Abstract
Background
BA.2.12.1, BA.4 and BA.5 subvariants of SARS-CoV-2 variant-of-concern (VOC) Omicron (B.1.1.529) are spreading globally. They demonstrate higher transmissibility and immune escape.
Objectives
Determine BA.2.12.1, BA.4 and BA.5 virus plaque reduction neutralization test (PRNT) antibody titres in individuals recently vaccinated with BNT162b2 (n = 20) or CoronaVac (n = 20) vaccines or those convalescent from ancestral wild- type (WT) SARS-CoV-2 (n = 20) or BA.2 infections with (n = 17) or without (n = 7) prior vaccination.
Results
Relative to neutralization of the WT virus, those vaccinated with BNT162b2 had 4.8, 3.4, 4.6, 11.3 and 15.5-fold reductions of geometric mean antibody titres (GMT) to BA.1, BA.2, BA.2.12.1, BA.4 and BA.5 viruses, respectively. Similarly, those vaccinated with CoronaVac had 8.0, 7.0, 11.8, 12.0 and 12.0 fold GMT reductions and those with two doses of CoronaVac boosted by BNT162b2 had 6.1, 6.7, 6,3, 13.0 and 21.2 fold GMT reductions to these viruses, respectively. Vaccinated individuals with BA.2 breakthrough infections had higher GMT antibody levels vs. BA.4 (36.9) and BA.5 (36.9) than unvaccinated individuals with BA.2 infections (BA.4 GMT 8.2; BA.5 GMT 11.0).
Conclusions
BA.4 and BA.5 subvariants were less susceptible to BNT162b2 or CoronaVac vaccine elicited antibody neutralization than subvariants BA.1, BA.2 and BA.2.12.1. Nevertheless, three doses BNT162b2 or booster of BNT162b2 following two doses of CoronaVac elicited detectable BA.4 and BA.5 neutralizing antibody responses while those vaccinated with three doses of CoronaVac largely fail to do so. BA.2 infections in vaccinated individuals led to higher levels of BA.4 or BA.5 neutralizing antibody compared to those who were vaccine-naive.
Keywords: COVID-19; SARS-CoV-2; Omicron, Neutralization; Vaccine
Abbreviations: SARS-CoV-2, SARS coronavirus 2
1. Introduction
The new variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) within the Pango lineage B.1.1.529 lineage was first recognised in countries in Southern Africa as a newly emerged variant and it was designated as a variant of concern (VOC) Omicron by the WHO on 26th November 2021 [22]. Multiple sub-variants of Omicron were quickly recognised, but BA.1 subvariant, followed by BA.2, were the ones that spread globally, initially [20]. Compared to the wild-type virus or other previously circulating VOCs, Omicron variants have over 35 amino acid changes in the spike protein in addition to other amino acid substitutions elsewhere in the virus genome [20]. Some of these amino acid substitutions in the spike protein resulted in these viruses evading neutralizing immunity elicited by prior infections or vaccination [5], [6], [7], contributing to greater transmissibility and competitive advantage to displace previously circulating virus variants [5]. BA.2.12.1 is a variant of BA.2 first recognised in the USA, now also seen elsewhere, and contains identical spike receptor binding domain (RBD) sequences to BA.2 but with the addition of amino acid substitutions L452Q and S704L which may have capacity for immune evasion [16]. Early in 2022, two other sub-variants of Omicron, BA.4 and BA.5 were recognised [18]. Both BA.4 and BA.5 had spike proteins similar to BA.2 with the exception of having a 69–70del (present in the Alpha variant and the BA.1 lineage), L452R (present in the Delta variant), F486V and the reversion to the wild-type amino acid at Q493 [18]. BA.4 and BA.5 have spike proteins identical to each other differ from each other elsewhere in the genome. The amino acid substitutions in spike protein of Omicron variants relative to the ancestral WT virus are summarised in the Supplementary table.
BA.5 is now rapidly increasing in prevalence in many parts of the world. It is important to assess the impact of these variants on further immune evasion from immunity elicited by prior infection or vaccination. In individuals vaccinated with RNA vaccines, studies using pseudovirus neutralization carrying the spike proteins of these respective viruses showed that BA.4/BA.5, and to lesser extent BA.2.12.1 neutralizing antibody titers, were even lower than BA.1 and BA.2 neutralizing antibody titers, suggesting that the SARS-CoV-2 BA.4/5 has continued to evolve with increasing neutralization escape [9,21]. However, pseudotype neutralization assays are known to give varying conclusions, depending upon the virus vector used. Pseudotype neutralization also ignores impact of mutations outside of the spike protein which may have impact on viral replication competence, immune innate evasion and neutralizability. Plaque reduction neutralization assays are a “gold-standard” method to define virus neutralization titres and we used this approach to address the relative WT and Omicron subvariant neutralization titres cohorts who were vaccinated and those convalescent from infection.
2. Materials and methods
2.1. Clinical specimens
Subsets of sera from those previously collected during 11 Aug 2020 to 29 Dec 2021 were randomly selected for a previous study comparing neutralizing antibody to WT, BA.1 and BA.2 subvariants in vaccinated and non-vaccinated individuals [7] and these sera are used in the present study. These cohorts include individuals who had no previous SARS-CoV-2 infection and received 3 doses BNT162b2, 3 doses CoronaVac or 2 doses CoronaVac boosted by a dose of BNT162b2, with blood collected 3–5 weeks after the last vaccine dose. They also included unvaccinated individuals who were convalescent from WT SARS-CoV-2 (143–196 days post infection). In addition, paired acute and convalescent sera were collected from patients with presumed BA.2 subvariant infection (n = 24) during the period 24 Jan 2022 and 29 Mar 2022, a period of predominant BA.2 infection in Hong Kong. Among the 24 patients with BA.2 infection, 17 were vaccine-breakthrough infection and 7 were previously unvaccinated. Of the 17 breakthrough infections, three had received 1 dose BNT162b2, seven received 2 doses BNT162b2, one received 1 dose CoronaVac, four received 2 doses CoronaVac and two received 3 doses CoronaVac vaccines.
2.2. Virus isolation
SARS-CoV-2 Omicron subvariants Pango lineage BA.2.12.1 (SARS-CoV-2/human/USA/COR-22–062,161/2022 ) and BA.5 (SARS-CoV-2/human/USA/COR-22–063,113/2022 ) were kindly provided by Dr Richard Webby, St Jude Children's Research Hospital, Memphis and BA.4 (hCoV-19/USA/MD-HP30386/2022) was kindly provided by Dr Andy Pekosz, The Johns Hopkins Bloomberg School of Public Health, Baltimore, MD. Ancestral (WT) SARS-CoV-2 (BetaCoV/Hong Kong/VM20001061/2020), subvariant BA.1 (hCoV-19/Hong Kong/VM21044713_WHP5047-S5/2021) and BA.2 (hCoV-19/Hong Kong/VM22000135_HKUVOC0588P2/2022) were isolated in Hong Kong [6, 7, 17]. Virus stocks were obtained by passage in Vero-E6 TMPRSS2 cell, aliquoted, stored frozen at −80°C, virus titres obtained in plaque titrations and used in the plaque reduction neutralization tests. Sequences of the viruses used are available in GISAID as EPI_ISL_412,028, EPI_ISL_6,716,902, EPI_ISL_9,570,707 and EPI_ISL_12,416,220.
2.3. Plaque reduction neutralization test (PRNT)
The live virus neutralization PRNT were performed in duplicate using 24-well tissue culture plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) in a biosafety level 3 facility using Vero E6 TMPRSS2 cells [14] as previously described [12]. Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) medium supplemented with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin-streptomycin. All sera were heat-inactivated at 56 °C for 30 min prior to testing. Serial two-fold dilutions from 1:10 to 1:320 of each serum sample were incubated with 30–40 plaque-forming units of virus for 1 h at 37 °C and the virus–serum mix was added onto pre-formed cell monolayers and incubated for 1 at 37 °C in a 5% CO2 incubator. The virus-antibody inoculum was then removed and the cell monolayer was overlaid with 1% agarose in cell culture medium. After 3 days incubation, the plates were fixed with 10% formalin in PBS overnight and stained with 1% crystal violet in ethanol. Antibody titres were defined as the highest serum dilution that resulted in ≥ 50% reduction in the number of virus plaques (PRNT50). The average plaque numbers observed in the duplicate dilution-series was used for this computation. Virus back titrations, positive and negative control sera were included in every experiment.
2.4. Statistical analysis
2.4.1. Sample size calculations
The maximum standard deviation (SD) of log PRNT50 titers for the uninfected vaccinated groups was previously observed to be 1.37. Assuming a 3-fold difference in GMT, a sample size of 10 in each group would have statistical power of >0.99 for detecting a difference between groups using the two-tailed Mann-Whitney U test. Comparisons between groups with larger sample size or smaller within-group variation would have larger statistical power.
2.4.2. Statistical methods
Categorical variables were summarized as proportions or percentage and continuous variables were summarized as geometric mean with standard deviation (SD). Sera with undetectable (<1:10) antibody titres were assigned an antibody titre of 1:5, for purposes of geometric mean titre calculations or statistical comparisons. Comparison of antibody titres to different viruses was done using the Wilcoxon signed-rank test when comparing paired antibody titres to different viruses in the same serum and the two tailed Mann-Whitney U test when comparisons were made between different groups of individuals. Absolute P values were provided. P values < 0.05 were considered statistically significant.
2.5. Ethical statement
This study was approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (Ref no: 2020.229) and Hong Kong West Cluster HKU/HA HKW IRB UW 20–169.
3. Results
The demographics of the vaccine or infection-convalescent groups investigated are shown in Table 1 . The mean ages of all the groups were comparable with the exception of the unvaccinated BA.2 infections who were older (median age 68.1).
Table 1.
Demographics of the study cohorts and geometric mean PRNT50 antibody titres to SARS-CoV-2 Omicron subvariants.
| Exposure group | n | Mean Age (SD) | Age range | Male: female | WT GMT (95% CI) | BA.1 GMT(95% CI) | BA.2 GMT(95% CI) | BA.2.12.1 GMT (95% CI) | BA.4 GMT (95% CI) | BA.5 GMT (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| BNT162b2 (3 doses) | 20 | 49.5 (14.8) | 22–72 | 11:9 | 320 (320–320) |
67.3 (45.5–99.6) |
95.1 (73.7–122.8) |
69.6 (52.1–93.1) |
28.3 (21.2–37.7) |
20.7 (17.0–25.2) |
| CoronaVac (3 doses) | 20 | 49.8 (7.9) | 36–68 | 4:16 | 65.0 (1.0) | 8.1 (8.0) | 9.3 (7.0) | 5.3 (12.3) | 5.4 (12.0) | 5.4 (12.0) |
| CoronaVac (2 dose + BNT162b2 third dose) | 20 | 48.4 (9.2) | 31–66 | 10:10 | 309.1 (287.5–332.4) |
51.0 (32.9–79.0) |
46.0 (29.2–72.3) |
49.3 (31.9–76.1) |
23.8 (15.9–35.7) |
14.6 (10.1–21.2) |
| SARS-CoV-2 (ancestral) Convalescent | 20 | 48.7 (15.0) | 20–70 | 7:13 | 82.8 (58.0–118.3) |
8.1 (5.5–12.1) |
9.0 (6.7–12.2) |
7.8 (5.9–10.4) |
6.8 (5.0–9.3) |
5.9 (5.0–7.1) |
| BA.2 breakthrough infection (convalescent) | 17 | 47.1 (17.8) | 24–73 | 9:8 | 166.7 (91.7–302.8) |
70.8 (32.2–155.9) |
98.1 (52.4–183.6) |
98.1 (49.9–192.8) |
36.9 (18.3–74.3) |
36.9 (19.7–69.1) |
| Unvaccinated + BA.2 infection (convalescent) | 7 | 68.1 (7.9) | 56–82 | 4:3 | 6.7 (4.8–9.5) |
9.1 (4.2–19.7) |
24.4 (9.3–63.6) |
32.8 (16.1–67.0) |
8.2 (5.1–13.3) |
11.0 (6.2–19.7) |
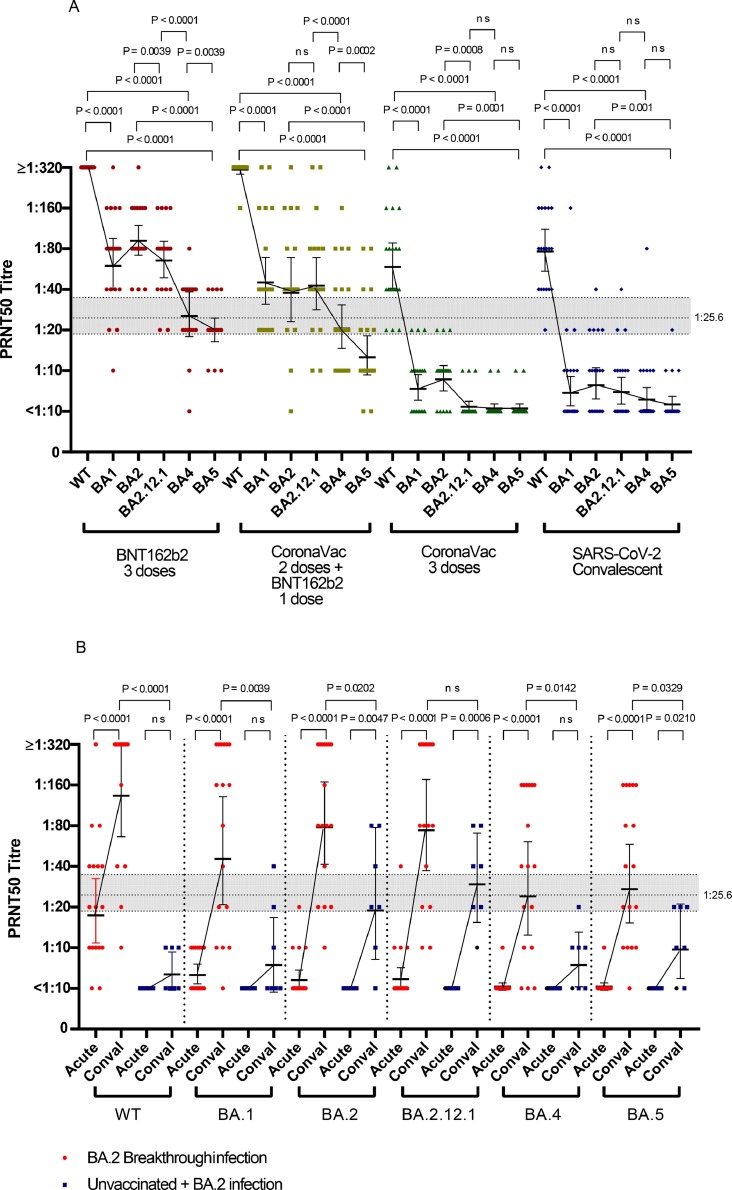
Omicron neutralization in individuals vaccinated with BNT162b2 or CoronaVac vaccines or unvaccinated individuals infected with ancestral WT SARS-CoV-2.
All individuals in the three vaccine groups (three doses of BNT162b2 or CoronaVac, or two doses of CoronaVac followed by a third dose of BNT162b2)(n = 20 in each group) had detectable (≥1:10) PRNT50 antibody to WT virus. While all or most (≥90%) individuals in the BNT162b2 or BNT162b2 boosted vaccine-groups had detectable PRNT50 antibody to all the viruses tested, only 55–75% of those vaccinated with CoronaVac had detectable PRNT50 antibody to BA.1 or BA.2 and ≤15% to BA.2.12.1, BA.4 or BA.5 (Table 2 , Fig. 1 A). While unvaccinated individuals naturally infected with the WT virus had detectable titres to the infecting virus, they had poor cross-reactive neutralization to Omicron subvariants (Table 1, Fig. 1). There was one 56-year-old woman in the group convalescent from WT virus infection who was an outlier having high PRNT50 titres to multiple subvariants of Omicron. Her infection occurred in late 2020 and she had illness of moderate severity (as did the majority of others in this group). The convalescent blood sample was collected in Jan 2021, before the COVID-19 vaccination program commenced in Hong Kong and she has no record of vaccination or of known reinfection. An unsuspected re-infection cannot be excluded.
Table 2.
PRNT50 antibody titres to wild type virus and Omicron subvariants in COVID-19 vaccinated or infected cohorts.
| Exposure group | Number (%) with PRNT50 titre ≥1:10 | Number (%) with PRNT50 titre ≥1:25.6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | BA1 | BA2 | BA2.12.1 | BA4 | BA5 | WT | BA1 | BA2 | BA2.12.1 | BA4 | BA5 | |
| BNT162b2 (3 doses) | 20 (100%) | 20 (100%) | 20 (100%) | 20 (100%) |
19 (95%) |
20 (100%) | 20 (100%) | 17 (85%) | 20 (100%) |
18 (90%) |
12 (60%) | 4 (20%) |
| CoronaVac (3 doses) | 20 (100%) | 11 (55%) |
15 (75%) |
3 (15%) |
2 (10%) |
2 (10%) |
17 (85%) |
0 (0%) |
0 (0%) |
0 (0%) |
0 (0%) |
0 (0%) |
| CoronaVac (2 dose + BNT162b2 third dose) |
20 (100%) | 20 (100%) | 19 (95%) |
20 (100%) |
20 (100%) | 18 (90%) |
20 (100%) | 14 (70%) | 16 (80%) |
16 (80%) |
7 (35%) |
4 (20%) |
| SARS-CoV-2 (ancestral) Convalescent |
20 (100%) | 8 (40%) |
11 (55%) |
9 (45%) |
6 (30%) |
4 (20%) |
19 (95%) |
1 (5%) |
1 (5%) |
1 (5%) |
1 (5%) |
0 (0%) |
Fig. 1.
PRNT50 antibody titres to wild-type (WT) SARS-CoV-2 and Omicron subvariants BA.1, BA.2, BA.2.12.1, BA.4 and BA.5. A: Uninfected BNT162b2 individuals or CoronaVac vaccinated and non-vaccinated convalescent individuals from WT SARS-CoV-2 infection. GMTs to each virus in the same cohort are connected by lines. B: BA.2 infections in vaccinated (n = 17) and unvaccinated individuals (n = 7). GMTs in acute and convalescent stages were connected by lines. Comparison of antibody titres to different viruses was done using the Wilcoxon signed-rank test when comparing paired antibody titres to different viruses in the same serum and the two tailed Mann-Whitney U test when comparisons were made between different groups of individuals. The horizontal dotted line at a titre 1:25.6 represents the 50% protective titre against symptomatic infection and the shaded area represents the 95% confidence interval of this protective threshold (see results and reference [11] for details).
It has been previously reported that the neutralizing antibody titer associated with protection from symptomatic infection in 50% of a group of individuals corresponded to 20% of the mean neutralizing titre observed in SARS-CoV-2 convalescent individuals [11]. Using convalescent sera from our previous cohort of RT-PCR confirmed SARS-CoV-2 infections, using the same PRNT50 methods used here, we previously estimated that this 20% convalescent antibody titer, and thus the threshold for 50% protection from symptomatic infection in our PRNT50 assay to be a titre of 1:25.6 (95% CI 18.3–36.0) [12]. For those with three doses of BNT162b2, the relative percentage of individuals achieving or exceeding this protective PRNT50 threshold of 1:25.6 to WT, BA.1, BA.2, BA.2.12.1, BA.4 and BA.5, were 100%, 85%, 100%, 90%, 60% and 20%, respectively (Table 2). However, none of those vaccinated with three doses of Coronavac had PRNT50 antibody titres of ≥1:25.6 for any of the Omicron subvariants. Of those vaccinated with two doses of CoronaVac and boosted with BNT162b2, ≥70% had BA.1, BA.2 or BA.2.12.1 titres above the protective threshold, while only 35% and 20% met this threshold for BA.4 and BA.5, respectively (Table 2).
In the three vaccine groups, BA.1 and BA.2 had 3.4 to 8-fold reduction in titres relative to WT virus, while BA.2.12.1 had 4.6–11.8 fold, and BA.4 and BA.5 viruses had 11.3–21.2 fold-reduction in PRNT50 titres relative to WT virus (Table 3 ). Relative to BA.2 PRNT50 titres in the BNT162b2 vaccinated and boosted group, BA.5 virus had a further 3.2–4.6 fold reduction in PRNT50 titres (Table 3). The BA.2 PRNT50 titres in the three-dose CoronaVac vaccinated group or those infected with WT SARS-CoV-2 were too low to allow assessment of further reduction with BA.5. It was of interest that BA.5 PRNT50 GMT was modestly but significantly lower than BA.4 in sera from those vaccinated with three doses of BNT162b2 or two doses of CoronaVac boosted with BNT162b2. The BA.2 antibody titres in the three-dose CoronaVac vaccinated and WT convalescent groups were too low to demonstrate any further reduction with BA.4 and BA.5 viruses.
Table 3.
Fold reduction in PRNT50 titres in comparison to wild type virus or to Omicron BA.2 virus in vaccine or infection cohorts.
| Exposure group | fold reduction vs WT | fold reduction vs BA.2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | BA1 | BA2 | BA2.12.1 | BA4 | BA5 | WT | BA1 | BA2 | BA2.12.1 | BA4 | BA5 | |
| BNT162b2 (3 doses) | 1.0 | 4.8 | 3.4 | 4.6 | 11.3 | 15.5 | 0.3 | 1.4 | 1.0 | 1.4 | 3.4 | 4.6 |
| CoronaVac (3 doses) | 1.0 | 8.0 | 7.0 | 11.8 | 12.0 | 12.0 | 0.1 | 1.1 | 1.0 | 1.7 | 1.7 | 1.7 |
| CoronaVac (2 dose + BNT162b2 third dose) |
1.0 | 6.1 | 6.7 | 6.3 | 13.0 | 21.2 | 0.1 | 0.9 | 1.0 | 0.9 | 1.9 | 3.2 |
| SARS-CoV-2 (ancestral) Convalescent |
1.0 | 10.2 | 9.2 | 10.6 | 12.2 | 14.0 | 0.1 | 1.1 | 1.0 | 1.2 | 1.3 | 1.5 |
Omicron neutralization in unvaccinated or vaccinated individuals with Omicron BA.2 infections.
We investigated 17 vaccinated individuals with BA.2 breakthrough infections and 7 unvaccinated individuals with BA.2 infection (Fig. 1B). The demographics and information on type and numbers of vaccine doses received by these individuals are shown in Tables 1 and 4 . Eleven of them had two doses of vaccine, 4 had one dose of vaccine and 2 had three doses of vaccine. In the vaccinated cohort, 15 of 17 had detectable PRNT50 to WT in the acute serum (GMT 22.6, 95% CI 12.8–39.8) but only 4 had cross-reactive antibody to BA.2 (GMT 6.1, 95% CI 5.0–7.6). As expected, BA.2 infection led to increase in antibody to BA.2 in all individuals in the convalescent serum (GMT 98.1, 95% CI 52.4–183.6). Interestingly, there was also an increase in antibody to WT (convalescent GMT 166.7, 95% CI 91.7–302.8) as well as to other subvariants of Omicron, with convalescent GMTs to Omicron subvariants ranging from 36.8 to 98.1 (Fig. 1B,Table 1).
Table 4.
Demographic and other relevant clinical details of previously vaccinated or unvaccinated individuals with BA.2 infections, Hong Kong, January 24th 2022- March 29th 2022, (n = 24).
| Case | Age (yrs), sex | Severity of illness | Vaccine | Doses | Days ill at collection of 1st serum* | Days ill at collection of 2nd serum* |
|---|---|---|---|---|---|---|
| BA.2 breakthrough infections in vaccinated | ||||||
| 1 | 31,F | Mild | BNT162b2 | 2 | 3 | 32 |
| 2 | 62,M | Mild | CoronaVac | 3 | 3 | 39 |
| 3 | 53,M | Mild | BNT162b2 | 2 | 3 | 32 |
| 4 | 24,F | Mild | CoronaVac | 2 | 1 | 29 |
| 5 | 45,M | Mild | BNT162b2 | 1 | 3 | 29 |
| 6 | 32,F | Mild | BNT162b2 | 2 | 2 | 35 |
| 7 | 32,F | Mild | BNT162b2 | 2 | 4 | 9 |
| 8 | 65,M | Mild | CoronaVac | 2 | 3 | 46 |
| 9 | 34,M | Mild | BNT162b2 | 2 | 2 | 52 |
| 10 | 26,F | Mild | BNT162b2 | 2 | 1 | 37 |
| 11 | 25,F | Mild | BNT162b2 | 2 | 2 | 38 |
| 12 | 66,M | Asymptomatic | CoronaVac | 1 | 1 | 51 |
| 13 | 48,F | Mild | CoronaVac | 3 | 3 | 46 |
| 14 | 70,F | Mild | CoronaVac | 2 | 5 | 48 |
| 15 | 73,M | Mild | BNT162b2 | 1 | 5 | 10 |
| 16 | 71,M | Asymptomatic | CoronaVac | 2 | 5 | 30 |
| 17 | 44,M | Mild | BNT162b2 | 1 | 5 | 48 |
| BA.2 infections in unvaccinated | ||||||
| 18 | 56,M | Mild | Unvaccinated | 0 | 5 | 18 |
| 19 | 67,M | Asymptomatic | Unvaccinated | 0 | 0 | 32 |
| 20 | 82,M | Mild | Unvaccinated | 0 | 2 | 45 |
| 21 | 66,F | Mild | Unvaccinated | 0 | 4 | 7 |
| 23 | 67,F | Mild | Unvaccinated | 0 | 2 | 52 |
| 24 | 66,M | Asymptomatic | Unvaccinated | 0 | 3 | 44 |
| 25 | 73,F | Mild | Unvaccinated | 0 | 3 | 23 |
*Days after onset of symptoms (or days after first RT-PCR positive in asymptomatic infections) at which first and second serum was collected.
None of the previously unvaccinated individuals with BA.2 infection had detectable antibody to WT or to any subvariants of Omicron in the acute serum collected early after BA.2 infection (Fig. 1B). The convalescent serum collected after BA.2 infection had GMT of 6.7, 9.1, 24.4, 32.8, 8.2 and 11.0 to WT, BA.1, BA.2, BA.2.12.1, BA.4 and BA.5 viruses (Table 1, Fig. 1B).
4. Discussion
It has been previously shown that Omicron variants BA.1 and BA.2 were less well neutralized by sera from individuals vaccinated with first generation vaccines based on the ancestral WT virus spike protein or from those naturally infected with WT SARS-CoV-2 [5], [6], [7]. The extent of evasion of neutralization was reduced with a third vaccine-dose but still remained substantial. We now find that Omicron subvariants BA.4 and BA.5 are even less well neutralized by three-dose vaccine sera than were the BA.1 or BA.2 variants. In BNT162b2 vaccinated and boosted individuals, PRNT50 titres to BA.2.12.1, BA.4 and BA.5 were reduced by 4.6, 11.3 and 15.5-fold relative to WT virus titres, respectively. PRNT50 titres to BA.2.12.1, BA.4 and BA.5 were 1.4, 3.4 and 4.6-fold reduced relative to BA.2 titres. Our assays were based on live virus neutralization which remains the reference methodology for quantitative assessment of neutralizing competence of a serum to different viruses. A study using fluorescence-focus neutralization in vaccinated (but not boosted) individuals found 19.8-fold, 19.6-fold and 20.9-fold reduction in neutralization of BA.1, BA.4 and BA.5 viruses relative to neutralization of WT virus in a cohort of vaccinated by not boosted individuals [10]. A number of previous studies of RNA (mRNA-1273 or BNT162b2) vaccinated individuals using virus pseudotype neutralization tests showed a similar trend, that BA.4/BA.5 virus spike demonstrate further evasion of neutralization compared with Omicron subvariants BA.1. and BA.2 [3,4,8,9,19,21].
Although subvariant BA.2.12.1 had substantial immune escape in comparison to the wild-type virus, PRNT50 titres were comparable with those observed with BA.2 and these was less immune evasion relative to BA.4 and BA.5. Omicron subvariants BA.4 and BA.5 have del69–70, L452R and F486V amino acid substitutions and Q493 reversion in spike protein compared to BA.2. BA.2.12.1 has L452Q and S704L that differs from BA.2. The greater evasion of neutralizing antibody shown by BA.4 and BA.5, in comparison with BA.2.12.1, may therefore largely be attributable to F486V which is an amino acid substitution not found in any of the other Omicron subvariants, as well as to L452R which differs from L452Q possessed by BA.2.12.1. While amino acid L has a hydrophobic side chain, Q is an uncharged amino acid while R is an amphipathic amino acid and these are likely to have a greater impact on interfering with antibody binding.
Although both BA.4 and BA.5 have identical spike protein sequences, it appears that BA.5 has modestly, but significantly, lower neutralization titres than does BA.4. Changes elsewhere in the genome may affect viral replication or innate immune evasion and may explain these differences in neutralizability. In comparison with BA.4, BA.5 has additional D3N substitution in the M protein, reversion of D61 in ORF6 protein and nt26858 and nt27259. It is relevant to note that Omicron subvariant BA.5 is outcompeting BA.4 and other Omicron subvariants globally, perhaps due to these differences between virus genomes. A direct comparison of BA.4 and BA.5 titres was not possible with the previously reported pseudotype virus neutralization assays, which only assess the role of antibody in blocking spike protein mediated viral entry.
As previously observed with BA.1 and BA.2 subvariants, three doses of BNT162b2 or two doses of CoronaVac followed by a BNT162b2 booster vaccine dose provided markedly better neutralizing antibody titres against BA.4, BA.5 and BA.2.12.1 than did three doses of CoronaVac vaccine. The percentages of individuals with detectable PRNT50 titres or meeting the estimated threshold titres for 50% protection from symptomatic infection for BA.2 and BA.2.12.1 were comparable. Of those vaccinated with three doses of BNT162b2 or two doses of CoronaVac followed by a booster with BNT162b2, 90–100% had detectable antibody titres to BA.5 but only 20% met the 50% protective threshold titre of 1:25.6. In those vaccinated with three doses of CoronaVac, only 10% had detectable PRNT50 antibody to BA.5 and none met the protective threshold. It should be noted that the sera tested were collected approximately three-five weeks after the last vaccine dose and it is known that antibody titres wane over time [13,23]. Thus, our neutralizing antibody findings may suggest that three doses of CoronaVac vaccine may not offer significant protection against symptomatic re-infection with Omicron variants. It is notable however, that three doses of either CoronaVac or BNT162b2 vaccines proved highly effective in reducing severe disease and mortality from a large Omicron BA.2 wave of infection in Hong Kong, even though neutralizing antibody responses following CoronaVac against BA.2 subvariant was very poor [15]. Thus, mechanisms other than neutralizing antibody, such as T-cell immunity, may have contributed to protection from severe disease against BA.2. CoronaVac vaccine may therefore still provide significant protection from severe disease against BA.4 and BA.5 but direct evidence is awaited.
In unvaccinated individuals with past infection with the wild-type virus, few (20%) had detectable PRNT50 antibody to BA.5 and none of these reached the threshold for protection from symptomatic infection with BA.5. Similar findings were reported by others [8]. While unvaccinated individuals with past BA.2 infection had detectable PRNT50 antibodies to BA.5, none of them reached the protective threshold. Thus, past infection with BA.2 in unvaccinated people may not be anticipated to provide much protection from reinfection with BA.5. However, 16 of 17 previously vaccinated (even two doses) individuals who had BA.2 breakthrough infection had detectable BA.5 antibodies and 9 attained protective threshold. Similar results were reported by others for BA.1 breakthrough infection [10]. Thus, vaccinated individuals with BA.1 or BA.2 breakthrough infections are likely to be protected from BA.4 or BA.5 re-infection at least for a period of time. These results may explain the recent vaccine efficacy data from a well vaccinated population in Qatar which suggested that prior infection with pre-Omicron viruses provided minimal protection against symptomatic re-infection with BA.4/BA.5 while prior infection with previous Omicron infection did provide good protection [1].
We had previously reported that BA.2 breakthrough infections led to high titres of neutralizing antibody to multiple VOCs [6]. We now observe that this extends to BA.2.12.1, BA.4 and BA.5, although the PRNT50 titres to BA.4 and BA.5 are lower than to variants BA.1 and BA.2. In general, our cohort of individuals with BA.2 breakthrough infections (most receiving two doses of vaccine prior to infection) and those with three doses of BNT162b2 vaccine without breakthrough infection had comparable PRNT50 titres to the different Omicron subvariants tested. However, three dose vaccinees had higher GMT to the wild-type virus. These findings are in concordance with vaccine effectiveness studies to BA.1 and BA.2 infections where three doses of BNT16b2 or mRNA-1273 vaccine provided comparable vaccine efficacy to two vaccine doses and previous infection in protection from symptomatic infection [2]. In those with three vaccine doses and infection, higher levels of protection were observed.
Interestingly, in the vaccinated individuals with breakthrough infections, all had PRNT50 titres below the protective threshold of 1:25.6 to BA.2 in the “acute” serum sample, although many had titres above the protective threshold to the WT virus in the acute serum. If we assume that the “acute” serum sample taken within a few days after onset of illness or first RT-PCR diagnosis reflects the pre-infection antibody titres, the presumed pre-infection titres explain why they were susceptible to infection with the BA.2 variant and is compatible with the concept of a protective threshold of antibody.
Limitations of the study included the lack of longitudinal follow-up to compare antibody waning in the vaccinated, infected and breakthrough groups. It will be important to investigate whether the duration of cross-reactive antibody in breakthrough infections is more prolonged than following booster vaccination. Only two vaccines were compared and other Omicron variants of recent interest such as BA.2.75 were not included.
In summary, we find that Omicron subvariants BA.4 and BA.5 have significantly greater capacity for evasion from neutralizing antibody compared to previous Omicron variants and to other variants of concern. Therefore, they are likely to continue to circulate even in populations that have been well vaccinated and populations that have both vaccination and high rates of prior natural infection. It is important to monitor vaccine effectiveness during these BA.4 and BA.5 outbreaks to establish whether these well vaccinated and infected populations are well protected from severe disease and death.
Author contributions statement
MP, DSCH, CKPM and SMSC conceptualized and supervised the study, TJ, AK, AP and RJW provided resources and reagents, SMSC and MP wrote original draft of manuscript, SMSC, CKPM, SSN, FWK, CC, KY, KKPC, BHSL, KCKC, LLHL, JKCL, LCHT and LLMP provided methodology, investigation, analysis and supervision. All authors were invovled with writing review and agreed with its submission.
Declaration of Competing Interest
None of the authors had competing financial or non-financial interests.
Funding
This research was supported by grants from the Health and Medical Research Fund Commissioned Research on the Novel Coronavirus Disease (COVID-19), Hong Kong SAR (COVID1903003; COVID190126) (CKPM, DSH and MP), US National Institute of Allergy and Infectious Diseases under contract nos. U01-Grant AI151810 and HHSN266200700005C (MP, RW), National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme (N_HKU737/18) (CKPM and MP), RGC theme-based research schemes (T11-712/19-N and T11-705/21-N) (LLMP, DSH, MP), Guangdong-Hong Kong-Macau Joint Laboratory of Respiratory Infectious Disease (20191205) (CKMP), C2i (LLMP, MP) administered by Innovation and Technology Commission of Hong Kong and visiting scientist scheme from Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore (CKPM).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105273.
Appendix. Supplementary materials
References
- 1.Altarawneh H.N., Chemaitelly H., Ayoub H., Hasan M.R., Coyle P., Yassine H.M., Al Khatib H.A., Benslimane F., Al-Kanaani Z., Al Kuwari E., Jeremijenko A., Kaleeckal A.H., Latif A.N., Shaik R.M., Abdul Rahim H.F., Nasrallah G., Al Kuwari M.G., Butt A.A., Al Romaihi H.E., Al-Thani M.H., Al Khal A., Bertollini R., Tang P., Abu-Raddad L.J. Protection of SARS-CoV-2 natural infection against reinfection with the BA.4 or BA.5 Omicron subvariants. medRxiv. 2022 2022.2007.2011.22277448. [Google Scholar]
- 2.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., Al-Kuwari E., Jeremijenko A., Kaleeckal A.H., Latif A.N., Shaik R.M., Abdul-Rahim H.F., Nasrallah G.K., Al-Kuwari M.G., Butt A.A., Al-Romaihi H.E., Al-Thani M.H., Al-Khal A., Bertollini R., Abu-Raddad L.J. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387(1):21–34. doi: 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowen J.E., Addetia A., Dang H.V., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., Franko N.M., Czudnochowski N., Powell A.E., Dellota E., Jr., Ahmed K., Ansari A.S., Cameroni E., Gori A., Bandera A., Posavad C.M., Dan J.M., Zhang Z., Weiskopf D., Sette A., Crotty S., Iqbal N.T., Corti D., Geffner J., Snell G., Grifantini R., Chu H.Y., Veesler D. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377(6608):890–894. doi: 10.1126/science.abq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., Yu Y., Wang P., Zhang Z., Liu P., An R., Hao X., Wang Y., Wang J., Feng R., Sun H., Zhao L., Zhang W., Zhao D., Zheng J., Yu L., Li C., Zhang N., Wang R., Niu X., Yang S., Song X., Chai Y., Hu Y., Shi Y., Zheng L., Li Z., Gu Q., Shao F., Huang W., Jin R., Shen Z., Wang Y., Wang X., Xiao J., Xie X.S. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608(7923):593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., Karim F., Bernstein M., Lustig G., Archary D., Smith M., Ganga Y., Jule Z., Reedoy K., Hwa S.H., Giandhari J., Blackburn J.M., Gosnell B.I., Abdool Karim S.S., Hanekom W., von Gottberg A., Bhiman J.N., Lessells R.J., Moosa M.S., Davenport M.P., de Oliveira T., Moore P.L., Sigal A. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng S.M., Mok C.K.P., Chan K.C., Ng S.S., Lam B.H., Luk L.L., Ko F.W., Chen C., Yiu K., Li J.K., Chan K.K., Tsang L.C., Poon L.L., Hui D.S., Peiris M. SARS-CoV-2 Omicron variant BA.2 neutralisation in sera of people with Comirnaty or CoronaVac vaccination, infection or breakthrough infection, Hong Kong, 2020 to 2022. Euro Surveill. 2022;27(18) doi: 10.2807/1560-7917.ES.2022.27.18.2200178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., Ng S.S., Chan K.C.K., Ko F.W., Chen C., Yiu K., Lam B.H.S., Lau E.H.Y., Chan K.K.P., Luk L.L.H., Li J.K.C., Tsang L.C.H., Poon L.L.M., Hui D.S.C., Peiris M. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022;28(3):486–489. doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruell H., Vanshylla K., Korenkov M., Tober-Lau P., Zehner M., Münn F., Janicki H., Augustin M., Schommers P., Sander L.E., Kurth F., Kreer C., Klein F. SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., Samsunder N., Sivro A., San J.E., Giandhari J., Tegally H., Pillay S., Naidoo Y., Mazibuko M., Miya Y., Ngcobo N., Manickchund N., Magula N., Karim Q.A., von Gottberg A., Abdool Karim S.S., Hanekom W., Gosnell B.I., Lessells R.J., de Oliveira T., Moosa M.S., Sigal A. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13(1):4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 12.Lau E.H., Hui D.S., Tsang O.T., Chan W.H., Kwan M.Y., Chiu S.S., Cheng S.M., Ko R.L., Li J.K., Chaothai S., Tsang C.H., Poon L.L., Peiris M. Long-term persistence of SARS-CoV-2 neutralizing antibody responses after infection and estimates of the duration of protection. EClinicalMedicine. 2021;41 doi: 10.1016/j.eclinm.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., Rubin C., Freedman L., Kreiss Y., Regev-Yochay G. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385(24):e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Proc Natl Acad Sci USA. Vol. 117. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells; pp. 7001–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMenamin M.E., N J., Lin Y., Wong J.Y., Cheung J.K., Lau E.H.Y., Wu P., Leung G.M., Cowling B.J. Vaccine effectiveness of one, two, and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong: a population-based observational study. Lancet Inf. Dis. 2022 doi: 10.1016/S1473-3099(22)00345-0. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.New York State Department of Health (2022). New York State Department of Health Announces Emergence of Recently Identified, Highly Contagious Omicron Subvariants in New York and Urges Continued Vigilance Against COVID-19.
- 17.Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., Baxter C., Althaus C.L., Anyaneji U.J., Kekana D., Viana R., Giandhari J., Lessells R.J., Maponga T., Maruapula D., Choga W., Matshaba M., Mbulawa M.B., Msomi N., Naidoo Y., Pillay S., Sanko T.J., San J.E., Scott L., Singh L., Magini N.A., Smith-Lawrence P., Stevens W., Dor G., Tshiabuila D., Wolter N., Preiser W., Treurnicht F.K., Venter M., Chiloane G., McIntyre C., O'Toole A., Ruis C., Peacock T.P., Roemer C., Pond S.L.K., Williamson C., Pybus O.G., Bhiman J.N., Glass A., Martin D.P., Jackson B., Rambaut A., Laguda-Akingba O., Gaseitsiwe S., von Gottberg A., de Oliveira T. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., Das R., Skelly D., Ritter T.G., Amini A., Bibi S., Adele S., Johnson S.A., Constantinides B., Webster H., Temperton N., Klenerman P., Barnes E., Dunachie S.J., Crook D., Pollard A.J., Lambe T., Goulder P., Paterson N.G., Williams M.A., Hall D.R., Fry E.E., Huo J., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422–2433. doi: 10.1016/j.cell.2022.06.005. e2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., Choga W.T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N.Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S.L., Kraemer M.U.G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R.J., Lockman S., Lucaci A.G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M.B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M.S., Mphoyakgosi T., Msomi N., Mwangi P.N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J.E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., van Wyk S., Weaver S., Wibmer C.K., Wilkinson E., Wolter N., Zarebski A.E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O.G., Bhiman J., Glass A., Martin D.P., Rambaut A., Gaseitsiwe S., von Gottberg A., de Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature. 2022;603(7902):679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., Shah J.G., Nguyen N., Chen Z., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D.D. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022 doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. (2021, 26 November 2021). "Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern.".
- 23.Zeng G., Wu Q., Pan H., Li M., Yang J., Wang L., Wu Z., Jiang D., Deng X., Chu K., Zheng W., Wang L., Lu W., Han B., Zhao Y., Zhu F., Yu H., Yin W. Immunogenicity and safety of a third dose of CoronaVac, and immune persistence of a two-dose schedule, in healthy adults: interim results from two single-centre, double-blind, randomised, placebo-controlled phase 2 clinical trials. Lancet Infect. Dis. 2022;22(4):483–495. doi: 10.1016/S1473-3099(21)00681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.