Abstract
Background
Mother-to-child-transmission (MTCT) is the main route of HIV-1 infection in children. Genetic studies suggest HLA-B alleles play an important role on HIV-1 transmission, progression, and control of HIV-1 infection.
Objective
To evaluate which polymorphisms of HLA-B are involved in HIV-1 MTCT.
Methods
Two independent reviewers performed a systematic review on search engines PubMed, Europe PMC, Cochrane, Scientific Electronic Library Online (SciELO), and Literatura Latino-americana e do Caribe em Ciências da Saúde (Lilacs), using the following key terms: “HIV infection”, “HIV newborn”, “HLA polymorphisms”, “HLA-B”, and “Mother to child transmission”. All studies focusing on evaluation of HIV-1 MTCT, HIV infection evolution, and molecular analyses of HLA-B in children were selected.
Results
Nine studies fulfilled the inclusion criteria. Sixteen HLA-B alleles groups were associated with HIV-1 infection; seven of them (43.8%) were related to slow disease progression or reduced risk of MTCT, while six (37.5%) alleles groups were linked to a faster progression of HIV infection in children and to increased risk of MTCT. The available evidence suggest that HLA-B*57 group allele is associated with slow disease progression, while HLA-B*35 group allele is associated to increased risk of MTCT and rapid disease progression in infected children. The role of HLA-B*18, B*58 and B*44 are still controversial because they were associated to both, protection against MTCT, and to higher HIV replicative capacity, in different studies.
Conclusion
HLA-B*57 group allele can be protective against MTCT while HLA-B*35 groups alleles are consistently associated with HIV-1 MTCT.
Keywords: “Mother to child transmission”, “HIV”, “HLA-B”
Introduction
Mother-to-child-transmission (MTCT) is the main route of HIV-1 infection in children.1 Although the magnitude of maternal HIV-1 RNA plasma viral load is the main risk factor in MTCT, genetic characteristics play an important role in HIV-1 transmission to children born to HIV-infected mothers.2, 3
The role of HLA genes on the control of HIV-1 replication has been studied since 1996.4 Currently, there is great interest on variants of HLA genes codifying regions that can affect the host immune response to HIV infection. HLA alleles are responsible for antigen presentation to CD4+ and CD8+ T cells.5, 6, 7, 8, 9 Despite strong evidence on the important role of HLA-B polymorphisms on HIV-1 MTCT risk, the available data are often contradictory.10, 11, 12
The most recent studies have focused on HLA genes codifying regions that can affect functional and structural gene's properties, modifying their capacity of presenting antigens to T cells and, in consequence, the effectiveness of immune response.5, 6, 7, 8, 9 Cytotoxic T lymphocytes are believed to promote a better control of HIV infection in response to epitopes presented by HLA-B alleles, due to the relative resistance of this allele to Nef-mediated negative regulation.13 Much has been learned about HLA-mediated influences on the evolution of HIV with the MTCT. However, several points remain unresolved, and there are controversial results on the effects of HLA on HIV MTCT.14
HLA-B is the most polymorphic gene, with a total of 5212 alleles (by August, 2018, IMGT-HLA).5 Although there are several published studies on HLA-B alleles and their association with HIV progression in infected adults, there is a lack of information on their role in MTCT.15
The goal of this systematic review was to evaluate the association between HLA-B reported polymorphisms, HIV-1 MTCT rate, and the progression of HIV disease in children.
Methods
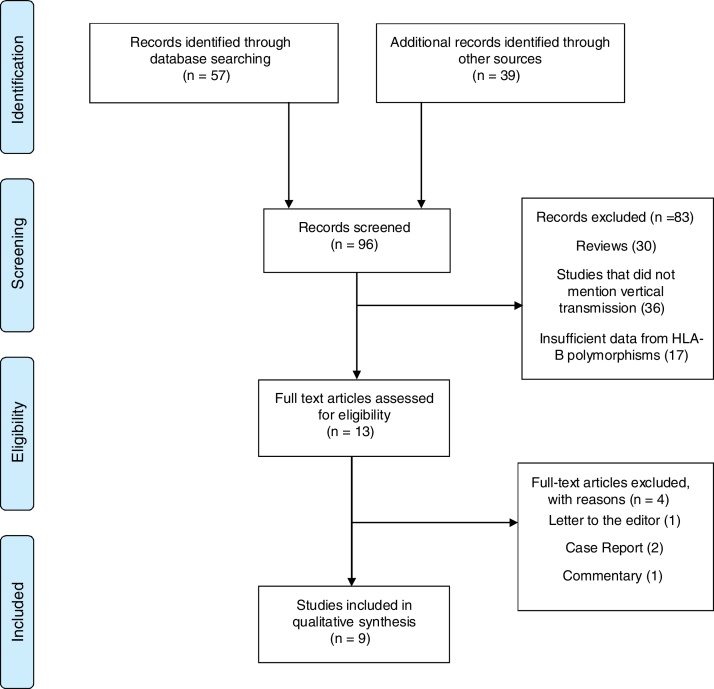
We performed a systematic review on HLA polymorphisms and their association with HIV-1 MTCT, according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) recommendations.16 Two independent reviewers (CAJM, CCT) performed the search in the following databases: PubMed (MEDLINE and EMBASE), Europe PMC, Cochrane, Scientific Electronic Library Online (SciELO), and Literatura Latino-americana e do Caribe em Ciências da Saúde (Lilacs). The key words used in the search were: “HIV infection”, “HIV newborn” as the problems, “HLA polymorphisms”, “HLA-B” as predictors, and “Mother to child transmission” as result. The search strategy in PUBMED is presented in supplementary file 1. All papers published in English, Spanish or Portuguese were searched, without time or study population restriction. Initially, titles and abstracts were read by reviewers (Fig. 1). Reviews, letters to the editor, and comments were not included, but were sources of additional references that did not appear in the first search.
Fig. 1.
Study selection of the current systematic review of the association of HLA-B and HIV mother-to-child transmission.
The inclusion criteria were: (1) cross-sectional, cohort, or case-control studies; (2) focusing on HIV-1 MTCT; (3) describing HLA-B polymorphisms; (4) using molecular methods (Restriction length fragment – RFLP, PCR-RFLP, specific sequence PCR- SSP, specific sequence of PCR oligonucleotides-SSO, PCR-Luminex, and PCR sequence-based typification-SBT) to evaluate HLA-B frequencies; and (5) comparing children exposed to HIV-1 during pregnancy or breast-feeding period. Papers that did not study HLA-B in HIV-1 MTCT, those duplicated or with no description of HLA-B polymorphisms were excluded.
All studies included maternal HIV-1 plasma RNA viral load (PVL) and CD4+ cell count as the main variables to evaluate the role of HLA-B on HIV infection and MTCT.
Data extraction was performed independently by two authors (Cubillos-Angulo JM and Castro-Cuesta T) and discrepancies between reviewers were solved by consensus. A table for data extraction was built by each reviewer before manuscript writing. The table included information on all relevant variables in each retrieved study, including socio-demographic data, time of maternal HIV diagnosis, duration of antiretroviral treatment (ART), use of peripartum prophylaxis, use of ART during pregnancy, type of delivery, presence of coinfections or other relevant comorbidity.
Results
Initially, 96 studies were selected in the main databases search. After detailed review, 83 were excluded: 30 reviews, 36 not related to MTCT, 17 had not enough information on HLA-B polymorphisms, or no sufficient data on HLA-B polymorphisms. In addition, four other studies were excluded for being case-reports,2 short communication,1 or letter to the editor.1, 17, 18, 19, 20 The remaining nine studies describing HLA-B polymorphisms in HIV-1 MTCT were included in this review. Fig. 1 summarizes the studies selection process.
Among the nine selected studies, three had a case-control design and six were cohort studies. In all studies, cases were HIV-1-infected children, and all studies were based on previously established cohorts of HIV-infected adults (both sexes). The studies were conducted in six different countries, three of them from South African cities (Johannesburg, Kimberley and Durban).
In six studies, the variables HIV-1 plasma RNA viral load (PVL) and CD4+ cell count, were evaluated only in mothers, two studies reported results for both children and mothers, and in a single study CD4+ count and PVL were evaluated only in children. In one study only HLA-B was evaluated, while in the remaining eight all class I HLA alleles were included in the evaluation, as shown in Table 1.
Table 1.
Summary and characteristics of the articles and results of the HLA-B alleles reported.
| Authors | Type of study | N | HLA-B alleles studied | Population | Main results |
|---|---|---|---|---|---|
| (Nkenfou, Nemes E, Mekue L, Grifoni A, Dambaya B, 2015)25 | Cohort | 28 = EU 34 = INF | HLA-B *44 | Cameroon | Associated with resistance to MTCT of HIV |
| (Adland et al., 2015)10 | Cohort | 47 = TR 84 = INF | HLA-B *45:01 HLA-B *18:01 HLA-B *58:02 | Kimberley, South Africa. | Viral replicative capacity was higher in children born to mothers expressing any of these alleles that increase risk of disease. |
| HLA-B *57 HLA-B *58:01 HLA-B *81:01 | A modest decrease in viral replicative capacity in children expressing these alleles. | ||||
| (Paximadis et al., 2011)39 | Cases and controls | 150 = NT 150 = EU 72 = INF 74 = TR | Homozygous for HLA-B1 | Johannesburg, South Africa | Greater representation in IP than in EU. |
| HLA-B *08:01 | Smaller representation in INF than in EU. | ||||
| HLA-B *14:02 | Greater representation in TR than in NT. | ||||
| HLA-B *42:01 | Smaller representation in TR than in NT. | ||||
| (Arnaiz-Villena et al., 2009)24 | Cases and controls | 63 = INF 57 = EU 31 = TR 36 = NT 175 = Controls | HLA-B *35 | Spain | Consistent association with allele expression and development of AIDS in children. |
| (Schneidewind et al., 2009)2 | Cohort | 13 = TR 13 = INF | HLA-B *57 | Jamaica Barbados Haiti | HLA-B expression confers a consistent benefit on viral control during childhood that is independent of parental inheritance (mothers or fathers). |
| (Thobakgale et al., 2009)11 | Cohort | 61 = TR 61 = INF 236 = NT | HLA-B *81:01 | Durban, South Africa | Slow progression of disease in INF, particularly when these protective HLA alleles were not shared with the mother. |
| HLA-B *58:01 | |||||
| HLA-B *57 | |||||
| (Winchester et al., 2004)12 | Cases and controls | 163 = NT 163 = EU 83 = INF 83 = TR | HLA-B *35:01 HLA-B *35:03 | Prospective Multicentric Cohort WITS | These alleles expression was more frequent among TR than in NT. Increased risk of transmission was found mainly among cases with low viral loads. |
| HLA-B *44:02 | Increased transmission rates among African-American and Hispanic mothers. | ||||
| HLA-B *14:02 | HLA-B*14:02 expression was associated with increased MTCT rate. | ||||
| HLA-B *13:02 | HLA-B *13:02 expression was associated with increased MTCT. | ||||
| HLA-B *50:01 | This allele expression was predominant among Hispanic mothers, and was associated with increased transmission in this ethnic group. | ||||
| HLA-B *49:01 HLA-B *53:01 | Lower risk of vertical transmission of HIV-1 in mothers with high viral loads. | ||||
| (Kuhn et al., 2004)22 | Cohort | 59 = INF 59 = TR | Homozygous for HLA-B1 | New York | Patients expressing this allele were three times more likely to develop AIDS or death in children. |
| HLA-B *27 HLA-B *57 | These alleles expression was associated with lower risk of AIDS or death when they were inherited from the father. | ||||
| (Farquhar et al., 2004)26 | Cohort | 76 = INF 357 = EU | HLA B *18 | Kenia, Nairobi | The allele expression was protective against early acquisition of HIV-1. No infants expressing the allele have acquired HIV-1 after one month of birth, suggesting that they may protect against late infection through breastfeeding. |
1, in this study, homozygotes were considered for the two HLA-B alleles equal to those of the mother.
NT, HIV (+) mothers who did not transmit HIV (+); EU, children not infected by HIV (+) from Mothers; INF, children infected with HIV (+) from mothers; TR, HIV (+) mothers who transmitted HIV (+) to their children; IP, HIV infected children (+) intrapartum.
In this review we detected 16 alleles groups significantly associated with risk of HIV MTCT and/or with progression of disease in HIV-infected children (Table 1). HLA-B homozygosis was assumed as one allele group, HLA-B*57 allele was the most frequent allele showing a protective effect against the risk for HIV infection in children. This protective effect was detected in four different studies.10, 11, 21, 22 Four alleles groups (HLA-B*27, B*57, B*58, B*81) were significantly associated with slower progression of HIV infection in children while six alleles groups (HLA-B*8, B*18, B*42, B*44, B*49, B*53) were associated with reduced risk of HIV-1 MTCT (Table 1). HLA-B*53:01 allele was associated with reduced risk of HIV-1 MTCT in the study by Winchester et al., but was also associated to rapid disease progression in the study by Gao et al.12, 23
On the other hand, five alleles groups (HLA-B*18, B*35, B*45, B*58, B*homozygosis) were related to rapid HIV progression in children, and six alleles groups (HLA-B*13, B*14, B*35, B*44, B*50, B*homozygosis) were associated with increased risk of MTCT (37.5%). The HLA-B*35 allele was associated with both, increased risk of MTCT and rapid progression to AIDS in HIV-infected children.24
Controversial evidence was found for three out 16 alleles groups: HLA-B*58:02 allele was associated with rapid HIV progression in the study by Adland et al.10; however HLA-B *58:01 allele was associated with slower progression of HIV infection in children in the studies by Adland et al. and Thobakgale et al.10, 11 HLA-B*44 allele group was linked to lower risk of HIV infection by Nkenfou et al.,25 while Winchester et al. detected a higher risk of HIV-1 MTCT in the presence of HLA-B*44:02 allele.12 HLA-B*18 allele was associated with protection against early HIV-1 infection in the study by Farquhar, while26 Adland detected association between HLA-B*18 and increased HIV-1 replication capacity.10
Discussion
HLA-B polymorphisms present evolutionary relationships and differences directly associated to epitopes processing and peptides linkage.27 This characteristic leads to induction of T and NK cells response to pathogens with distinct specificities.7 Knowing the associations between HLA-B polymorphic variants and HIV-1 MTCT can add to the understanding of the pathogeny of HIV-1 infection. This systematic review showed that there are 16 HLA-B alleles groups related to HIV-1 MTCT, the main cause of HIV infection in children.28
Eight out of nine included studies performed molecular analysis of all class I HLA alleles and found stronger associations between MTCT and HLA-B allelic groups. According to Carrington and Walker the specificity in HIV-1 control is determined by polymorphisms in the nucleotides sequence that codify the antigen ligation region. Once HLA-B is the most polymorphic alleles in class I HLA, its amino acids have a stronger association to HIV-1 control than any other HLA group.8
Our systematic review identified a significant association between HLA-B*53:01 and a lower risk of HIV-1 MTCT in mothers presenting high HIV-1 plasma viral load.12 However, Gao et al. demonstrated that adult patients presenting with HLA-B*53:01 allele had a faster progression to AIDS than patients without it.23 Apparently, HLA-B*53:01 allele is protective against HIV-1 MTCT, although involved in progression to AIDS in a similar way to that of HLA-B*35 allele, since they are phylogenetically related.23
In addition, this review showed an association between alleles HLA-B *35:01 or HLA-B *35:03, with increased risk of HIV-1 MTCT and faster disease progression in children.24 The available evidence demonstrates that HLA-B*35 group allele expression is associated to faster AIDS progression, probably because it is not able to establish a ligation to HIV-1 peptides, failing in building a protective immune response.29, 30
On the other hand, slower progression to AIDS and minimal impact on CD4+ cells count are consistently associated with HLA-B*57 expression.10, 11, 21, 22 According to Košmrlj, thymic T-cells restriction caused by HLA molecules like HLA-B*57 present less auto-peptides and, in consequence, are more efficient in recognizing point mutations in viral peptides.31
Some associations between specific alleles and MTCT are not detected in distinct geographic regions, because HLA alleles profiles are different, and association between MTCT and HLA alleles could vary in different populations, as concluded by Zhang et al.32 For instance, Nkenfou et al. detected association between expression of B*44 allelic group and resistance against HIV infection in Cameroonian children.25 However, Winchester et al. detected association between HLA-B*44:02 allelic and increased risk of MTCT in Afro-American and Hispanic mothers in Puerto Rico and United States.12
The available evidence allowed us to conclude that both, HLA-B*57 and HLA-B*81, can be grouped as classic protective alleles against MTCT. The HLA-B*57 allele was associated to lower HIV-1 plasma viremia in several studies, in different populations, and slower HIV disease progression,33, 34 while HLA-B*81 allele was associated to lower decrease in CD4+ cells count in HIV-infected patients.35
On the other hand, HLA-B*35 allele can be placed in the group of HLA alleles that increase the risk of HIV-1 MTCT, although it is also associated to lower HIV-1 plasma viral load in infected mothers.12
Some HLA-B*35 subtypes can be included in two major groups, according to their ability to link to peptides, B*35Py e B*35Px alleles.36 B*35-Px group expression is associated to a faster progression to AIDS and greater decline of CD4+ cells count, and includes the B*35:03 subtype.37 It is also important to note that HLA homozygous mothers presented higher HIV-1 plasma viral load, a clear risk factor to both, MTCT and faster AIDS progression in children. It is consistently reported the association between mothers’ HLA homozygosis and these poor outcomes, regardless of alleles’ type, or mothers’ HIV-1 plasma viral load, as a result of poor alloimmune response in children and consequent prolonged survival of HIV-infected cells.38 The available data reinforce that HLA-B coincidence of mothers and children increases the risk of in uterus HIV infection, due to the reduced ability of children's immune system in recognizing HIV-1 through maternal immune response, which results in lower likelihood of alloimmune response.24, 25, 38, 39
Sharing the same HLA-B subtypes of mothers decrease the T-lymphocytes cytotoxic ability, since the cells surface molecules are not recognized as self-ones.40 Mapping HLA-C, E, G, and HLA-B alleles in trophoblast could identify children at higher risk of MTCT, as described by Meuleman et al.40
Some limitations of included studies are also clear: studies on the association of HLA allele frequency and specific diseases are sometimes hard to replicate, due to variations in different populations and HLA polymorphisms.22 Adland et al. also pointed out that distinct age range of children included as faster or slower progressions contributed to limit the generalization of results.10
In this review, a clear limitation is the concentration of studies (53%) in African populations, making it difficult to extend the conclusions to other population groups. The small sample size of the majority of studies also limits the reach of our conclusions and does not allow us to perform a meta-analysis.
However, taken together, the data obtained in this review provide a greater insight on the genetic mechanisms involved in HIV-1 MTCT, especially its association with HLA-B allele expression. Our findings suggest that HLA-B polymorphisms can impact the risk of MTCT and the evolution of HIV-1 infection in children. Larger studies in different populations are required to shed more light on the genetic factors involved in MTCT.
Conclusions
This systematic review showed a significant association between HLA-B alleles expression and HIV-1 MTCT, with at least 16 alleles groups linked to HIV-1 transmission and disease progression in HIV-infected children.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Fouda G.G., Moody M.A., Permar S.R. Antibodies for prevention of mother-to-child transmission of HIV. Curr Opin VIH SIDA. 2015;2:177–182. doi: 10.1097/COH.0000000000000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy J.A. In: 3rd ed. Levy J.A., editor. vol. 1. Address Editorial; 2007. (HIV and the pathogenesis of AIDS). 680 p. [Google Scholar]
- 3.Milligan C., Overbaugh J. The role of cell-associated virus in mother-to-child HIV transmission. J Infect Dis. 2014;210(Suppl 3):S631–S640. doi: 10.1093/infdis/jiu344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaslow R.A., Carrington M., Apple R., et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 5.Goulder P.J.R., Watkins D.I. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiepiela P., Leslie A.J., Honeyborne I., et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–774. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 7.Raghavan M., Geng J. HLA-B polymorphisms and intracellular assembly modes. Mol Immunol. 2015;68:89–93. doi: 10.1016/j.molimm.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrington M., Walker B.D. Immunogenetics of spontaneous control of HIV. Annu Rev Med. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jan Klein A.S. The HLA system. Nejm. 2000;343:702–709. doi: 10.1056/NEJM200009073431006. [DOI] [PubMed] [Google Scholar]
- 10.Adland E., Paioni P., Thobakgale C., et al. Discordant impact of HLA on viral replicative capacity and disease progression in pediatric and adult HIV infection. PLoS Pathog. 2015;11:1–26. doi: 10.1371/journal.ppat.1004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thobakgale C.F., Prendergast A., Crawford H., et al. Impact of HLA in mother and child on disease progression of pediatric human immunodeficiency virus type 1 infection. J Virol. 2009;83:10234–10244. doi: 10.1128/JVI.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winchester R., Pitt J., Charurat M., et al. Mother-to-child transmission of HIV-1: strong association with certain maternal HLA-B alleles independent of viral load implicates innate immune mechanisms. J Acquir Immune Defic Syndr. 2004;36:659–670. doi: 10.1097/00126334-200406010-00002. [DOI] [PubMed] [Google Scholar]
- 13.Rajapaksa U.S., Li D., Peng Y.-C., McMichael A.J., Dong T., Xu X.-N. HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc Natl Acad Sci. 2012;109:13353–13358. doi: 10.1073/pnas.1204199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kløverpris H.N., Leslie A., Goulder P. Role of HLA adaptation in HIV evolution. Front Immunol. 2016;6 doi: 10.3389/fimmu.2015.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulder P.J.R., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberati A., Altman D.G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Child L., Feeney M.E., Tang Y., et al. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-L. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackelprang R.D., Carrington M., John-Stewart G., et al. Maternal human leukocyte antigen A*2301 is associated with increased mother-to-child HIV-1 transmission. J Infect Dis. 2010;202:1273–1277. doi: 10.1086/656318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feeney M.E., Tang Y., Rathod A., Kneut C., McIntosh K. Absence of detectable viremia in a perinatally HIV-1-infected teenager after discontinuation of antiretroviral therapy. J Allergy Clin Immunol. 2006;118:324–330. doi: 10.1016/j.jaci.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Goulder P.J.R., Brander C., Tang Y., et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 21.Schneidewind A., Tang Y., Brockman M.A., et al. Maternal transmission of human immunodeficiency virus escape mutations subverts HLA-B57 immunodominance but facilitates viral control in the haploidentical infant. J Virol. 2009;83:8616–8627. doi: 10.1128/JVI.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn L., Abrams E.J., Palumbo P., et al. Maternal versus paternal inheritance of HLA class I alleles among HIV-infected children: consequences for clinical disease progression. Aids. 2004;18:1281–1289. doi: 10.1097/00002030-200406180-00006. [DOI] [PubMed] [Google Scholar]
- 23.Gao X., Nelson G.W., Karacki P., et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 24.Arnaiz-Villena A., Martín-Villa J.M., Amador J.T.R., et al. Risk of vertical HIV transmission combines the “B35-Cw4 disadvantage” and the “pattern of inheritance” theories of progression. Curr HIV Res. 2009;7:314–319. doi: 10.2174/157016209788348029. [DOI] [PubMed] [Google Scholar]
- 25.Nkenfou, Nemes E., Mekue L., et al. Human leucocyte antigen class I diversity among human immunodeficiency virus exposed negative and positive children in Cameroon. J AIDS Clin Res. 2015;06 [Google Scholar]
- 26.Farquhar C., Rowland-Jones S., Mbori-Ngacha D., et al. Human leukocyte antigen (HLA) B*18 and protection against mother-to-child HIV type 1 transmission. AIDS Res Hum Retroviruses. 2004;20:692–697. doi: 10.1089/0889222041524616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1–15. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UNAIDS . Jt United Nations Program HIV/AIDS; 2017. UNAIDS data 2017; pp. 1–248. [Google Scholar]
- 29.O’Brien S.J., Gao X., Carrington M. HLA and AIDS: a cautionary tale. Trends Mol Med. 2001;7:379–381. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- 30.Carrington M., O’Brien S.J. The influence of HLA genotype on AIDS*. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 31.Košmrlj A., Read E.L., Qi Y., et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Huang X.J., Xia W., et al. HLA-B*44 is associated with a lower viral set point and slow CD4 decline in a cohort of Chinese homosexual men acutely infected with HIV-1. Clin Vaccine Immunol. 2013;20:1048–1054. doi: 10.1128/CVI.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coloccini R.S., Dilernia D., Ghiglione Y., et al. Host genetic factors associated with symptomatic primary HIV infection and disease progression among argentinean seroconverters. PLOS ONE. 2014;9:1–10. doi: 10.1371/journal.pone.0113146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Price M.A., He D., et al. Host genetics and viral load in primary HIV-1 infection: clear evidence for gene by sex interactions. Hum Genet. 2014;133:1187–1197. doi: 10.1007/s00439-014-1465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prentice H.A., Porter T.R., Price M.A., et al. HLA-B*57 versus HLA-B*81 in HIV-1 infection: slow and steady wins the race? J Virol. 2013;87:4043–4051. doi: 10.1128/JVI.03302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur G., Mehra N. Genetic determinants of HIV-1 infection and progression to AIDS: immune response genes. Tissue Antigens. 2009;74:373–385. doi: 10.1111/j.1399-0039.2009.01337.x. [DOI] [PubMed] [Google Scholar]
- 37.Gao X., Bashirova A., Iversen A.K.N., et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 38.Mackelprang R.D., John-Stewart G., Carrington M., et al. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J Infect Dis. 2008;197:1156–1161. doi: 10.1086/529528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paximadis M., Minevich G., Winchester R., et al. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meuleman T., Lashley L.E.L.O., Dekkers O.M., van Lith J.M.M., Claas F.H.J., Bloemenkamp K.W.M. HLA associations and HLA sharing in recurrent miscarriage: a systematic review and meta-analysis. Hum Immunol. 2015;76:362–373. doi: 10.1016/j.humimm.2015.02.004. [DOI] [PubMed] [Google Scholar]