Abstract
Introduction
Staphylococcus spp. – both S. aureus, including methicillin-resistant strains (MRSA) and coagulase negative staphylococci (CoNS) – are relevant agents of healthcare-associated infections. Therefore, the rapid recognition of MRSA and methicillin-resistant CoNS from blood stream infections is critically important for patient management. It is worth noting that inappropriate empiric therapy has been associated with higher in-hospital mortality.
Material and methods
In this study we evaluated a multiplex polymerase chain reaction (multiplex PCR) standardized to detect Staphylococcus spp., S. aureus, and mecA gene-encoded oxacillin resistance directly from blood culture bottles. A total of 371 blood cultures with Gram-positive microorganisms confirmed by Gram-stain were analyzed. Results from multiplex PCR were compared to phenotypic characterization of isolates.
Results
Staphylococcus aureus was detected in 85 (23.0%) blood cultures and CoNS in 286 (77.0%). There was 100% agreement between phenotypic and multiplex PCR identification. Forty-three (50.6%) of the 85 S. aureus carried the mecA gene and among the 286 CoNS, 225 (78.7%) were positive for the mecA gene.
Conclusions
The multiplex PCR assay developed here was found to be sensitive, specific, rapid, and showed good agreement with the phenotypic results besides being less expensive. This PCR method could be used in clinical laboratories for rapid identification and initiation of specific and effective treatment, reducing patient mortality and morbidity. Furthermore, this method may reduce misuse of antimicrobial classes that are more expensive and toxic, thus contributing to the selection of antibiotic-resistant Staphylococcus spp.
Keywords: Staphylococcus spp., Staphylococcus aureus, MRSA, mecA gene, Blood cultures, Multiplex PCR
Introduction
Sepsis is the systemic response to infection and is the result of the complex interaction between the host's immune system and the infecting microorganism.1 Many bacteria are isolated from bloodstream infections, including Gram-positive and Gram-negative bacteria. According to the US Centers for Disease Control and Prevention (CDC),2 a clear change in the nature of the infectious microorganisms has occurred over the past decade, when Gram-positive cocci have exceeded Gram-negative bacilli as the main etiological agents of sepsis and Staphylococcus spp. have become the most prevalent agents.
The genus Staphylococcus comprises 52 species3; of these, 17 can be isolated from human biological samples. The genus can be divided into two main groups: coagulase-positive staphylococci, whose main member is S. aureus, and coagulase-negative staphylococci (CoNS). Staphylococcus aureus possesses a wide variety of virulence factors, including peptidoglycan, teichoic acid, enzymes and toxins, adhesion molecules, and four types of cytotoxins.4 They can cause different types of infection, many of them acquired in the hospital environment. About 20% to 30% of the human population is colonized with this bacterium and the nasal mucosa is the main ecological niche.5
A relevant factor that complicates treatment of staphylococcal infections is the high resistance rate to β-lactam antibiotics observed among Staphylococcus spp., which requires large-scale use of expensive or toxic antibiotics. Staphylococcus spp. resistance to β-lactam antibiotics is mainly due to two different mechanisms which, however, can interact. The first mechanism consists of the production of β-lactamase, an enzyme that hydrolyzes the antibiotic. The second mechanism is related to a change in the site of action of β-lactams by producing a new penicillin-binding protein, PBP2a, which has low affinity for antibiotics and is absent in susceptible staphylococci.6 PBP2a is encoded by the mecA gene. This gene is located on a mobile genetic element called SCCmec (staphylococcal cassette chromosome mec), which is widely distributed among staphylococci.7 PBP2a acts as a transpeptidase that resumes cell wall synthesis functions when other PBPs are inhibited, thus ensuring the integrity of the bacterial cell in the presence of β-lactams.8
In view of the above considerations, rapid detection of bacteremia and the subsequent rapid identification of the bacteria involved and their susceptibility to antibiotics are of great diagnostic and prognostic importance. Multiplex polymerase chain reaction (PCR) allows to amplify genes of multiple pathogens and more than one specific DNA sequence of each pathogen using multiple sets of primers in a single reaction. This technique permits rapid detection and is effective in identifying pathogenic microorganisms in clinical samples, especially those that require urgent initiation of treatment.
The aim of the present study was to evaluate the efficacy, accuracy and sensitivity of multiplex PCR in detecting Staphylococcus spp., S. aureus, and mecA gene-encoded oxacillin resistance directly from blood culture bottles.
Material and methods
Isolates
A total of 371 Staphylococcus spp. isolated from positive blood cultures between May 2011 and May 2012 were provided by the Laboratory of Microbiology at the Teaching Hospital of Botucatu Medical School, UNESP - Univ Estadual Paulista (City of Botucatu, São Paulo State, Brazil).
Sample collection
The isolates included in the study were obtained on the day following growth detection in a Bactec 9120 system and after confirmation of containing probable Staphylococcus spp. by Gram staining (Gram-positive cocci) and positive catalase tests.
The blood culture bottles were disinfected with 70% alcohol and 3 mL of blood was collected with a 5 mL syringe and 70 × 25-mm needle into two microtubes (1.5 mL per microtube). One microtube was used for seeding onto blood agar for phenotypic identification and the other microtube was used for extraction of bacterial DNA for multiplex PCR.
Phenotypic identification of S. aureus and coagulase-negative staphylococci
Phenotypic identification was performed at the Laboratory of Bacteriology, Department of Microbiology and Immunology, Botucatu Biosciences Institute, UNESP. The samples were seeded into blood agar and incubated for 24 h at 37 °C. After isolation from blood agar, the bacteria were submitted to Gram staining for confirmation of morphological staining characteristics, catalase test for characterization of the genus Staphylococcus, and coagulase test for differentiation between S. aureus and CoNS.
DNA extraction directly from blood culture
Before DNA extraction, the samples were treated to remove interfering elements in the blood culture. The sample was centrifuged at 850 × g for 2 min.9 The supernatant was transferred to another microtube and again centrifuged at 11,000 × g for 1 min. The supernatant was carefully discarded, leaving only the pellet. Five hundred microliters of lysozyme buffer (1 M EDTA, 0.1 M NaCl, 10 mM Tris, pH 8.0, and 5% Triton X-100) was then added to the pellet and the mixture homogenized in a vortex. Next, 800 μL benzyl alcohol (Sigma) was added, the mixture homogenized in a vortex, and the microtube centrifuged at 7000 × g for 5 min.10 Two phases formed in the microtube, the lower phase contained the alcohol with interfering substances (inhibitors) and the upper phase the bacteria.
For DNA extraction, 320 μL of the liquid was removed from the upper phase and transferred to another microtube and 10 μL lysozyme was added (10 mg lysozyme in 1 mL 10 mM Tris–HCl). The microtube was homogenized in a vortex and incubated at room temperature for 15 min, homogenizing the microtube at five-minute intervals. After this period, 10 μL proteinase K was added and the mixture was incubated for 15 min at 56 °C, homogenizing the microtube at five-minute intervals in a vortex. The liquid (330 μL) was transferred to a silica column provided by the Illustra extraction kit (GE Healthcare). This column was placed inside a tube and centrifuged at 11,000 × g for 1 min. Lysis solution (500 μL) was added and the column centrifuged at 11,000 × g for 1 min. Next, 500 μL washing solution was added and the column was again centrifuged at 11,000 × g for 3 min. The tube was discarded and the column was transferred to a microtube. Elution solution (200 μL) pre-warmed to 70 °C in water bath was then added to the column. After incubation at room temperature for 1 min, the microtube was centrifuged at 11,000 × g for 1 min and the DNA obtained was stored in a freezer at −20 °C.
Antimicrobial susceptibility testing
The isolates were submitted to in vitro susceptibility testing by the disk diffusion method on Mueller-Hinton agar plates at the Laboratory of Microbiology, Department of Internal Medicine, University Hospital, Botucatu Medical School, UNESP, according to the Clinical and Laboratory Standards Institute criteria.11
Standardization of multiplex PCR for the detection of Staphylococcus spp., S. aureus, and mecA gene
First, PCR was standardized using a range of annealing temperatures to establish the adequate annealing temperature for all primers (Table 1). Amplification was performed in an Eppendorf thermocycler. The amplification conditions were: 92 °C for 3 min, followed by 30 cycles of DNA denaturation at 92 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 3 min. The final reaction volume was 35 μL containing 10.2 μL autoclaved Milli-Q water, 3.5 μL deoxyribonucleotide triphosphates (dNTP) (Life Technologies), 0.8 μL Taq DNA polymerase (Biotools), 2.5 μL MgCl2-free buffer (Biotools), 2.5 μL MgCl2 (Biotools), 0.14 μM of each 16S primer, 0.86 μM of each MRS primer, 0.35 μM of each SAU primer, 0.57 μM of each COA primer, and 3.5 μL of bacterial DNA. The amplification products were analyzed by electrophoresis on 3% agarose gel at 70 V. The following controls were included in all amplification reactions: ATCC 33591 (mecA-positive S. aureus), ATCC 25923 (mecA-negative S. aureus), ATCC 12228 (mecA-negative S. epidermidis), and negative control (water).
Table 1.
Primers used for the detection of Staphylococcus spp., S. aureus and the mecA gene by multiplex PCR.
| Gene | Primer | Amplicon (bp) |
|---|---|---|
| S. aureus24 | SAU327 – GGA CGA CAT TAG ACG AAT CA SAU1645 – CGG GCA CCT ATT TTC TAT CT |
1250 |
| 16S rRNA25 | 16S1 – 5′ CCTATAAGACTGGGATAACTTCGGG 3′ 16S2 – 3′ CTTTGAGTTTCAACCTTGCGGTCG 5′ |
791 |
| Coa26 | COA1 – 5′ GTA GAT TGG GCA ATT ACATTT TGG AGG 3′ COA2 – 5′ CGC ATCAGC TTT GTT ATC CCA TGT A 3′ |
117 |
| mecA27 | MRS1 – 5′ AAAATCGATGGTAAAGGTTGGC 3′ MRS2 – 3′ AGTTCTGCAGTACCGGATTTGC 5′ |
533 |
Sensitivity test
The sensitivity of multiplex PCR was evaluated by amplifying serial dilutions of S. aureus and CoNS control strains, starting with a dilution of 10−1 (corresponding to 107 CFU/mL) until 10−7 (10 CFU/mL) based on a 0.5 McFarland standard (1.5 × 108 CFU/mL).
Specificity test
The specificity of multiplex PCR was evaluated by PCR using strains of other microbial species isolated directly from a culture and from positive blood culture bottles containing other bacterial species. The following species were analyzed: Enterococcus faecalis, Enterococcus faecium, Streptococcus viridans, Streptococcus pneumoniae, Candida spp., Gram-positive bacilli, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Enterobacter cloacae, Enterobacter aerogenes, Salmonella spp., Morganella morganii, and Proteus mirabilis.
Detection of β-lactamase hyperproduction and resistance profile confirmation
The isolates not carrying mecA gene although resistant to oxacillin by the disk diffusion test were submitted to the fowling tests to confirm if they were β-lactamase hyperproducers, and to confirm the resistance profile: nitrocefin disk test for β-lactamase hyperproduction; simplex PCR for mecA gene detection, cefoxitin by disk diffusion, and amoxicillin/clavulanic acid disk test for resistance confirmation.
Statistical analysis
The kappa statistic was calculated to assess agreement between the methods used for the identification of Staphylococcus sp. A level of significance of 5% was adopted for all tests. Statistical analyses were performed using SSPS software version 19.0 (SSPS Inc., Chicago, IL).
Results
Phenotypic identification and susceptibility tests
Identification by the phenotypic method detected 85 (23.0%) S. aureus strains and 286 (77.0%) CoNS strains in the 371 blood cultures analyzed. The disk diffusion test revealed oxacillin resistance in 43 (50.6%) S. aureus isolates and in 236 (82.5%) CoNS isolates.
Multiplex PCR
Multiplex PCR was standardized for genotypic identification of the genus Staphylococcus (16S rRNA gene), to differentiate coagulase-positive staphylococci and CoNS (coa gene), to identify S. aureus (S. aureus-specific gene), and to detect oxacillin resistance (mecA gene) directly from blood culture bottles (Fig. 1).
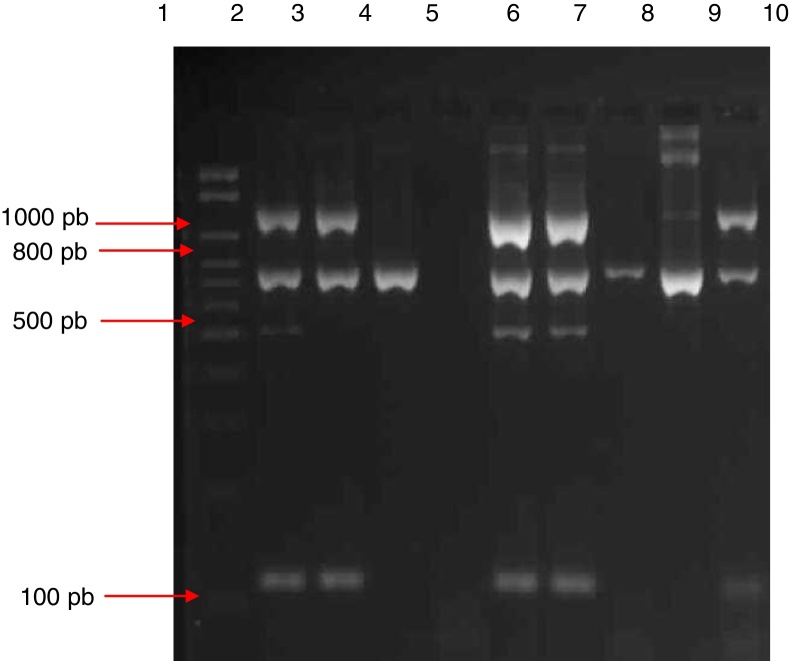
Fig. 1.
Electrophoresis gel (3% agarose) of the products of multiplex amplification in blood cultures for the detection of Staphylococcus spp. and the mecA gene. Lane 1: 100–2000 bp Ladder; lane 2: ATCC 33591 (mecA-positive S. aureus); lane 3: ATCC 25923 (mecA-negative S. aureus); lane 4: ATCC 12228 (mecA-negative S. epidermidis); lane 5: negative control; lanes 6 and 7: mecA-positive S. aureus isolate; lanes 8 and 9: mecA-negative CoNS isolate; lane 10: mecA-negative S. aureus isolate.
Amplification was performed on all 371 isolates included in the study. In the case of the 85 S. aureus isolates identified phenotypically, there was 100% agreement with the multiplex PCR results, with amplification of the 16S rRNA, S. aureus and coa genes. Among the 286 CoNS isolates included, 100% agreement between the two methods was only observed in terms of amplification of the 16S rRNA gene.
Amplification of the mecA gene was observed in 43 (50.6%) of the 85 S. aureus isolates; of these, 42 were resistant to oxacillin by the disk diffusion test and one isolate was susceptible. Among the 42 (49.4%) mecA gene-negative isolates, 41 were susceptible by the disk diffusion method and one isolate was resistant. The mecA gene was amplified in 225 (78.7%) of the 286 CoNS isolates; of these, 218 were resistant to oxacillin by the disk diffusion test and seven were susceptible. Among the 61 (21.3%) mecA gene-negative CoNS isolates, 43 were classified as susceptible by the disk diffusion test and 18 were resistant.
Detection of β-lactamase hyperproduction and resistance profile confirmation
Among the 19 isolates not carrying mecA gene although resistant to oxacillin by disk diffusion test, all were positive by nitrocefin disk test and cefoxitin disk diffusion test. On the other hand, six isolates were resistant to amoxicillin/clavulanic acid by disk diffusion test.
Sensitivity and specificity of the multiplex PCR
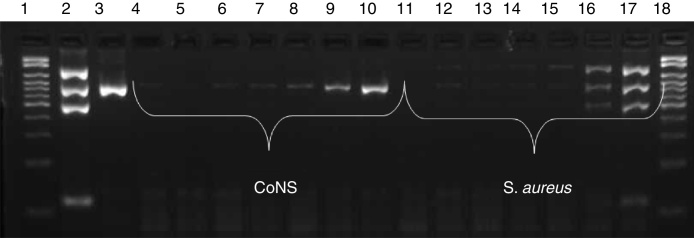
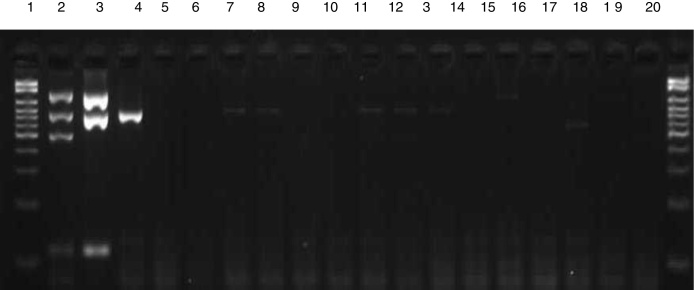
In the sensitivity test, amplification of fragments was observed up to a dilution of 10−5 (103 CFU/mL) for CoNS and of 10−2 (106 CFU/mL) for S. aureus, as shown in Fig. 2. Analysis of specificity using isolates of other species revealed no amplification of any fragments of the genes investigated by multiplex PCR (Fig. 3).
Fig. 2.
Sensitivity of multiplex PCR. Electrophoresis gel showing the amplification of different dilutions of CoNS and S. aureus. Lane 1: 100–2000 bp molecular weight marker; 2: mecA-positive S. aureus ATCC; 3: S. epidermidis ATCC; 4: CoNS 10−7 dilution (10 CFU/mL); 5: 10−6 (102 CFU/mL); 6: 10−5 (103 CFU/mL); 7: 10−4 (104 CFU/mL); 8: 10−3 (105 CFU/mL); 9: 10−2 (106 CFU/mL); 10: 10−1 (107 CFU/mL), 11: S. aureus 10−7 dilution (10 CFU/mL); 12: 10−6 (102 CFU/mL); 13: 10−5 (103 CFU/mL); 14: 10−4 (104 CFU/mL); 15: 10−3 (105 CFU/mL); 16: 10−2 (106 CFU/mL); 17: 10−1 (107 CFU/mL); 18: 100–2000 bp molecular weight marker.
Fig. 3.
Electrophoresis gel illustrating the specificity of multiplex PCR. Lane 1: 100–2000 bp Ladder; 2: mecA-positive S. aureus ATCC; 3: mecA-negative S. aureus ATCC; 4: CoNS ATCC; 5: Salmonella spp.; 6: Proteus mirabilis; 7: Morganella morganii; 8: Klebsiella pneumoniae; 9: Pseudomonas aeruginosa; 10: Acinetobacter baumannii; 11: Enterobacter cloacae; 12: Enterobacter aerogenes; 13: E. coli; 14: E. faecalis; 15: E. faecium; 16: Streptococcus viridans; 17: Streptococcus pneumoniae; 18: Candida spp.; 19: Gram-positive bacilli.
Costs analysis
The cost of multiplex PCR and the automated Vitek 2 system, the system used for direct bacterial identification in blood cultures at the Laboratory of Microbiology, Department of Internal Medicine, University Hospital, Botucatu Medical School, UNESP, was lower than the phenotypic method (Table 2).
Table 2.
Comparison of the identification costs with the Vitek 2 system and multiplex PCR.
| Method | Cost |
|---|---|
| Vitek identification Total: US$ 6.76 per isolate |
Blood agar: US$ 0.5 MacConkey: US$ 0.21 Vitek: US$ 6.05 |
| Multiplex PCR identification Total: US$ 4.35 per isolate |
Extraction: US$ 2.85 Multiplex PCR: US$ 1.21 Electrophoresis gel: US$ 0.29 |
Discussion
Among the 371 Staphylococcus spp. positive blood culture samples included in this study, 23% were positive for S. aureus and 77% were positive for CoNS; of these, 50.6% S. aureus isolates and 82.5% CoNS isolates were resistant to oxacillin by the disk diffusion test. In studies conducted with patients from the hospital where the current study was performed,12 45.1% S. aureus isolates and 72.5% CoNS isolates were resistant to this drug. Other studies also reported high rates of oxacillin-resistant Staphylococcus spp. isolates.13, 14 In view of this increase in resistant isolates, treatment options have narrowed down to glycopeptides and new synthetic drugs such as linezolid.15 In addition to being expensive and toxic, these drugs select resistant strains. In some countries there are reports of Staphylococcus spp. resistant to vancomycin, which increasingly limits the therapeutic options.16, 17
In view of these considerations, the faster the results of culture and antimicrobial susceptibility testing are released, the faster the physician can adjust treatment, reducing the risks and harm caused by the organism to the patient. In the present study, a multiplex PCR assay that permitted direct detection of oxacillin-resistant S. aureus (MRSA) in blood cultures was developed using an extraction technique that was effective and able to eliminate interfering elements present in blood. Bloodstream infections caused by MRSA complicate the prognosis of the patient, increasing the length-of-hospital stay, mortality rate, number of invasive procedures, and hospitalization costs.18
Investigation of the coa gene included in the multiplex assay for detection of all coagulase-positive staphylococci and the use of the S. aureus-specific SAU primer permit to establish whether the isolate is S. aureus or not.
In addition to the detection of Staphylococcus spp., the multiplex assay proposed in this study also included investigation of the mecA gene to detect oxacillin resistance, in order to guide patient treatment at the time of identification. Pereira et al.19 developed a multiplex PCR assay that directly identified three Staphylococcus spp. species, S. aureus, S. epidermidis and S. haemolyticus, and the mecA gene in blood cultures, but this identification was restricted to these species. The multiplex assay standardized in this study permits the detection of all staphylococcal species isolated from bacteremias. The multiplex PCR was tested using 371 samples, 13 blood culture samples containing other bacterial species and bacterial DNA of other species directly from bacterial culture, showed good sensitivity and specificity.
In addition to being specific, our multiplex PCR assay exhibited a detection threshold of MRSA of 106 CFU/mL. Similar thresholds have been reported in other studies.20 The threshold may decrease with increasing number of amplified genes, but even so the threshold found in this study was within the proposed range of detection since, according to Pereira et al.,20 positive blood cultures present a growth of about 109 CFU/mL. The only discrepancy observed in this study was the detection of the mecA gene by the standardized multiplex PCR assay and susceptibility to oxacillin in the phenotypic test, with the mecA gene not being detected in 19 isolates resistant to oxacillin (one S. aureus isolate and 18 CoNS isolates). In these isolates, the mecA gene was detected by simplex PCR, confirming the results of multiplex PCR. This discrepancy has also been reported in other studies20, 21, 22, 23 and may be related to other types of resistance such as β-lactamase hyperproduction or modification of another PBP. Furthermore, eight isolates were positive for the mecA gene and were susceptible in the phenotypic test (one S. aureus isolate and 7 CoNS isolates). These findings highlight the importance of genotypic techniques for the detection of oxacillin resistance, but do not rule out the need for other phenotypic techniques to initiate more specific and effective treatment.
The implementation of a molecular biology technique such as multiplex PCR to assist in the rapid diagnosis of bloodstream infections may be associated with high costs for the institution. A cost analysis performed in the present study showed that the identification of species in culture, i.e., after detection of a positive blood culture with the Bactec system, its seeding on culture plates and identification, which is done with the Vitek 2 system at the Laboratory of Microbiology in Botucatu Medical School has an average cost of US$ 6.76. In the case of multiplex PCR, which does not require seeding of the blood culture for isolation of the microorganisms since it uses direct identification from blood culture bottles, DNA extraction and PCR amplification for detecting Staphylococcus spp., S. aureus, and the mecA gene were estimated to cost US$ 4.35 per isolate.
The multiplex PCR assay developed here was found to be sensitive, specific, rapid and less expensive than the phenotypic technique used, and showed good agreement with the phenotypic results. This method could be used in clinical laboratories for rapid identification and initiation of specific and effective treatment, reducing patient mortality and morbidity. Furthermore, this method may reduce the misuse of other antimicrobial classes that are more expensive and toxic, thus contributing to the selection of antibiotic-resistant Staphylococcus spp.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
São Paulo Research Foundation (FAPESP Grant 2010/14250-0) and National Council for Scientific and Technological Development (CNPq) for financial support.
References
- 1.Russel J.A. Management of sepsis. N Engl J Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 2.Magill S.S., Edwards J.R., Bamberg W., et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Euzeby J. 2017. List of Prokaryotic Names with Standing in Nomenclature: LPSN. Available from: http://www.bacterio.net/s/staphylococcus.html. [Google Scholar]
- 4.Bannerman T.L. In: Manual of clinical microbiology. 8th ed. Murray P.R., Baron E.J., Jorgensen J.H., Pfaller M.A., Yolken R.H., editors. ASM Press; Washington, DC: 2003. Staphylococcus, Micrococcus and the other catalase-positive cocci that grow aerobically. [Google Scholar]
- 5.Springer B., Orendi U., Much P., et al. Methicillin-resistant Staphylococcus aureus: a new zoonotic agent? Wien Klin Wochenschr. 2009;121:86–90. doi: 10.1007/s00508-008-1126-y. [DOI] [PubMed] [Google Scholar]
- 6.Hackbarth C.J., Chambers H.F. Methicillin-resistant Staphylococci: genetics and mechanisms of resistance. Antimicrob Agents Chemother. 1989;33:991–994. doi: 10.1128/aac.33.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim T.T., Coombs G.W., Grubb W.B. Genetic organization of mecA and mecA-regulatory genes in epidemic methicillin-resistance S. aureus from Australia and England. J Antimicrob Chemother. 2002;50:819–824. doi: 10.1093/jac/dkf238. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser T.D.L., Pacheco F.C., Lima A.A., Pereira E.M., Santos K.R.N., Nunes A.P.F. Avaliação de métodos comumente usados em laboratório para a determinação da suscetibilidade à oxacilina entre amostras de Staphylococcus spp., isoladas de um Hospital de Vitória, estado do Espírito Santo. Soc Bras Med Trop. 2010;43:298–303. doi: 10.1590/s0037-86822010000300017. [DOI] [PubMed] [Google Scholar]
- 9.Hogg G.M., Mckenna J.P., Ong G. Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert® blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of 4 DNA extraction methods. Diagn Microbiol Infect Dis. 2008;61:446–452. doi: 10.1016/j.diagmicrobio.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Fredricks D.N., Relman D.A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2012. Performance standards for antimicrobial susceptibility testing. Approved standards: M100-S22. [Google Scholar]
- 12.Martins A., Pereira V.C., Cunha M.L.R.S. Oxacillin resistance of Staphylococcus aureus isolated from the university hospital of Botucatu Medical School in Brazil. Chemotherapy. 2010;56:112–119. doi: 10.1159/000313523. [DOI] [PubMed] [Google Scholar]
- 13.Cuevas O., Cercenado E., Goyanes M.J., et al. Staphylococcus spp. in Spain: present situation and evolution of antimicrobial resistance (1986–2006) Enferm Infecc Microbiol Clin. 2008;26:269–277. doi: 10.1157/13120413. [DOI] [PubMed] [Google Scholar]
- 14.Cunha M.L.R.S., Lopes C.A.M. Estudo da produção de beta-lactamases e sensibilidade às drogas em linhagens de Estafilococos coagulase-negativos isolados de recém-nascidos. J Bras Patol Med Lab. 2002;38:281–290. [Google Scholar]
- 15.Woods W., Ramotar K., Lem P., Toye B. Oxacillin susceptibility testing of coagulase-negative staphylococci using the disk diffusion method and the Vitek GPS card. Diagn Microbiol Infect Dis. 2002;42:291–294. doi: 10.1016/s0732-8893(02)00365-6. [DOI] [PubMed] [Google Scholar]
- 16.Chang S., Sievert D.M., Hageman J.C., et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:342–1347. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 17.Cunha M.L.R.S. Nova Science Publishers; New York: 2014. Staphylococcus aureus and coagulase-negative staphylococci: virulence antimicrobial resistance and molecular epidemiology. [Google Scholar]
- 18.McHugh C.G., Riley L.W. Risk factors and costs associated with methicillin-resistant Staphylococcus aureus bloodstream infections. Infect Control Hosp Epidemiol. 2004;25:425–430. doi: 10.1086/502417. [DOI] [PubMed] [Google Scholar]
- 19.Pereira E.M., Schuenck R.P., Malvar K.L., et al. Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus haemolyticus: methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiol Res. 2010;165:243–249. doi: 10.1016/j.micres.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Pereira V.C., Martins A., Rugolo L.M.S.S., Cunha M.L.R.S. Detection of oxacillin resistance in Staphylococcus aureus isolated from the neonatal and pediatric units of a Brazilian teaching hospital. Clin Med Pediatr. 2009;3:23–31. doi: 10.4137/cmped.s2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimitz F.J., Mackenzie C.R., Hofman B., et al. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by multiplex PCR. J Med Microbiol. 1997;46:773–778. doi: 10.1099/00222615-46-9-773. [DOI] [PubMed] [Google Scholar]
- 22.Caierão J., Superti S., Dias C.A., d’Azevedo P.A. Automated systems in the identification and determination of methicillin resistance among coagulase negative Staphylococci. Mem Inst Oswaldo Cruz. 2006;101:277–280. doi: 10.1590/s0074-02762006000300009. [DOI] [PubMed] [Google Scholar]
- 23.Kilic A., Muldrew K.L., Tang Y.W., Baustaoglu A.C. Triplex real-time polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci and determination of methicillin resistance directly from positive blood culture bottles. Diagn Microbiol Infect Dis. 2010;66:349–355. doi: 10.1016/j.diagmicrobio.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Riffon R., Sayasith K., Khalil H., Dubreuil P., Drolet M., Lagacé J. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J Clin Microbiol. 2001;39:2584–2589. doi: 10.1128/JCM.39.7.2584-2589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason W.J., Blevins J.S., Beenken K., Wibowo N., Ojha N., Smeltzer M.S. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J Clin Microbiol. 2001;39:3332. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearns A.M., Siders P.R., Wheeler J., Freeman R., Steward M. Rapid detection of methicillin-resistant Staphylococci by multiplex PCR. J Hosp Infect. 1999;53:33–37. doi: 10.1053/jhin.1999.0631. [DOI] [PubMed] [Google Scholar]
- 27.Murakami K., Minamide W., Wada K., Nakamura E., Teraoka H., Watanabe S. Identification of methicillin-resistant strains of Staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]