − RA, − RC and − RD transcripts appear always, only the − RB transcript shows expression in the larva but does not express in embryo and pupa. In the adult stage the Ntan1-RB transcript is possibly specific to the brain, as it does not appear in neither the third antennal segment nor the eye. As for the other transcripts Ntan1-RA, Ntan1-RC and Ntan1-RD show generalized expression in the adult organs with the only exception of − RA that does not show expression in the eye. This means that the eye lacks isoform 1 as it expresses neither − RA, nor − RB. This is not unprecedented, as previous reports in Drosophila on other protein families such as G-proteins have shown specificities associated with eye-related tissues20–22.
However, it is important to highlight that although the coding sequence of the Ntan1-RB transcript is identical to the sequence of Ntan1-RA transcript (Fig. 1A) they differ in their regulatory regions, 5′ UTR and 3′ UTR. Thus, these differences in transcript expression might account for the different expression levels of the Ntan1 gene that drive different cell growth mechanisms in PNG cells and the SPG cells1. In vertebrates a similar behavior has been reported for this gene, as variable expression of an approximately 1.4-kb transcript was reported in all 8 mouse tissues examined, with lowest expression in testis. However, a transcript of about 1.1 kb was prominently expressed in testis only2.
By attending to the two characteristics, transcript and isoform type, qualitative and quantitative determination seems to play an important role in gene function. In Drosophila there are some organs such as the compound eyes that express only one Ntan1 isoform, while in the other cases analyzed both are expressed, but including either one or both transcripts. However, it should not be forgotten that both the different developmental stages studied and the adult organs include a variable number of cell types, so that the data obtained as a whole may be masking specificities at the cellular level. Some data in this direction was reported in mouse, where the Ntan1 expression decreased following differentiation in C2C12 mouse myoblasts, but not in MEL mouse leukemia cells2.
Detailed studies at the cellular level are difficult to address in small organisms by RNA expression analysis when the gene studied shows widespread expression in different cell types and quantitative issues play an important role. In such cases, immunohistochemical analysis with the gene product does not usually give consistent results either. In Drosophila, thanks to the development of numerous genetic tools such as the Gal4/UAS system and its variants, this analysis can be done by selective labeling with reporter genes such as GFP whose expression is directed with drivers that recapitulate the expression pattern of the gene under study23,24. In many cases, this is obtained by insertions within the gene. These tools can even serve to follow gene expression in different cell types throughout development like in the G-TRACE method25,26, as well as to direct the generation of functional mutants restricted to some cell types. However, the validity and significance of the data obtained depends on the accuracy and reliability with which these drivers recreate the expression of the genes of interest.
In this sense, we have located a new driver within the Ntan1 gene (Mz317-Gal4 line) and we have studied the expression of the gene in this line compared to wildtype flies. Moreover, we have described the expression patterns in glia and neurons of other driver lines that had already been described by their insertion within this gene (Mz97-Gal4 and Ntan1NP1111), in order to characterize how they recapitulate the expression of the original gene and can be used for further studies.
In this work we report for the first time that the Mz317-Gal4 line16 has an insert within the Ntan1 gene. According to Sen et al.15 Mz317::GFP is a marker for perineural glia that labels the majority of peripheral glia. We characterized the expression of Ntan1 in the Mz317-Gal4 line both at the molecular and cellular level. RT-PCR analysis of the adult organs in the Mz317-Gal4 line recapitulates the same expression pattern that the wildtype individuals.
At the cellular level, immunohistochemistry in the third antennal segment and brain of Mz317-Gal4;UAS-mCD8::GFP individuals reveals the cell types marked in both structures. Although we detected specific expression of perineural glia in the third antennal segment, in agreement with what has been previously reported15, the labeling in the brain includes mostly glial cells and a reduced number of neurons.
We also compared Ntan1 gene expression in wildtype and Mz317-Gal4 individuals with that obtained with other drivers previously mapped to the Ntan1 gene, both at the molecular and cellular levels. The transcript Ntan1-RB is not expressed neither in the embryo nor in the pupa of the wildtype genotype whereas there is expression of this transcript in the Gal4 lines studied. In adults, the expression pattern of the Mz317-Gal4 line matches that of wildtype individuals while the Mz97-Gal4,UAS-Stinger line shows no expression of the -RA or -RC transcript in the brain. In a previous work, it was indicated that the Mz97-Gal4 insertion might have led to a mutant Ntan1 allele1. In homozygous Mz97-Gal4 flies there is still expression of various transcripts of the Ntan1 gene, although quantitative differences in gene expression might not be detected by the methodology used. Our data suggest that a mutant phenotype may be generated if it depends on the quantitative expression of either or both Ntan1 isoforms in the brain.
At the cellular level, the expression patterns of the different Gal-4 lines mapped to the Ntan1 gene are very similar. In the third antennal segment, expression is restricted to PNG glia, whereas in the brain staining shows as a glial wrap with the exception that Ntan1NP1111-Gal4;UAS-mCD8::GFP and Mz97-Gal4,UAS-Stinger;UAS-mCD8::GFP show a lower number of neurons than Mz317-Gal4;UAS-mCD8::GFP.
The differences in gene expression among the different lines, both at the molecular and cellular level, relate to the different positions of the Gal4 insertion that may lead to mutations in some cases. This indicates the importance of performing previous expression studies if we want to use Gal4 insertion lines as tools for the study of gene expression as well as to generate cell type-specific mutations. However, the difficulties in targeting a certain cell type can be solved in some cases with additional elements like the Gal80 inserts.
Summarizing, we consider that the Ntan1 gene could be easily used as a marker and/or driver of just perineural glia, by restricting neuronal expression with a Gal80 insertion. Therefore, with this work we show that the Gal4 lines associated to the Ntan1 gene in Drosophila will add new genetic tools for studying the regulation and function of glia.
Glia is essential in the development and homeostasis of the nervous system due to its interaction with neurons and its modulatory role in neuronal activation27. In mammals, glia can constitute between 33 and 66% of the brain mass28,29. However, in Drosophila, glia makes up approximately 10% of the cells in the nervous system6. The low percentage of glial cells relative to other species and the similarity of its nervous system to vertebrates27,30,31, make Drosophila melanogaster an excellent model organism to investigate the biological mechanisms behind glial regulation of the nervous system.
Finally, it has been shown that the Ntan1 gene performs numerous functions in vertebrates and defects in the gene can have serious effects at the level of the organism. Thus, alterations in the Ntan1 gene expression have been associated with different types of memory and social behavior32,33, sporadic childhood cancer34 and risk of schizophrenia35. Therefore, studies in Drosophila using all the new tools available achieve a greater projection in determining the cellular basis of these phenotypes, which is necessary for their possible reversion.
Materials and methods
Drosophila lines
Flies were maintained in a thermoregulated chamber at 25 ± 1 °C in 12 h light:12 h dark conditions. The following fly lines were used in this work: Canton-S (Bloomington Drosophila Stock Center, BDSC 64349), Mz97-Gal4,UAS-Stinger (BDSC 9488), UAS-mCD8::GFP (BDSC 5130), Ntan1NP1111-Gal4 (Kyoto Stock Center, DGRC 103887), Mz317-Gal416, repo-Gal80 and elav-Gal80 (kindy provided by Leiserson, W., Yale University, USA; Lee, T. Janelia Research Campus, USA; Casas-Tintó, S. Instituto Cajal CSIC, Madrid, Spain, respectively).
Phylogenetic analysis
Aminoacid sequences of NTAN1 of Drosophila melanogaster (Dm), Ceratitis capitata (Cc), Bactrocera dorsalis (Bd), Anopheles gambiae (Ag), Aedes aegypti (Aa), Culex quinquefasciatus (Cq), Bombyx mori (Bm), Apis mellifera (Am), Nasonia vitripennis (Nv), Mus musculus (Mm) and Homo sapiens (Hm) were obtained from the National Center for Biotechnology Information (NCBI) database (DmNTAN1-1: NP_611355.2; DmNTAN1-2: NP_001163199.1; BdNTAN1: XP_011208810.1; AgNTAN1: XP_001230643.2; AaNTAN1: XP_021711016.1; CqNTAN1-X1: XP_038110105.1; CqNTAN1-X2: XP_038110106.1; BmNTAN1: XP_037869646.1; AmNTAN1: XP_016769476.1; NvNTAN1-X1: XP_032456886.1; NvNTAN1-X2: XP_032456890.1; MmNTAN1-1: NP_035076.1; MmNTAN1-2: NP_001333035.1; HsNTAN1-1: NP_775745.1; HsNTAN1-2: NP_001257695.1). All sequences were aligned with MUSCLE using default parameters. The alignment was used to build a Neighbor Joining tree with evolucionary distances computed using the Jones, Taylor and Thorton matrix-based method36, 1000 bootstrap replicates and gaps handled with pairwise deletion. Phylogenetic analyses were conducted in MEGA1137.
Immunostaining
Immunofluorescence on antennal cryosections were performed as described in38. Briefly, 14 µm antennal cryosections were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, Pennsylvania) in Phosphate Buffered Saline (PBS) for 7 min. Antennal cryosections were washed two times with PBS and incubated with PBST (PBS with 0.1% Triton X-100, Sigma-Aldrich, Saint Louis, USA) for 30 min. Then, the samples were incubated in PTS (5% heat inactivated normal goat serum (Biowest, Nuaillé, France) in PBST) for 30 min. After that, samples were incubated overnight at 4 °C in dark with anti-GFP rabbit primary antibody (1:5000; Invitrogen, Eugene, USA) diluted in PTS. Samples were washed 10 min for three times in PBST and incubated in PTS for 30 min. Subsequently, cryosections were incubated with Alexa 488 anti-rabbit secondary antibody (1:1000; Invitrogen, Eugene, USA) diluted in PTS for 2 h at room temperature in dark. Finally, after several washes with PBST, samples were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, USA).
To detect GFP expression in the brain, immunohistochemical experiments were carried out on whole brain dissections previously fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, Pennsylvania) in PBS with 0.2% Triton X-100 (Sigma-Aldrich, Saint Louis, USA) for 3 h at 4 °C following the protocol described in39. After several washes in PBST 0.2% (PBS with 0.2% Triton X-100, (Sigma-Aldrich, Saint Louis, USA), brains were dissected in blocking solution (5% heat inactivated normal goat serum (Biowest, Nuaillé, France) in PBST 0.2%). Dissected brains were incubated with primary antibodies diluted in blocking solution: anti-GFP rabbit (1:2500; Invitrogen, Eugene, USA) and anti-nc82 mouse 1:10 (1:10; Developmental Studies Hybridoma Bank, lowa City, USA) overnight at 4 °C in dark. Samples were washed 30 min for 6 times with PBST and they were incubated with secondary antibodies diluted in blocking solution: Alexa 488 goat anti-rabbit (1:100; Invitrogen, Eugene, USA) and Cy3 donkey anti-mouse (1:100; Jackson ImmunoResearch Laboratories, Ely, UK). Finally, after several washes with PBST0.2%, samples were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, USA). Immunostainings were performed in both males and females with no obvious expression differences between the sexes.
Images of antennal cryosections and brain dissections were collected on a Leica TCS-SP8X Confocal Laser Microscope (Leica Microsystems) and analysed with free ImageJ software (https://imagej.nih.gov/ij/)40. Six brains were used to count neuron cell bodies on the Mz317-Gal4/repo-Gal80;UAS-mCD8::GFP genotype.
RNA extraction and RT-PCR
Reverse transcription (RT) experiments were carried out on different developemental stages and adult tissues. Total RNA from ten embryos, one third instar larva, two 20–30 h APF pupas, 50 third antennal segments, five brains and 50 eyes, from the wildtype genotype (Canton-S), Mz317-Gal4 and Mz97-Gal4,UAS-Stinger lines were isolated with the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cDNA was synthesised from the isolated RNA using SuperScript™ First Strand Synthesis System Kit for RT-PCR (Invitrogen, Carlsbad, USA) with oligo dTs following the manufacturer´s instructions. Total RNA from 10 embryos, 10 larvae and 10 pupae at different time points and 3 pupae at 24 h, 48 h and 72 h APF from Canton-S were isolated for performing RT-PCRs in Fig. S4. The exact numbers of RT-PCR experiments are given in Table S1.
Polymerase chain reaction (PCR) and gel electrophoresis
All primers used for the PCRs were designed using Primer 3 Plus software specifically for amplyfing different size fragments for cDNA and gDNA (Table 4). Primers for the four transcripts of the Ntan1 gene were also designed specifically in the 5'UTR and 3'UTR regions of each transcript, being the transcripts: Ntan1-RA, Ntan1-RB, Ntan1-RC and Ntan1-RD, which were distinguished by size, two by two (Table 4).
PCR was carried out in a final volume of 20 µl using 2 µl of template cDNA and Taq polymerase (Promega, Madison, USA) according to the manufacturer's recommendations. Samples were subjected to 35 cycles of PCR. Each cycle included 30 s of denaturation at 95 °C, 45 s of banding at 58 °C and 30 s of elongation at 72 °C for the Ntan1 gene and 90 s of elongation at 72 °C for the transcripts. After amplification, 20 μl of the amplification products were analysed by gel electrophoresis on a 2% agarose gel with a DNA ladder according to the amplified DNA size of the sample (Table 4).
Splinkerette method
The insertion site of the Gal4 element of the Mz317-Gal4 line was inferred using the Splinkerette PCR method previously described17 with slight modifications. In short, a standard phenol–chloroform genomic DNA extraction was performed using 20 adult flies from each genotype: Mz317-Gal4 and GH146-Gal4 (used as a positive control). Purified DNA was digested with PsuI (BstYI) enzyme (Thermo Scientific, Lithuania, EU) at 37 °C overnight, and then inactivated for 20 min at 80 °C. The digested DNA was ligated (T4 DNA Ligase, New England BioLabs, Ipswich, USA) to double stranded splinkerette oligonucleotide17 for 2 h at room temperature in a total volume of 50 µl. Then, ligated DNA was used as a template to amplify both the 3′ and 5′ P-element insertion sites using primers 3′SPLNK#1 (CACTCAGACTCAATACGACAC) and 3′SPLNK#2 (GGATGTCTCTTGCCGAC) and primers 5′SPLNK#1-GAWB (TGGGAGAGTAGCGACACTCC) and 5′SPLNK#2-GAWB (GAGCTTTTTAAGTCGGCAAATATCG) respectively. After amplification, 20 μl of the amplification products were analysed by gel electrophoresis on a 2% agarose gel with a DNA ladder according to the amplified DNA size of the sample (Fig. S1). The gel band obtained from the 3′ P-element insertion site of Mz317-Gal4 was extracted, purified and Sanger sequenced using the 3′SPLNK-SEQ (CGGGACCACCTTATG) primer by the Servicios Científico-Técnicos from the University of Oviedo using standard methods (Fig. S2). The sequence generated is available in the NCBI repository (GenBank OP158015).
Supplementary Information
Acknowledgements
Authors would like to acknowledge the technical support provided by Servicios Científico-Técnicos from the Universidad de Oviedo and the Bloomington Stock Center for providing fly lines.
Author contributions
Conceptualization: E.A., C.G.D.; Methodology: C.G.D., F.M., E.A.; Formal analysis: E.A., C.G.D., F.M., A.C.S., L.C.C.; Investigation: A.C.S., L.C.C.; Writing—original draft: C.G.D., A.C.S.; Writing—review & editing: C.G.D., E.A., A.C.S.; Supervision: C.G.D., F.M., E.A.; Project administration: E.A., C.G.D.; Funding acquisition: E.A., C.G.D.
Funding
This work was funded by the following sources, Spanish Ministry of Economy, Industry and Competitiveness [SAF2013-48759-P to E.A. and BFU2017-85882-P to E.A. and C.G.D.], the University of Oviedo [PAPI-17-PEMERG-2 to C.G.D.], and the Principality of Asturias Government [IDI-2018–000182 to E.A.]. L.C.C. and A.C.S. were funded by a Severo Ochoa PhD fellowship from the Principality of Asturias Government [PA-20-PF-BP19-040 to A.C.S. and PA-20-PF-BP19-042 to L.C.C.].
Data availability
All data supporting the results in this work are available from the corresponding author on request. The Mz317-Gal4 3′ insertion site sequence generated is available in the NCBI repository with the accession number GenBank OP158015 (https://www.ncbi.nlm.nih.gov/genbank/). Not-cropped original images of all the gel electrophoresis are provided in the Supplemental Data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-18999-8.
References
- 1.Zülbahar S, Sieglitz F, Kottmeier R, Altenhein B, Rumpf S, Klämbt C. Differential expression of Öbek controls ploidy in the Drosophila blood-brain barrier. Development. 2018;145:164111. doi: 10.1242/dev.164111. [DOI] [PubMed] [Google Scholar]
- 2.Grigoryev S, Stewart AE, Kwon YT, Arfin SM, Bradshaw RA, Jenkins NA, et al. A mouse amidase specific for N-terminal asparagine: the gene, the enzyme and their function in the N-end rule pathway. J. Biol. Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 3.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 4.Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 5.Wood E, Nurse P. Sizing up to divide: Mitotic cell-size control in fission yeast. Annu. Rev. Cell. Dev. Biol. 2015;31:11–29. doi: 10.1146/annurev-cellbio-100814-125601. [DOI] [PubMed] [Google Scholar]
- 6.Kremer MC, Jung C, Batelli S, Rubin GM, Gaul U. The glia of the adult Drosophila nervous system. Glia. 2017;65:606–638. doi: 10.1002/glia.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klämbt C. Organization and function of the blood–brain barrier in Drosophila. J. Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildirim K, Petri J, Kottmeier R, Klämbt C. Drosophila glia: Few cell types and many conserved functions. Glia. 2019;67:5–26. doi: 10.1002/glia.23459. [DOI] [PubMed] [Google Scholar]
- 9.Avet-Rochex A, Kaul AK, Gatt AP, McNeill H, Bateman JM. Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development. 2012;139:2763–2772. doi: 10.1242/dev.074179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Awasaki T, Lai S-L, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr-Weaver TL. When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 2015;31:307–315. doi: 10.1016/j.tig.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jhaveri D, Sen A, Rodrigues V. Mechanisms underlying olfactory neuronal connectivity in Drosophila-the atonal lineage organizes the periphery while sensory neurons and glia pattern the olfactory lobe. Dev. Biol. 2000;226:73–87. doi: 10.1006/dbio.2000.9855. [DOI] [PubMed] [Google Scholar]
- 14.Jhaveri D, Rodrigues V. Sensory neurons of the Atonal lineage pioneer the formation of glomeruli within the adult Drosophila olfactory lobe. Development. 2002;129:1251–1260. doi: 10.1242/dev.129.5.1251. [DOI] [PubMed] [Google Scholar]
- 15.Sen A, Shetty C, Jhaveri D, Rodrigues V. Distinct types of glial cells populate the Drosophila antenna. BMC Dev. Biol. 2005;5:25. doi: 10.1186/1471-213X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K, Urban J, Technau GM. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch. Dev. Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 17.Potter CJ, Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rein K, Zöckler M, Heisenberg M. A quantitative three-dimensional model of the Drosophila optic lobes. Curr. Biol. 1999;9:93–96. doi: 10.1016/S0960-9822(99)80021-9. [DOI] [PubMed] [Google Scholar]
- 19.Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Boto T, Gomez-Diaz C, Alcorta E. Expression analysis of the 3 G-protein subunits, Galpha, Gbeta, and Ggamma, in the olfactory receptor organs of adult Drosophila melanogaster. Chem. Senses. 2010;35:183–193. doi: 10.1093/chemse/bjp095. [DOI] [PubMed] [Google Scholar]
- 21.Lee YJ, Dobbs MB, Verardi ML, Hyde DR. dgq: a drosophila gene encoding a visual system-specific G alpha molecule. Neuron. 1990;5:889–898. doi: 10.1016/0896-6273(90)90349-K. [DOI] [PubMed] [Google Scholar]
- 22.Yarfitz S, Niemi GA, McConnell JL, Fitch CL, Hurley JB. A G beta protein in the Drosophila compound eye is different from that in the brain. Neuron. 1991;7:429–438. doi: 10.1016/0896-6273(91)90295-B. [DOI] [PubMed] [Google Scholar]
- 23.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 24.Martín F, Alcorta E. Novel genetic approaches to behavior in Drosophila. J. Neurogenet. 2017;31:288–299. doi: 10.1080/01677063.2017.1395875. [DOI] [PubMed] [Google Scholar]
- 25.Olson JM, Evans CJ, Ngo KT, Kim HJ, Nguyen JD, Gurley KGH, et al. Expression-based cell lineage analysis in Drosophila through a course-based research experience for early undergraduates. G3. 2019;9:3791–3800. doi: 10.1534/g3.119.400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, et al. G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bittern J, Pogodalla N, Ohm H, Brüser L, Kottmeier R, Schirmeier S, et al. Neuron-glia interaction in the Drosophila nervous system. Dev. Neurobiol. 2020;81:438–452. doi: 10.1002/dneu.22737. [DOI] [PubMed] [Google Scholar]
- 28.Azevedo FAC, Carvalho LRB, Grinberg LT, Farfel JM, Ferretti REL, Leite REP, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 29.Herculano-Houzel S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 2014;62:1377–1391. doi: 10.1002/glia.22683. [DOI] [PubMed] [Google Scholar]
- 30.Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Stork T, Bernardos R, Freeman MR. Analysis of glial cell development and function in Drosophila. Cold Spring Harb. Protoc. 2012;2012:1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balogh SA, Kwon YT, Denenberg VH. Varying intertrial interval reveals temporally defined memory deficits and enhancements in NTAN1-deficient mice. Learn. Mem. 2000;7:279–286. doi: 10.1101/lm.33500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, et al. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 2000;20:4135–4148. doi: 10.1128/MCB.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danuta G, Tobias M, Marcus D, Miriam E, Nergiz K, Olesja S, et al. Molecular karyotyping and gene expression analysis in childhood cancer patients. J. Mol. Med. 2020;98:1107–1123. doi: 10.1007/s00109-020-01937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietiläinen O, et al. Copy number variations of chromosome 16p131 region associated with schizophrenia. Mol. Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 40.Rasband, W. S. ImageJ (U.S. National Institutes of Health, 1997–2018). https://imagej.nih.gov/ij/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results in this work are available from the corresponding author on request. The Mz317-Gal4 3′ insertion site sequence generated is available in the NCBI repository with the accession number GenBank OP158015 (https://www.ncbi.nlm.nih.gov/genbank/). Not-cropped original images of all the gel electrophoresis are provided in the Supplemental Data.