Abstract
We have cloned a Candida albicans gene (CaMIG1) that encodes a protein homologous to the DNA-binding protein Mig1 from Saccharomyces cerevisiae (ScMig1). The C. albicans Mig1 protein (CaMig1) differs from ScMig1, in that, among other things, it lacks a putative phosphorylation site for Snf1 and presents several long stretches rich in glutamine or in asparagine, serine, and threonine and has the effector domain located at some distance (50 amino acids) from the carboxy terminus. Expression of CaMIG1 was low and was similar in glucose-, sucrose-, or ethanol-containing media. Disruption of the two CaMIG1 genomic copies had no effect in filamentation or infectivity. Levels of a glucose-repressible α-glucosidase, implicated in both sucrose and maltose utilization, were similar in wild-type or mig1/mig1 cells. Disruption of CaMIG1 had also no effect on the expression of the glucose-repressed gene CaGAL1. CaMIG1 was functional in S. cerevisiae, as judged by its ability to suppress the phenotypes produced by mig1 or tps1 mutations. In addition, CaMig1 formed specific complexes with the URS1 region of the S. cerevisiae FBP1 gene. The existence of a possible functional analogue of CaMIG1 in C. albicans was suggested by the results of band shift experiments.
Candida albicans is an opportunistic pathogen able to produce a variety of lesions in cutaneous surfaces or life-threatening systemic infections in immunocompromised hosts. The organism can grow as a yeast or in filamentous form; although this point has not been definitely established, the filamentous form seems to be implicated in the invasiveness of the organism (35). A number of mutations that produce changes in morphology have been described; among those, disruptions in the TUP1 gene give rise to filamentous morphology in all growth conditions (3). Tup1 has been characterized in Saccharomyces cerevisiae as a repressor of the transcription of several genes regulated by glucose, oxygen, or cell type (13, 25, 49). In S. cerevisiae, Tup1 forms a complex with Cyc8 that is recruited by Mig1 to glucose-sensitive promoters (44, 47). Mig1 is a C2H2 zinc finger protein (34) which in S. cerevisiae binds to the promoters of many genes repressed by glucose (27). In the absence of glucose, Mig1 is localized in the cytosol; in its presence, it migrates to the nucleus (8). It is thought that this change in localization is due to changes in phosphorylation, which in turn are regulated by the protein kinase Snf1, whose activity is necessary for derepression of glucose repressible genes. Curiously, SNF1 is apparently essential for the viability of C. albicans (40) or C. tropicalis (24), in contrast with the situation in S. cerevisiae (5) or C. glabrata (41). Due to the differences in phenotype produced by mutations in similar genes in C. albicans and S. cerevisiae and taking into account the relationship between Mig1, Tup1, and Snf1, we became interested in the MIG1 gene from C. albicans (CaMIG1). During a study of the CaTPS1 gene, encoding trehalose-6-phosphate synthase (51), we fortuitously isolated a fragment of DNA whose sequence presented similarity with that of the MIG1 gene from S. cerevisiae (ScMIG1). We report in this work the isolation and characterization of the CaMIG1 gene and show that its disruption has no effects on filamentation and infectivity or on the expression of the CaMAL2 and CaGAL1 genes, encoding an α-glucosidase implicated in the utilization of sucrose and maltose (15) and galactokinase (30), respectively. Our results also suggest the existence of a CaMIG1 analogue.
MATERIALS AND METHODS
Yeast strains, growth, and transformation.
The following strains were used in this work: S. cerevisiae WDC-3A (MATa ade2-1 his3-11,15 ura3-1 leu2-1 trp1-1 tps1::HIS3) (2), S. cerevisiae H190 (MATa ade2 ura3 leu2 trp1 his3 can1 mig1::LEU2) (34), C. albicans SC5314 (17), C. albicans RM1000 (ura3::imm434/ura3::imm434 his1::hisG/his1::hisG) (32), C. albicans LOZ123 (ura3::imm434/ura3::imm434 his1::hisG/his1::hisG MIG1/mig1::HIS1) (this work), and C. albicans LOZ124 (ura3::imm434/ura3::imm434 his1::hisG/his1::hisG mig1::hisG-CaURA3-hisG/mig1::HIS1) (this work). The yeasts were grown with shaking at 30°C in 1% yeast extract–2% peptone (YP) or in a synthetic medium (Difco yeast nitrogen base) with adequate auxotrophic requirements. As carbon source, 2% glucose, galactose, sucrose, or ethanol, 3% raffinose or glycerol, or a mixture of 2% glucose plus 2% sucrose was added. For formation of hyphae, C. albicans strains were grown at 30°C in YP containing glucose until stationary phase and then shifted to the same medium containing 10% newborn calf serum (Gibco BRL) at 37°C. S. cerevisiae was transformed by the lithium acetate method (23), and C. albicans was transformed by electroporation (26).
Bacterial strains and plasmids.
Escherichia coli DH5α was used for transformations and preparation of plasmid DNA. Plasmids pUC18 (50) and pGEM-T (Promega) were used in E. coli, and YEp352 (20) was used for constructions in S. cerevisiae. A genomic library from C. albicans in vector YEp352 was provided by C. Nombela and J. Pla (Madrid, Spain). Plasmid pOZ2-20, containing the 5′ noncoding region and the entire open reading frame (ORF) of CaMIG1, was constructed as follows. A 1,071-bp fragment from the 3′ region of CaMIG1 (880 bp from the ORF and 191-bp noncoding region) was obtained by PCR using primers O2 5′CTTCAACTAGCCTATATTCCGATGG3′ and O8 5′-CTTTCTGTAGGTACCAACAACTAC3′ and plasmid pOZ2-14 (Fig. 1A) as the template. O8 introduces a KpnI site (underlined). The PCR product was digested with BglII and KpnI, and the 841-bp fragment containing 650 bp of the coding region and 191 bp of the 3′ noncoding region was subcloned in pOZ2-8 digested with the same enzymes. Plasmid pOZ2-8 is a derivative of pOZ2-2 (Fig. 1) which lacks the 600-bp XbaI-ClaI fragment and contains only the BglII site internal to the ORF. Plasmid pOZ7, containing the ScMIG1 gene, was constructed by cloning the 2.2-kb SacI-SacI fragment from pMIG1 (34) in the SacI site of YEp352.
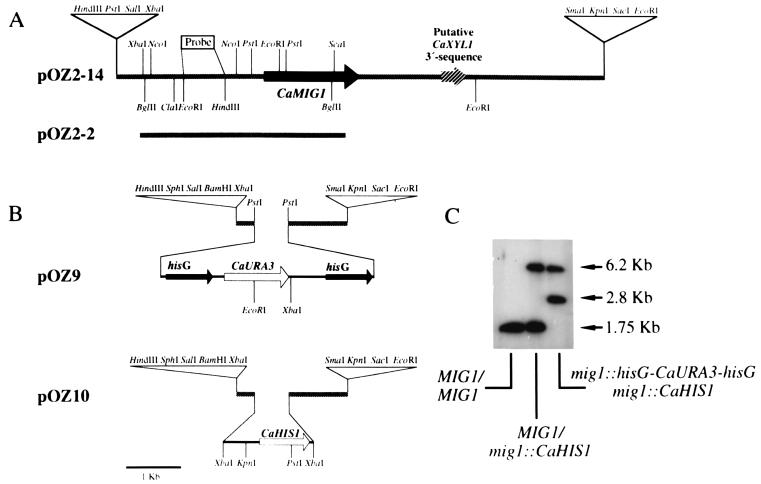
FIG. 1.
Map of the genomic region around CaMIG1, disruption strategy, and Southern blot of the CaMIG1 disruptants. (A) Restriction map of inserts of C. albicans DNA that contain entire or truncated versions of CaMIG1. The CaMIG1 gene and direction of transcription are indicated by the arrow in pOZ2-14. Plasmid pOZ2-2 lacks 288 bp of the 3′ region of the gene. pOZ2-14 and pOZ2-2 have identical polylinkers. Thick lines indicate genomic DNA from C. albicans. A region with high similarity with the 3′ regions of genes encoding xylose reductases is indicated without details of restriction sites. (B) Disruption of the CaMIG1 gene with CaURA3 and CaHIS1 (for details, see Materials and Methods). (C) Southern blot of the mig1/mig1 disruptants. Genomic DNA was digested with EcoRI. As probe, the 0.75-kb EcoRI-HindIII fragment indicated in panel A was used. Sizes of the bands are indicated at the right. Relevant genotypes of the strains used for the Southern analysis are also indicated.
DNA and RNA manipulations.
Recombinant DNA manipulations were done by standard techniques. DNA probes were labelled as described elsewhere (11). Genomic DNA was obtained as described previously (21). Total RNA from C. albicans was extracted from 50-mg (wet weight) samples with the Gibco BRL Trizol reagent (7). The RNA samples were heated at 65°C for 15 min, fractionated on 1.5% agarose gels containing 2.2 M formaldehyde, and transferred to a nylon membrane. rRNA in the membrane was visualized by staining with 0.02% methylene blue in 0.3 M sodium acetate and subsequent washing with 2 mM Na2HPO4 (pH 7.4)–30 mM NaCl–0.25 mM EDTA–1% sodium dodecyl sulfate.
The GAL1 probe used was isolated by PCR using the nucleotides 5′GGTAATGTACCTACAGGAGG3′ and 5′GCTGCTGGTCTGTGTCGTTG3′ as primers and C. albicans genomic DNA as the template.
Colony screening.
To screen the genomic library for DNA fragments with the complete CaMIG1 gene, E. coli was transformed with DNA from the library and the transformed cells were treated basically as described elsewhere (18). The probe used was a 300-bp BglII-EcoRI fragment from pOZ2-2 (Fig. 1A).
DNA sequencing.
Sequencing was performed by the dideoxy-chain termination method (42). Computer analyses were carried out with the Wisconsin Package (Genetics Computer Group) software on a Digital 5000/200 workstation.
Chromosomal disruption of the CaMIG1 gene.
To interrupt the CaMIG1 gene with CaURA3 and CaHIS1, we first constructed plasmid pOZ2-10, derived from a plasmid (pOZ2-9) developed for other purposes. To obtain pOZ2-9, plasmid pOZ2-2 (Fig. 1A) was digested with NcoI, blunt-ended, and digested with SacI; the resulting 2-kb fragment was ligated into YEp352 digested with SacI and SmaI. CaMIG1 was excised from pOZ2-9 as a 2-kb SacI-XbaI fragment and cloned into a derivative of pUC18 lacking the PstI site, to produce pOZ2-10. Disruption of CaMIG1 with CaURA3 was done as follows. A 4-kb BamHI-BglII fragment from pCUB6K1 (a derivative of pCUB6 [12]) containing a hisG-CaURA3-hisG cassette was cloned in the BamHI site of plasmid Sac-Kiss Lambda (46). The resulting plasmid was digested with PstI, and the 4-kb fragment containing the hisG-CaURA3-hisG region was inserted into pOZ2-10 digested with PstI to give plasmid pOZ9 (Fig. 1B). To disrupt CaMIG1 with CaHIS1, the 1.5-kb SmaI-SmaI fragment from p34HHIS1 (provided by J. Pla), containing the CaHIS1 gene, was subcloned into pOZ2-10 digested with PstI and blunt ended. The resulting plasmid was called pOZ10 (Fig. 1B). To integrate the disruptions in the genome of C. albicans RM1000, plasmids pOZ9 and pOZ10 were digested with SacI and HindIII, and the digestion products were introduced into the yeast by electroporation. Transformants were selected by growth in the absence of uracil and histidine. Correct insertion of the disruption cassette was checked by PCR using appropriate primers. Colonies producing the expected PCR pattern were checked by Southern analysis (Fig. 1C).
Band shift assays.
Band shift experiments were performed as described elsewhere (48). Nuclear extracts from S. cerevisiae were prepared as described previously (43). Total extracts from C. albicans were prepared as described previously (6). As labelled probe, we used an oligonucleotide that contains the Mig1 binding site located between positions −201 and −184 in the promoter of the FBP1 gene from S. cerevisiae (URS1FBP1) (31). When indicated, a 100-fold excess of unlabelled oligonucleotide was added as competitor to the incubation mixture. For nonspecific competition, we used an oligonucleotide that contains the UAS1FBP1 region from −432 to −415 (31). The probe was labelled with the Klenow fragment. A total of 40,000 cpm was used in each incubation.
Enzymatic assays.
α-Glucosidase was assayed spectrophotometrically in cell extracts with an enzymatic coupled system. The assay mixture, at pH 7, consisted of 50 mM imidazole, 50 mM KCl, 1 mM MgCl2, 0.4 mM NADP, 0.5 U each of glucose-6-phosphate dehydrogenase and hexokinase per ml, and the needed amount of extract. The reaction was started by the addition of 50 mM sucrose, and the increase in absorbance at 340 nm was monitored. Protein was determined by using the Pierce reagent.
Infectivity test.
Male Swiss CD-1 mice (specific pathogen free; Charles River), 6 weeks old and weighing approximately 25 to 30 g, were used. They were inoculated in the lateral caudal vein with 200 μl of a cell suspension containing the strain being tested (107 viable C. albicans cells/ml) and were monitored for 1 week.
Nucleotide sequence accession number.
The sequence obtained for CaMIG1 has been submitted to the EMBL databank and assigned accession no. AJ238242.
RESULTS
Isolation and characterization of the CaMIG1 gene.
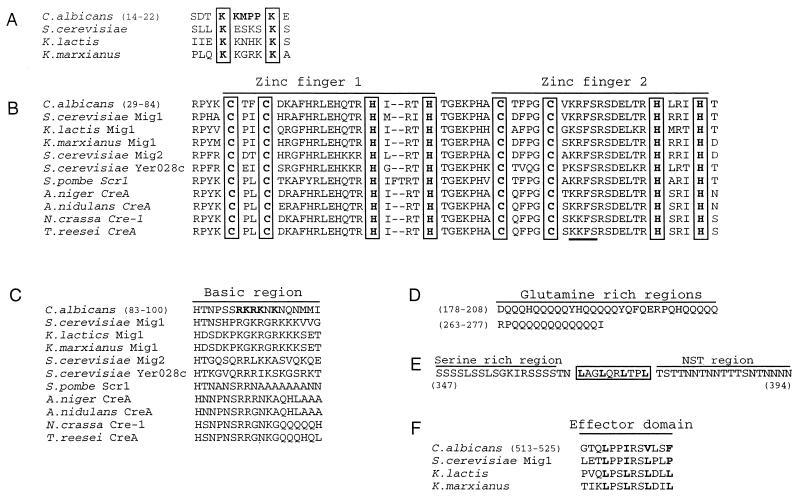
S. cerevisiae tps1 mutants, deficient in trehalose-6-phosphate synthase, do not grow in glucose (1). In a screen to isolate from C. albicans genes that complemented this phenotype (51), we isolated one plasmid, pOZ2-1, with a 7.7-kb insert of genomic DNA from C. albicans. A smaller insert of 3.8 kb (plasmid pOZ2-2) also complemented the growth defect of the tps1 mutant. The levels of trehalose in the yeast carrying this plasmid remained as low as in the original tps1 mutant, indicating that the plasmid carried a yeast DNA encoding a phenotypic suppressor of the tps1 mutation. The sequence of the C. albicans DNA in pOZ2-2 revealed similarity to the sequence of the ScMIG1 gene (34), but no putative stop codon was found. Moreover, it lacked a 3′ sequence that encodes a region of ScMIG1 apparently important for function (36). Therefore we screened the C. albicans library for a complete gene. In this way plasmid pOZ2-14 was isolated. This plasmid contained a complete ORF with similarity to ScMIG1 and also a fragment with high sequence similarity to the 3′ ends of genes encoding xylose reductase from other organisms (Fig. 1A). The similarity of sequence and the fact that a truncated version of ScMIG1 in single copy or, more efficiently, in multicopy suppresses the phenotype of a byp1-3 mutant, an allelic form of TPS1, in S. cerevisiae (22) made it likely that the cloned gene was the MIG1 gene from C. albicans. The nucleotide sequence revealed an ORF encoding a putative protein of 574 amino acids with a calculated molecular mass of 63 kDa. At positions 37, 941, and 1357 we found the triplet CTG, which usually encodes leucine but in C. albicans encodes serine. The amino acid sequence presents a series of characteristics that appear conserved among the Mig1 analogues from different organisms but shows some distinctive features that deserve consideration. The most characteristic feature is the existence of two zinc finger motifs of the type C2H2 located between positions 29 and 84, similar to those present in other analogues (Fig. 2B). Like in other proteins of this type, a putative phosphorylation sequence for the cyclic AMP-dependent protein kinase (KRFS) is located in the second zinc finger. Near the N terminus, the known Mig1 proteins present a basic region with two lysines separated by four amino acids. In the C. albicans protein this sequence is KMPPK, which appears in several proteins with presumed nuclear localization (19) (Fig. 2A). Close to the Zn fingers there is a region rich in basic amino acids, but CaMig1 has less positive charges than proteins of other yeasts (Fig. 2C). This region appears more similar to S. cerevisiae Mig2 and to the Mig1 homologues from Schizosaccharomyces pombe and other fungi. Another peculiarity of CaMig1 is the absence of a putative phosphorylation site for Snf1 that is found in the proteins of S. cerevisiae or Kluyveromyces lactis in the so-called regulatory domains (4). CaMig1 presents several stretches rich in glutamine, around amino acids 179 and 265, and in serine (around 347) or asparagine, serine, and threonine (NST region), around 380 to 400 (Fig. 2D). Although in S. cerevisiae stretches of polyglutamine and glutamine alternating with asparagine have been described (34), the frequency of glutamine residues is higher in the protein from C. albicans. Between the serine-rich and NTS regions, the sequence LAGLQRLTPL has a structure reminiscent of that proposed for the Mig1 effector domain at the C terminus (Fig. 2F). The position of a putative effector domain at the 3′ end also differs between the protein of C. albicans and those of other organisms. In S. cerevisiae and in two Kluyveromyces species, this effector domain, important for catabolite repression (36), is located within the 26 carboxy-terminal amino acids, whereas in CaMig1 this region is located 50 amino acids before the carboxy terminus. The 5′ upstream noncoding region of CaMIG1 did not show any conspicuous feature.
FIG. 2.
Multiple alignment of important regions of CaMig1. The numbers in parentheses indicate amino acid positions. Boldface letters highlight identities or differences in the sequences. (A) Basic residues in the N-terminal region. (B) Zinc finger regions. Cysteine and histidine residues are in boxes. A putative phosphorylation site for cyclic cAMP-dependent protein kinase is underlined. (C) Differences in the basic region. (D) The two glutamine-rich stretches. (E) Between the serine-rich and NTS regions, a potential effector domain with the sequence LxxLxxLxxL is boxed. (F) Effector domain. A putative effector domain in CaMig1 is situated 50 amino acids before the C-terminal amino acid.
Expression of the CaMIG1 gene.
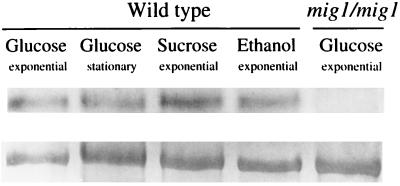
Since the capacity of Mig1 to repress transcription in S. cerevisiae varies depending on the growth conditions (44), we studied the expression of CaMIG1 in C. albicans grown in different media. As shown in Fig. 3, expression was not significantly affected by the carbon source or growth phase. RNA from a strain with both chromosomal copies of MIG1 disrupted did not show hybridization with the MIG1 probe (Fig. 3).
FIG. 3.
Expression of CaMIG1 during growth in different conditions. C. albicans SC5314 (wild type) and LOZ124 (mig1/mig1) were grown in the indicated carbon sources and harvested at the phases of growth shown; 15 μg of RNA was applied to each lane. The probe used was the 0.8-kb fragment PstI-EcoRI (the latter site from the polylinker) from pOZ2-2 (Fig. 1A). Bands in the lower row correspond to the 26S rRNA used as a control for the charge of RNA.
Effects of disruption of the CaMIG1 gene.
To determine the role of CaMIG1 in the physiology of C. albicans, we disrupted both chromosomal copies of the gene. The disruption was checked by Southern analysis (Fig. 1B and C; Materials and Methods), and additional proof for its correctness was provided by the results of the Northern analysis (Fig. 3). Growth rates of the wild type and the double disruptant in YP containing glucose were not significantly different (generation time of around 80 min). Also, no significant differences in growth rate were found when glycerol or galactose was used as a carbon source. When challenged with serum, the wild-type and mig1/mig1 strains formed hyphae in similar manners. Also, no significant changes in mortality were seen when mice were injected with the same amount of viable cells of a wild-type or a mig1/mig1 strain.
CaMIG1 is not required for repression of α-glucosidase or CaGAL1.
The most conspicuous effects of mig1 mutations in S. cerevisiae are seen in the pathways involved in sucrose (34) or galactose utilization (33). Therefore we examined the effects of CaMIG1 disruption in these pathways in C. albicans. Sucrose and maltose are utilized in C. albicans after hydrolysis by an α-glucosidase repressed by glucose (15). As shown in Table 1, the levels of α-glucosidase were strongly repressed by glucose, both in the wild type and in a CaMIG1-disrupted strain.
TABLE 1.
α-Glucosidase activity in C. albicans strains grown in different conditionsa
| Relevant genotype | Sp act (mU/mg of protein)
|
||
|---|---|---|---|
| Glucose | Glucose + sucrose | Sucrose | |
| MIG1/MIG1 | 2 | 2 | 400 |
| mig1/mig1 | 3 | 4 | 390 |
The yeasts were grown in YP with glucose, glucose plus sucrose, or sucrose and harvested at 15 to 20 mg/ml. α-Glucosidase activity was measured as described in Materials and Methods.
Expression of CaGAL1 was induced by galactose and repressed by glucose (Fig. 4). A low basal level of CaGAL1 mRNA was observed in glucose- or glycerol-grown cells. Disruption of CaMIG1 did not relieve significantly the repression by glucose.
FIG. 4.
Effect of disruption of CaMIG1 on the expression of CaGAL1. Total RNA was extracted (see Materials and Methods) from C. albicans SC5314 (wild type) or LOZ124 (mig1/mig1) grown in the indicated carbon sources and harvested at mid-log phase. The CaGAL1 probe was a 746-bp fragment isolated by PCR as described in Materials and Methods. The lower row shows bands corresponding to the 26S rRNA.
These results indicate either that expression of CaMAL2 and CaGAL1 is independent of CaMIG1 or that there is a functional analogue of it.
Functionality of CaMIG1 in S. cerevisiae.
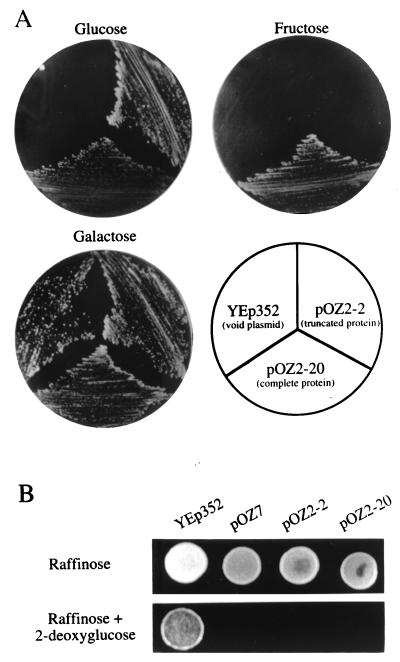
CaMIG1 appears to be functional in S. cerevisiae, as shown by its complementation of the tps1 mutation. We observed that a truncated form of CaMig1 lacking the 89 C-terminal amino acids (insert of plasmid pOZ2-2) complemented a tps1 mutant for growth on glucose but not for growth on fructose. Transformation of the tps1 mutant with plasmid pOZ2-20, carrying the complete gene, allowed growth on both sugars (Fig. 5A).
FIG. 5.
Functionality of truncated and complete versions of CaMIG1 in S. cerevisiae. (A) S. cerevisiae tps1 strain WDC-3A was transformed with plasmid pOZ2-2, carrying a truncated version of CaMIG1 encoding a protein lacking the 89 C-terminal amino acids, or with plasmid pOZ2-20, carrying a complete version of the gene, and spread on plates with glucose or fructose as the carbon source. As controls, growth of the tps1 mutant strain on galactose plates and of the same mutant transformed with a void plasmid are shown. (B) S. cerevisiae mig1 strain H190 was transformed with plasmid pOZ2-2 or pOZ2-20 and streaked on plates with 2% raffinose and with 2% raffinose–0.04% 2-deoxyglucose. As a control, the strain was transformed with plasmid pOZ7 carrying the complete ScMIG1.
To test more broadly the functionality of CaMIG1 in S. cerevisiae, we used two different approaches. In one, we examined the ability to restore catabolite repression of the SUC2 gene to an Scmig1 mutant. In the other, we observed whether CaMig1 was able to form specific complexes with the URS1FBP1 region of S. cerevisiae, which binds ScMig1 (31).
An Scmig1 mutant transformed with a plasmid carrying either the complete CaMIG1 gene or the truncated version did not grow on raffinose–2-deoxyglucose, indicating that in both cases, repression of SUC2 by the glucose analogue was restored (Fig. 5B). This result together with those for growth complementation of the tps1 mutant indicates that the C-terminal fragment is not absolutely required for all of its functions.
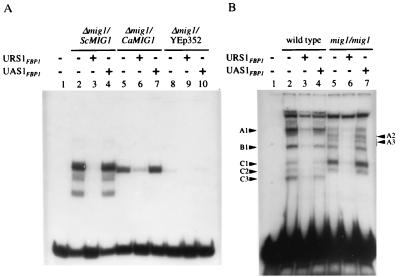
As shown in Fig. 6A, nuclear extracts from an S. cerevisiae strain expressing ScMIG1 formed one main specific complex and two weaker ones with the URS1 of ScFBP1. Nuclear extracts from an Scmig1 mutant transformed with CaMIG1 produced a major complex. This complex was specific, as shown by competition assays (Fig. 6A).
FIG. 6.
Band shift analysis using the URS1 fragment from the ScFBP1 promoter. (A) Five-microgram aliquots of protein of nuclear extracts from an Scmig1 mutant transformed with a void plasmid (YEp352) or the same plasmid carrying the ScMIG1 or CaMIG1 gene were used. (B) Fifteen-microgram aliquots of protein of total extracts from either C. albicans SC5314 (wild type) or LOZ124 (mig1/mig1) were applied to the gel. Labelling and experimental conditions were as described in Materials and Methods. The samples in lanes 1 were incubated without added protein. In lanes 3, 6, and 9, an excess of unlabelled URS1 was added; in lanes 4, 7 and 10, the DNA fragment UAS1FBP1 (see Materials and Methods) was used as the nonspecific competitor.
These results from two different approaches indicate that CaMIG1 is active in S. cerevisiae in a variety of functions.
Is there a functional CaMIG1 analogue?
If there is some analogue of CaMig1 in C. albicans, it could likely form specific complexes with the URS1 of ScFBP1 as CaMig1 does. As shown in Fig. 6B, in assays using extracts of wild-type C. albicans cells, five specific complexes are seen. Disruption of CaMIG1 did not abolish the formation of specific complexes but changed their pattern. The intensity of the complexes A1 and B1 decreased, that of C1 increased, and C3 disappeared; in contrast, two new complexes, A2 and A3, were formed. This result shows the existence in C. albicans of a protein able to bind to the same DNA sequence as CaMig1, which may function as a CaMig1 analogue.
DISCUSSION
We have isolated and characterized the gene encoding the DNA-binding protein Mig1 from the opportunistic pathogen C. albicans. This protein has been implicated in the glucose repression of several genes in S. cerevisiae (14) and in the repression of lactose metabolism in K. lactis (9). Although the sequence of the putative protein encoded by CaMIG1 bears an overall resemblance to those of the yeasts mentioned and other analogues from fungi (10), some particular characteristics merit consideration. One of them is the lack of a putative phosphorylation sequence for the protein kinase Snf1. In S. cerevisiae, Mig1 switches its localization between the nucleus and the cytoplasm in response to a phosphorylation (8) controlled by the protein kinase Snf1 (38, 45). The lack of a phosphorylation sequence for Snf1 in CaMig1 suggests that either its control is different or Mig1 has different roles in C. albicans than in S. cerevisiae. A peculiar characteristic is the existence of the sequence KMPPK, which is identical to one in the N-terminal region of hexokinase 2 of S. cerevisiae, involved in targeting the protein to the nucleus and instrumental in catabolite repression (19). Another sequence that could direct CaMig1 to the nucleus is HKKSR, found at position 428. In S. cerevisiae a similar motif, RKKSR, in position 364 is implicated in this function (M. Johnston, personal communication). However, in the case of CaMig1, deletion of this sequence did not reduce markedly the complementation of the growth on glucose of an S. cerevisiae tps1 mutant (unpublished results). Therefore, this sequence seems in this case to be not absolutely required for nuclear import.
We found that the expression of CaMIG1 was unchanged in different conditions, a result different from that reported for S. cerevisiae (28). Using a fusion of the ScMIG1 promoter to lacZ, these authors reported that the expression of MIG1 was decreased during growth in glucose in that yeast. This discrepancy could be due to differences in the regulation of MIG1 expression between the two species. To our knowledge there are no reports on levels of MIG1 mRNA in S. cerevisiae or on the expression of MIG1 in other yeasts.
Disruption of MIG1 in S. cerevisiae relieves glucose repression of the GAL genes (33) and partially relieves that of SUC2 (29) but has little or no effect on other genes whose promoters contain Mig1 binding sites (14). Disruption of CaMIG1 had no effect on the levels of the α-glucosidase-hydrolyzing sucrose in C. albicans or upon the expression of CaGAL1. An analysis of the promoters of these genes revealed that they do not present consensus sites for Mig1 binding. Expression of the K. lactis gene INV1, encoding invertase, is also insensitive to the functionality of the corresponding MIG1 (16). Expression of CaTPS1, which has a putative Mig1 binding site in its promoter (51), was also unaffected by the disruption of CaMIG1 (results not shown).
In S. cerevisiae, there are two other genes encoding homologues of Mig1: MIG2 and Yer028c (28, 29). SUC2 is repressed by both Mig1 and Mig2, but no role has been established for Yer028c (28). Our results of band shift experiments show that in C. albicans, proteins different from CaMig1 are able to bind to a Mig1 binding site, suggesting the existence of functional analogues of CaMIG1.
In S. cerevisiae, the Tup1-Cyc8 complex is recruited by Mig1 to repress glucose-sensitive promoters. Since in C. albicans disruption of TUP1 gives rise to filamentous growth, disruption of CaMIG1 could have produced a similar phenotype. However this was not the case, indicating that the filamentous phenotype of Catup1 mutants cannot be accounted for by a relief of inhibition by Mig1. Tup1 may therefore allow growth in the yeast form through its interaction with proteins different from CaMig1. In fact, in S. cerevisiae, Tup1 is also involved in Mig1-independent pathways (47).
It is not clearly understood how overexpression of MIG1 suppresses phenotypically the growth defects of the tps1 mutation in S. cerevisiae. In a tps1 mutant, an excessive flux in the initial steps of sugar utilization produces a metabolic imbalance and depletes the cell of ATP. Lack of inhibition of hexokinase by trehalose-6-phosphate is one important cause of this imbalance (2). Overexpression of MIG1 could decrease glucose influx by repressing some of the genes encoding glucose transporters (39). The differences in complementation for growth on glucose or fructose observed with a truncated and a full version of CaMIG1 contrast with the similar abilities of both versions for repression of SUC2. These differences indicate that the C-terminal region is important for some functions but dispensable for others. The importance of this region was highlighted by the observation that the 24 C-terminal amino acids of ScMig1 fused to a DNA-binding fragment could substitute for the Mig1 protein in S. cerevisiae (36). However, recent results indicate that mutations in this region decrease but do not abolish repression (37); another part of the protein must therefore provide the ability to block transcription. A potential candidate could be the region where a stretch of several leucines mimics the sequence in the effector domain (4, 37). In the case of CaMig1, a stretch with leucines is located around position 366 and could be the reason for the functionality of the truncated version in S. cerevisiae.
Although C. albicans SNF1 and MIG1 can complement snf1 and mig1 mutants in S. cerevisiae (reference 40 and this work), their roles in C. albicans are not completely equivalent to the roles of the corresponding analogues in S. cerevisiae. Thus, the ability of a regulatory protein to complement functions in heterologous organisms does not allow conclusions to be drawn directly about its mode of action in the original organism.
ACKNOWLEDGMENTS
We thank J. Pla and C. Nombela (Madrid, Spain) for strains and plasmids, M. A. Blázquez (La Jolla, Calif.) for critical reading of the manuscript, and Juana M. Gancedo for her interest in this work and help in composition of the manuscript.
This work was supported by grant PB97-1213-CO2-01 from the Spanish Dirección General de Enseñanza Superior e Investigación Científica. O.Z. had a fellowship from the Spanish PFPI, and C.R. had a fellowship from the Fundación Ramón Areces (Spain).
REFERENCES
- 1.Bell W, Klaasen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, van der Zee P, Wiemkem A. Characterization of the 56 kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem. 1992;209:951–959. doi: 10.1111/j.1432-1033.1992.tb17368.x. [DOI] [PubMed] [Google Scholar]
- 2.Blázquez M A, Lagunas R, Gancedo C, Gancedo J M. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993;329:51–54. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- 3.Bukhard R B, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–108. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 4.Cassart J P, Östling J, Ronne H, Vandenhaute J. Comparative analysis in three fungi reveals structurally and functionally conserved regions in the Mig1 repressor. Mol Gen Genet. 1997;255:9–18. doi: 10.1007/s004380050469. [DOI] [PubMed] [Google Scholar]
- 5.Celenza J L, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 6.Chaves R S, Herrero P, Moreno F. Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem Biophys Res Commun. 1999;254:345–350. doi: 10.1006/bbrc.1998.9954. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynscki P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.DeVit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Dickson R C. Glucose represses the lactose-galactose regulon in Kluyveromyces lactis through a SNF1 and MIG1-dependent pathway that modulates galactokinase (GAL1) expression. Nucleic Acids Res. 1997;25:3657–3664. doi: 10.1093/nar/25.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 12.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita A, Matsumoto S, Kuhara S, Misumi Y, Kobayashi H. Cloning of the yeast SFL2 gene: its disruption results in pleiotropic phenotypes characteristic for tup1 mutants. Gene. 1990;89:93–99. doi: 10.1016/0378-1119(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 14.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geber A, Williamson J H R, Sweeney E C, Bennett J E. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol. 1992;174:6992–6996. doi: 10.1128/jb.174.21.6992-6996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georis I, Cassart J P, Breuning K D, Vandenhaute J. Glucose repression of the Kluyveromyces lactis invertase gene KIINV1 does not require Mig1p. Mol Gen Genet. 1999;261:862–870. doi: 10.1007/s004380050030. [DOI] [PubMed] [Google Scholar]
- 17.Gillum A M, Tsay E Y, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 18.Grunstein M, Hogness D S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci USA. 1975;72:3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero P, Martínez-Campa C, Moreno F. The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 1998;434:71–76. doi: 10.1016/s0014-5793(98)00872-2. [DOI] [PubMed] [Google Scholar]
- 20.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. Gene. 1987;57:266–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 22.Hohmann S, Huse K, Valentín E, Mbonyi K, Thevelein J M, Zimmermann F K. Glucose-induced regulatory defects in the Saccharomyces cerevisiae byp1 growth inhibition mutant and identification of MIG1 as a partial suppressor. J Bacteriol. 1992;174:4183–4188. doi: 10.1128/jb.174.12.4183-4188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanai T, Ogawa K, Ueda M, Tanaka A. Expression of the SNF1 gene from Candida tropicalis is required for growth on various carbon sources, including glucose. Arch Microbiol. 1999;172:256–263. doi: 10.1007/s002030050768. [DOI] [PubMed] [Google Scholar]
- 25.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 26.Kohler G A, White T C, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutfiyya L L, Iyer V R, DeRisi J, DeVit M J, Brown P O, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magee B B, Koltin Y, Gorman J A, Magee P T. Assignment of cloned genes to the seven electrophoretically separated Candida albicans chromosomes. Mol Cell Biol. 1988;8:4721–4726. doi: 10.1128/mcb.8.11.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercado J J, Vincent O, Gancedo J M. Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 1991;291:97–100. doi: 10.1016/0014-5793(91)81112-l. [DOI] [PubMed] [Google Scholar]
- 32.Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology. 1997;143:297–302. doi: 10.1099/00221287-143-2-297. [DOI] [PubMed] [Google Scholar]
- 33.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilm's tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odds F C. Candida and candidosis. London, England: Baillière-Tindall; 1988. [Google Scholar]
- 36.Östling J, Carlberg M, Ronne H. Functional domains in the Mig1 repressor. Mol Cell Biol. 1996;16:753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Östling J, Cassart J P, Vandenhaute J, Ronne H. Four hydrophobic amino acid residues in the C-terminal effector domain of the yeast Mig1p repressor are important for its in vivo activity. Mol Gen Genet. 1998;260:269–279. doi: 10.1007/s004380050895. [DOI] [PubMed] [Google Scholar]
- 38.Östling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 39.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petter R, Chang Y C, Kwon-Chung K J. A gene homologous to Saccharomyces cerevisiae SNF1 appears to be essential for the viability of Candida albicans. Infect Immun. 1997;65:4909–4917. doi: 10.1128/iai.65.12.4909-4917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petter R, Kwon-Chung K J. Disruption of the SNF1 gene abolishes trehalose utilization in the pathogenic yeast Candida glabrata. Infect Immun. 1996;64:5269–5273. doi: 10.1128/iai.64.12.5269-5273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider R, Gander I, Müller V, Mertz R, Winnacker E L. A sensitive and rapid assay for nuclear factor I and other DNA-binding proteins in crude nuclear extracts. Nucleic Acids Res. 1986;14:1303–1317. doi: 10.1093/nar/14.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treitel M A, Carlson M. Repression of SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treitel M A, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang T C, Harris D T, Akporiaye E T, Schluter S F, Bowden G T, Hersh E H. Simple method for adapting DNA fragments and PCR products to all of the commonly used restriction sites. BioTechniques. 1996;20:51–52. doi: 10.2144/96201bm11. [DOI] [PubMed] [Google Scholar]
- 47.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 48.Vincent O, Gancedo J M. Analysis of positive elements sensitive to glucose in the promoter of the FBP1 gene from yeast. J Biol Chem. 1995;270:12832–12838. doi: 10.1074/jbc.270.21.12832. [DOI] [PubMed] [Google Scholar]
- 49.Williams F E, Trumbly R J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J M. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 51.Zaragoza O, Blázquez M A, Gancedo C. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J Bacteriol. 1998;180:3809–3815. doi: 10.1128/jb.180.15.3809-3815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]