Abstract
B cell-mediated immune responses play important roles in controlling SARS-CoV infection. Here, we performed the single-cell B cell receptor sequencing (scBCR-seq) of the PBMC samples from eleven healthy controls, five asymptomatic subjects and 33 symptomatic COVID-19 patients with various clinical presentations, and subsequently analyzed the abundance and diversity of the BCR repertoires in different groups, respectively. We revealed the skewed usage of the IGHV, IGLV and IGKV genes and identified a number of heavy or light chain VDJ gene pairs and combinational preference in each group, such as IGKV3–7 and IGKV2–24 enriched in the asymptomatic subjects, whereas IGHV3–13, IGHV3–23-IGHJ4, IGHV1–18–IGLV3–19, IGHV1–18–IGLV3–21, and IGHV1–18–IGLV3–25 enriched in the recovery patients with severe diseases. We also observed the differential expression of IGHV3–23 in various B cell clusters by analysis of the scRNA-seq data. Additional dock analysis indicated that IGHV3–13 could bind to the spike protein of SARS-CoV-2. These findings may advance our understanding of the humoral immune responses in COVID-19 patients and help develop novel vaccine candidates as well as therapeutical antibodies against SASR-CoV-2 infections.
Keywords: SARS-COV-2, COVID-19, Single-cell B cell receptor (BCR) sequencing, B cell receptor repertoire, Asymptomatic infection, Antibody
1. Introduction
The pandemic of the coronavirus disease 2019 (COVID-19) has posed unprecedented challenges to the international communities. As of August 5, 2022, the cumulative number of coronavirus disease-19 (COVID-19) has exceeded 579 million confirmed cases, including over 6.4 million deaths worldwide [1]. In addition, the emerging variants of SARS-CoV-2, such as the Alpha, Beta, Gamma, Delta, and Omicron have generated great concerns across the international communities [2], [3], [4], [5], [6]. In particular, the existing vaccines and convalescent serums have shown reduced neutralization activities against these SARS-CoV-2 variants [7,8].
SARS-CoV-2 infection may exhibit diverse clinical presentations, ranging from asymptomatic infection (AS) to critical conditions. While many studies have been conducted to understand the pathogenesis of the moderate disease (MD) and the severe disease (SD), the AS remains poorly characterized [9], [10], [11], [12], [13], [14], [15]. Further characterization of the immune responses in AS may help us understand the protective immunity against SARS-CoV-2 infection [16]. Previous studies have shown that asymptomatic individuals developed specific IgM, memory B cells and T cells against the S1 and N proteins of SARS-CoV2, whereas the recovered patients developed SARS-CoV-2-specific IgG antibodies as well as memory B and T cells [17], [18], [19], [20]. Since convalescent plasma has been successfully used for treatment of COVID-19 patients, additional study of convalescent patients may help us understand the humoral immunity and develop novel strategies against COVID-19 [21,22].
The B-cell receptor (BCR) is an immunoglobulin molecule to recognize specific antigen. The variable (V), diversity (D) and joining (J) gene segments of the immunoglobulin variable region undergo random rearrangements to give different antigenic characteristics of B cells, and thus form a huge BCR repertoires. Therefore, investigation of the abundance and diversity of the BCR repertoires in different type of COVID-19 patients may help develop B cell-based therapeutic antibodies and vaccines. Previous studies have demonstrated the crucial roles of the B cell-mediated immune responses against SARS-COV-2 infection and the BCR bias in the recovery patients [23,24]. However, a comparative study of BCR repertoires in COVID-19 patients with various clinical presentations has not reported yet.
Here, we performed the single-cell BCR sequencing (scBCR-seq) and analyzed the BCR repertoires in the asymptomatic (AS) and symptomatic (SM) COVID-19 patients as well as the healthy controls (HC). We revealed the skewed usage of the IGHV, IGLV and IGKV genes in AS and SM, respectively. Moreover, we identified a number of unique heavy or light chain VDJ gene pairs and combinational preferences in each group. These findings may advance our understanding of the humoral immune responses in COVID-19 patients and help develop novel vaccine candidates and therapeutical antibodies against SASR-CoV-2 infections.
2. Materials and method
2.1. Study cohort and blood sample collection
This study was approved by the Ethics Committee of The First affiliated Hospital of Xi'an Jiaotong University (XJTU1AF2020LSK-015) and The Renmin Hospital of Wuhan University (WDRY2020-K130). All participants enrolled in this study offered the written informed consent, or the oral consent from the patients or their family members, whereas written informed consent was waived by the Provincial and National Health Commissions in China under the exceptional circumstances for investigation of an ongoing disease outbreak.
The definition and classification of all COVID-19 patients in this study follow the Guidelines of the World Health Organization and the “Guidelines on the Diagnosis and Treatment of the Novel Coronavirus Infected Pneumonia” developed by the National Health Commission of People's Republic of China [25], [26], [27]. The study cohort included eleven healthy controls (HC, n = 11) and 38 SARS-CoV-2-infected individuals (COV, n = 38) consisting of five asymptomatic subjects (AS, n = 5) and 33 symptomatic patients (SM, n = 33) (Table 1 , Supplementary Table S1). Moreover, severe disease (SD, n = 10) and SD recovery (SDR, n = 10) group could be further classified from SM. The peripheral blood was collected into the standard EDTA-K2 Vacuette Blood Collection Tubes (Jiangsu Yuli Medical Equipment Co., Ltd, China; Cat.Y09012282) and stored at room temperature or 4 ℃ until processed. The peripheral blood mononuclear cells (PBMCs) were prepared from these study subjects, including two patients (ID: C19 and C26) with blood samples collected at two different time points during this study. PBMCs were stored in the liquid nitrogen storage tank (-196 ℃) until being used for the studies. All experimental procedures were completed inside a biosafety level 2 (BSL-2) laboratory at the Department of Clinical Diagnostic Laboratories, Renmin Hospital of Wuhan University.
Table 1.
Characteristics of the study cohort and main laboratory tests.
| Entire cohort (n = 49) | HC(n = 11) | AS(n = 5) | SM(n = 33) | p value | SD(n = 10) | SDR(n = 11) | p value | |
|---|---|---|---|---|---|---|---|---|
| Age, years (sd) | 54.2(14.3) | 43.1(7.9) | 41.6(20.5) | 59.8(11.6) | <0.01 | 65.0(9.9) | 63.6(11.5) | 0.76 |
| Gender | 0.86 | 0.16 | ||||||
| Female | 25(51.0%) | 6(54.5%) | 2(40.0%) | 17(51.5%) | NA | 1(10%) | 4(36.4%) | NA |
| Male | 24(49.0%) | 5(45.5%) | 3(60.0%) | 16(48.5%) | NA | 9(90%) | 7(63.6%) | NA |
| Mortality | 1(2.0%) | 0 | 0 | 1(3.0%) | 0.78 | 1(10%) | 0 | 0.28 |
| Diagnostic tests | ||||||||
| Chest CT abnormal | 33(67.3%) | 0 | 0 | 33(100%) | <0.01 | 10(100%) | 11(100%) | NA |
| Nucleic acid test (+) | 38(77.6%) | 0 | 5(100%) | 33(100%) | <0.01 | 10(100%) | 11(100%) | NA |
| SARS-COV-2 IgM (+) | 22(50%) | 0 | 0 | 22(66.7%) | <0.01 | 9(90%) | 6(54.5%) | 0.07 |
| SARS-COV-2 IgG (+) | 36(73.5%) | 0 | 3(60.0%) | 33(100%) | <0.01 | 10(100%) | 11(100%) | NA |
| Laboratory tests (sd) | ||||||||
| WBC × 10⁹ /L | 6.73(2.84) | NA | 7.66(2.93) | 6.58(2.85) | 0.23 | 7.46(4.49) | 5.74(1.88) | 0.60 |
| Neutrophil × 10⁹ /L | 4.70(2.72) | NA | 4.90(2.19) | 4.67(2.82) | 0.65 | 6.25(4.37) | 4.5(1.82) | 0.41 |
| Lymphocyte × 10⁹ /L | 1.30(0.69) | NA | 1.79(0.81) | 1.22(0.66) | 0.12 | 0.71(0.31) | 0.99(0.55) | 0.19 |
| CRP, mg/L | 53.13(36.10) | NA | 26.3(n = 1) | 54.4(36.48) | 0.58 | 59.2(39.6) | 63.7(36.5) | 0.82 |
| IL-2, pg/mL | 4.25(1.90) | NA | 3.80(0.46) | 4.33(2.07) | 0.65 | 3.90(0.29) | 6.77(4.32) | NA |
| IL-4, pg/mL | 3.07(0.51) | NA | 2.99(0.14) | 3.09(0.56) | 0.77 | 3.58(0.50) | 3.32(1.05) | NA |
| IL-6, pg/mL | 22.74(52.43) | NA | 14.05(16.69) | 24.12(56.23) | 0.57 | 87.6(110.0) | 6.38(5.15) | NA |
| IL-10, pg/mL | 8.02(6.17) | NA | 10.26(9.43) | 7.60(5.72) | 0.82 | 15.8(10.2) | 5.7(2.8) | NA |
| CD16+56+, /μL | 160.7(125.5) | NA | 158.0(75.8) | 161.1(131.7) | 0.96 | 84.4(49.9) | 97.0(38.7) | 0.56 |
| CD19+, /μL | 220.2(155.1) | NA | 383.8(116.5) | 199.1(147.9) | 0.04 | 124.4(88.6) | 244.6(181.0) | 0.09 |
| IgM, g/L | 0.95(0.35) | NA | 0.93(0.54) | 0.95(0.34) | 0.92 | 0.97(0.47) | 0.98(0.34) | 0.94 |
| IgG, g/L | 12.95(4.86) | NA | 11.68(2.43) | 13.04(4.99) | 0.70 | 14.77(7.14) | 12.97(4.41) | 0.48 |
| Complement C3, g/L | 1.084(0.184) | NA | 1.260(0.424) | 1.073(0.184) | 0.14 | 1.093(0.135) | 1.007(0.201) | 0.21 |
| Complement C4, g/L | 0.280(0.106) | NA | 0.284(0.117) | 0.279(0.108) | 1.00 | 0.329(0.666) | 0.240(0.095) | 0.04 |
Notes: NA: not available. Data are n (%) or mean±standard deviation (SD), unless otherwise stated. For statistical analyses, the Mann-Whitney U test was done for continuous variables that didn't conform to normal distribution and for homogeneity of variance, whereas Pearson's χ2 test was done for categorical variables. The p value less than 0.05 was considered statistically significant. For laboratory tests, some of the data were not available (see Supplementary Table S1 for more information). HC: Healthy control; AS: Asymptomatic subjects; SM: Symptomatic patients, including the patients with severe disease (SD) and the patients recovered from the severe disease recovery (SDR).
2.2. Preparation of single-cell suspensions
Retrieving the PBMC-containing cryo-tubes from the liquid nitrogen storage tank and placing them in a 37 ℃ water bath for rapid thawing. PBMCs were mixed with 10 mL washing medium (90% DMEM+10% FBS) in a 15-mL polypropylene tube and centrifuged at 500 g for 20 min. The supernatant was then aspirated (repeat twice). The cell pellets were resuspended with 500 μl× PBS (0.04% BSA) in the sterile RNase-free vacutainer tubes and added with 5 ml 1× red blood cell lysis buffer (MACS 130–094–183, 10×) and incubated at room temperature for 10 min to lyse remaining red blood cells. After incubation, the suspension was centrifuged at 500 g for 20 min at room temperature. The suspension was resuspended in 100 μl Dead Cell Removal MicroBeads (MACS 130–090–101) and remove dead cells using Miltenyi Dead Cell Removal Kit (MACS 130–090–101). Then the suspension was resuspended in 1× PBS (0.04% BSA) and centrifuged at 300 g for 3 min at 4 °C (repeat twice). The cell pellet was resuspended in 50 μl of 1× PBS (0.04% BSA). The overall cell viability was confirmed by trypan blue exclusion, which needed to be above 85%, and the single-cell suspensions were counted using a Countess II Automated Cell Counter and the cell concentration was adjusted to 700–1200 cells/μl.
2.3. Chromium 10x genomics library construction and sequencing
Approximately 5000 single cells each sample were captured using the Chromium Single-Cell 5′ kit (V1) according to the manufacturer's instructions (10x Genomics), followed by cDNA amplification and library construction performed according to the standard protocols. The libraries were sequenced on an Illumina NovaSeq 6000 sequencing platform (paired-end multiplexing run, 150 bp) by LC-Bio Technology Co. Ltd., (Hangzhou, China) at a minimum depth of 20,000 reads per cell. To avoid batch effects, the scRNA-seq data sets were generated by the same operators at the same laboratories using the standard operation protocols (SOPs) for cell dissociation, library preparation and sequencing.
2.4. Single-cell RNA-seq data processing
The sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. Sample demultiplexing, barcode processing and single-cell 5′ gene counting were completed by using the Cell Ranger pipeline (https://support.10xgenomics.com/single-cell geneexpression/software/pipelines/latest/what-is-cell-ranger, version 3.1.0) and the scRNA-seq data were aligned to Ensembl genome GRCh38 reference genome. A total of 222,457 single cells captured from eleven healthy controls and 42 COVID-19 patient samples were processed using 10x Genomics Chromium Single Cell 5′ Solution. The Cell Ranger Seurat (version 3.1.1) was used for dimensional reduction, clustering, and analysis of scRNA-seq data. Overall, 207,718 cells passed the quality control threshold: all genes expressed in less than one cell were removed, number of genes expressed per cell >500 as low cut-off, UMI counts less than 500, and the percentage of mitochondrial-DNA derived gene-expression < 25%. To visualize the data, we further reduced the dimensionality of all 207,718 cells by Seurat and used t-Distributed Stochastic Neighbor Embedding (t-SNE) to project the cells into 2D space. The steps included: (1) Using the LogNormalize method of the "Normalization" function of the Seurat software to calculate the expression level of genes; (2) The principal component analysis (PCA) was performed using the normalized expression level, within all the PCs, the top 10 PCs were used to do clustering and t-SNE analysis; (3) Using weighted Shared Nearest Neighbor (SNN) graph-based clustering method to find clusters. The marker genes for each cluster were identified with the "bimod"(Likelihood-ratio test) with default parameters via the FindAllMarkers function in Seurat. This selects marker genes that were expressed in more than 10% of the cells in a cluster and the average log (Fold Change) of greater than 0.26. To further avoid interference of putative multiplets (where more than one cell was loaded into a given well on an array), cells in a defined cluster that had high expression of more than one cell type canonical marker gene were filtered to ensure the data quality. As a result, a total of 119,799 cells were used for the final analysis in this study. The nine cell types were integrated for further sub-clustering. After integration, genes were scaled to unit variance. Scaling, principal component analysis and clustering were performed as described above.
2.5. BCR sequencing and analysis
The Barcoded, full-length BCR V(D)J segments were enriched from the amplified 5′ cDNA libraries using the Chromium Single-Cell V(D)J Enrichment kit according to the manufacturer's protocol (10X Genomics). If both T cells were expected to be present in the partitioned cell population, BCR transcripts were enriched in separate reactions from the same amplified cDNA material. Cell Ranger pipeline (https://support.10xgenomics.com/single-cell-vdj) that processes Chromium single cell 5′ RNA-seq output for V(D)J was employed to assemble, quantify, and annotate paired V(D)J transcript sequences. The workflow of Cell Ranger started by demultiplexing the Illumina sequencer's base call files (BCLs) for each flow cell directory into FASTQ files. Cell calling was performed independently of V(D)J read filtering and assembly. A clonotype was a set of cells that shared the same paired receptor sequences (presumably derived from the same progenitor cell). Cell barcodes were grouped into clonotypes if they shared the same set of productive CDR3 nucleotides sequences by exact match. The repertoire information of all samples was exhibited using a set of basic features: CDR3 abundance, CDR3 bases length, Variable (V) and Joining (J) segment usage (the frequency of associated reads for each V/J gene presented in the samples), the length of V/J gene in CDR3 region, and V-J gene paired frequency in CDR3 junctions. We used the hierarchical cluster (https://stat.ethz.ch/R-manual/Rdevel/library/stats/html/hclust.html) and Kruskal's Non-metric Multidimensional Scaling (isoMDS, https://cran.r-project.org/web/ packages/MASS/MASS.pdf) to estimate Immune Repertoires similarity. To avoid batch effects, as recommended by the standard 10X protocol, the data sets were generated with the same operators and were also produced in the same laboratories using the same cell dissociation protocols, library preparation approaches and/or sequencing platforms.
2.6. Dock analysis
The homology modeling of IGHV3–13 was performed based on the experimental structure (P01766) using SWISS-MODEL (https://www.swissmodel.expasy.org). The S protein of SARS-CoV-2 and IGHV3–13 were docked using HADDOCK server (https://bianca.science.uu.nl/haddock2.4/) with the default parameters [28,29]. The detailed statistical data are displayed for each cluster, representing the average values calculated over the top four best-scoring structures within each cluster. The most reliable cluster was selected by the Z-score (the lower the better) according to HADDOCK. The Z-score indicates how many standard deviations from the average this cluster is located.
2.7. Laboratory tests
All laboratory tests were conducted in the Renmin Hospital of Wuhan University, including white blood cells, neutrophils and lymphocytes (XN2800, SYSMEX, Japan); CRP, IL-2, IL-4, IL-6, IL-10 (AU5400, Beckman Coulter, Inc. USA); CD16/56 NK cells, CD19 B cells (CytoFLEX LX, Beckman Coulter, Inc. USA), and the specific IgM antibodies (Cat#20,203,400,769, YHLO Biotech) and IgG antibodies (Cat#20,203,400,770, YHLO Biotech) against the S and/or N proteins of SARS-COV-2.
2.8. Statistical analysis
All statistical analyses were performed by SPSS (Statistical Package for the Social Sciences) version 23.0 software (SPSS Inc) unless otherwise stated. Diagrams plotting were performed using GraphPad Prism 8.0.2 and R [30]. R packages ggplot2, pheatmap, UpSetR and ggseqlogo were used. Categorical variables were described as frequency rates and percentages, and continuous variables were described using mean with standard deviation. For two groups, using independent group t-tests when the data were normally distributed; otherwise, the Mann-Whitney test was used. For three groups, One-way Anova test conducted which was normally distributed and homoscedasticity, otherwise, Kruskal-Wallis was used. Besides, for the multiple comparisons, Bonferrion correction was used. Proportions for categorical variables were compared using the χ2 test, although the Fisher exact test was used when the data were limited. The statistical details of the experiments were provided in the respective table/figure legends.
3. Data availability
The raw sequence data reported in this paper have been deposited in GEO, under accession code GSE165080 and GSE180118 are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080 [31] and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180118. Other supporting raw data are available from the corresponding author upon reasonable request. Source data are provided with this paper.
4. Results
4.1. The study cohort and the single-cell BCR sequencing of PBMCs
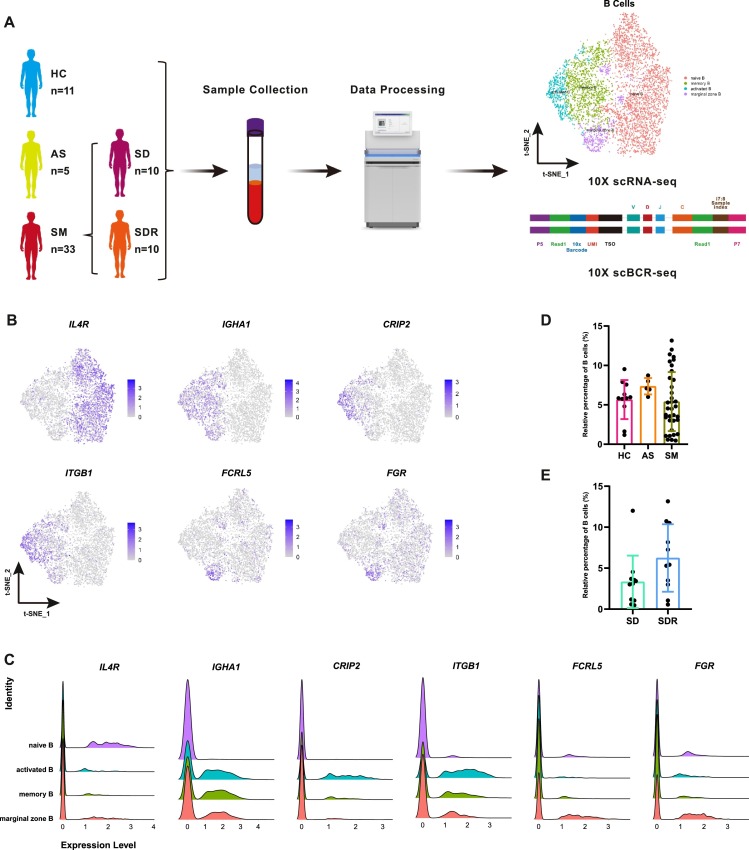
To identify the immune cell alternations in COVID-19 patients, we first performed deep single-cell RNA sequencing on peripheral blood mononuclear cells (PBMCs) and illustrated the immune landscape of B cells (Fig. 1 A). We subsequently completed the scBCR-seq and analyzed the abundance and diversity of the BCR repertoires in eleven health controls (HC), five asymptomatic subjects (AS), and 33 symptomatic COVID-19 patients (SM) that included ten patients with severe disease (SD) and ten recovery patients from the severe disease (SDR) (Fig. 1A, Supplementary Table S1). The T-distributed stochastic neighbor embedding (t-SNE) plots showed that four clusters of B cells: naïve B, memory B, activated B and marginal zone B cells. Moreover, representative marker genes included IL4R (naïve B), IGHA1(memory B), CRIP2 and ITGB1(activated B) as well as FCRL5 and FGR (marginal zone B) (Fig. 1B) [31]. The ridge plot shows the distribution identity of these marker genes in four clusters of B cells (Fig. 1C). Furthermore, more than 90% BCRs were matched to the B cells, and there was no biased proportion of BCRs from a single donor here (Supplementary Fig. S1A, 1B, Supplementary Table S1). We then calculated the relative percentage of B cells in each group. While some of these groups (e.g., SM vs AS, SDR vs SD) had relatively higher percentage (mean) of B cells, there were no significant difference among these groups (Fig. 1D, 1E). However, when we compared the routine laboratory testing results, we found that CD19+ B cells were significantly higher in AS than SM (383.8 ± 116.5, 199.1 ± 147.9, p = 0.036), and the count of CD19+ B cells decreased in SD compared with SDR patients (244.6 ± 181.0, 124.4 ± 88.6, p = 0.09) (Table 1). These results suggested that B cell alternations were associated with SARS-CoV-2 infections.
Fig. 1.
The Study cohort and overview of the single-cell BCR sequencing.
(A) schematic diagram showing the study cohort and the workflow of the single-cell B cell receptor sequencing (BCR-seq). This cohort consisted of eleven health controls (HC), five asymptomatic subjects (AS) and 33 symptomatic COVID-19 patients (SM). The SM included ten patients with severe disease (SD, n = 10) and ten recovery patients from SD (SDR, n = 10). Peripheral blood mononuclear cells (PBMCs) were collected and followed by scRNA-seq and BCR-seq using the 10xGenomics platform. (B) The T-distributed stochastic neighbor embedding (t-SNE) plots showing the marker genes used for identification of the B cell types described above. naïve B (IL4R), memory B (IGHA1), activated B (CRIP2, ITGB1) and marginal zone B (FCRL5, FGR). The plots are color-labelled based on the expression level of the respective marker gene in log scale, which was calculated via LogNormalize method of the "NormalizeData" function of the Seurat software. (C) The ridge plot shows the distribution identity of these marker genes in four clusters of B cells. Cell clusters were showed in different colors. (D) The relative percentage of B cells in HC, AS and SM groups. (E) The relative percentage of B cells in SD and SDR groups. The bars in Fig. 1D and E showing the mean value ± standard deviation (sd), p value ≤ 0.05 was considered significant.
4.2. Identification of BCR clonal expansions in SM and SDR patients
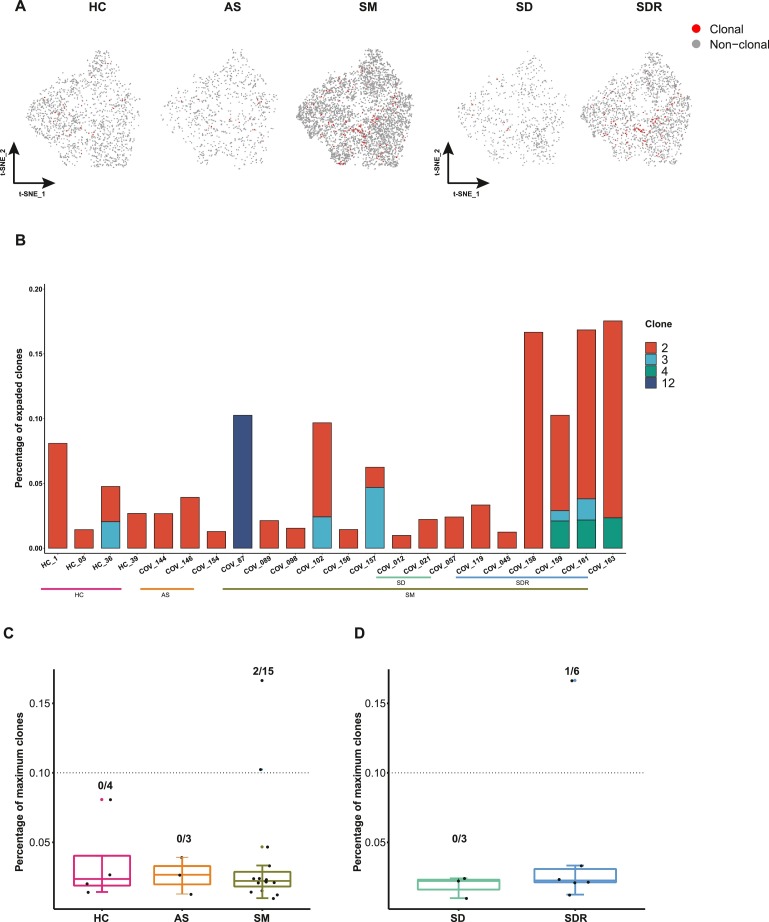
Here, we also performed scBCR-seq to assess the status of clonal expansions in PBMCs derived from the COVID-19 patients and the healthy controls. We found that more clonal expansions occurred in SM group compared with HC and AS. Of note, SDR had much more clonal cells than SD patients. In addition, memory B and activated B cells exhibited high clonal expansions in SM and SDR (Fig. 2 A, Supplementary Fig. S1C). We revealed that most SM patients exhibited highly expanded clones at the individual level (more than 10%) compared with the healthy or asymptomatic subjects (less than 5%) (Fig. 2B, Supplementary Table S2). We also observed that the recovery patients (SDR) had higher expanded clones than the SD patients (Fig. 2B, Supplementary Table S2). Moreover, quantification of the most highly expanded clones for each patient and healthy control found that SM had the highest percentages of the maximum clones (2/15) compared with HC and AS group. Furthermore, we revealed that the maximum clone numbers were higher in SDR (1/6) than SD group (Fig. 2C, D).
Fig. 2.
Detection of BCR clonal expansions in various groups.
(A) The t-SNE plot shows the B cell expansions in the HC, AS, SM, SD and SDR, clonal was marked with red color. (B) The BCR clonal expansion status and frequency of the clonotype in each sample. The number of color blocks represents the complexity of the clonal states. Some individuals’ data were not available due to the limitation of the sequencing technology. (C) The number of highly expanded clones (above the dotted line) in HC, AS, and SM, respectively. The highly expanded clone was defined as the clone comprising 10% or more of all BCR sequences. (D) The number of highly expanded clones (above the dotted line) in SD and SDR, respectively. In Fig. 2C and 2D, the bars showing mean ± sd, HC (n = 4), AS (n = 3), SM (n = 15), SD (n = 3) and SDR (n = 6). Samples with the percentage of expanded clones<0 were not shown.
4.3. B-cell immune repertoire in asymptomatic and symptomatic COVID-19 patients
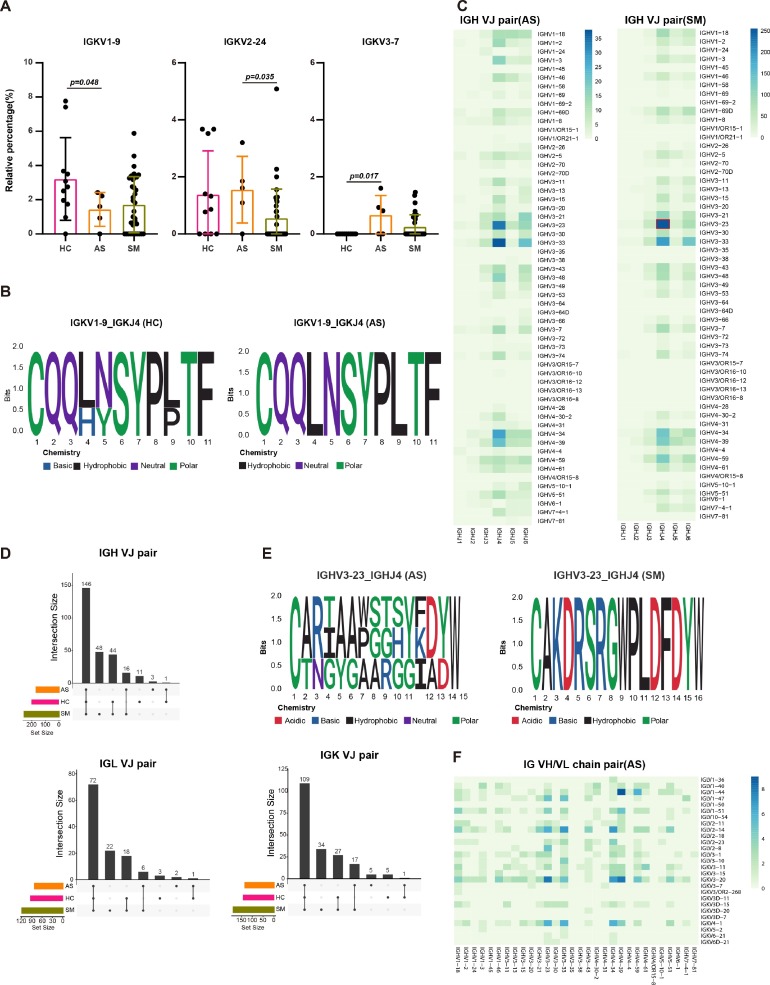
To study the specific changes and preference gene usages of BCR in COVID-19 patients with various clinical presentations, we compared the usage of VDJ genes in asymptomatic subjects (AS), symptomatic patients (SM) and health controls (HC). We found that IGKV2–24 and IGKV3–7 were highly enriched in the AS subjects compared with the SM and HC groups, whereas IGKV1–9 was preferred in the HC group (Fig. 3 A, Supplementary Fig. S2A-C, Supplementary Table S3). CDR3 (complementary determining region 3) is the most important region of an antibody structure, which interacts with the antigen. Usually, CDR3 region has a conserved length of amino acids with antigen specificity. To explore the relationship of gene usage and V-J pairs with the clonal expansions resulting from SARS-CoV-2 infection, we identified the CDR3 structures of IgG-BCR expressed on IGKV1–9. We found that eleven amino acids in IGKV1–9 had highest frequencies at specific lengths from HC and AS (Fig. 3B). Furthermore, IGHV3–23-IGHJ4, IGLV1–44-IGLJ3 and IGKV3–20-IGKJ1 were the top V-J pairs in all groups, and the highest frequency was observed in SM (Fig. 3C, Supplementary Fig. S2D-F, Supplementary Table S4–6). The Upset View showed the common and unique VJ pairs in HC, AS and SM group. There were three IGH, five IGK and two IGL VJ pairs in AS patients, respectively. However, the frequency of these VJ pairs was low (Fig. 3D, Supplementary Table S4–6). We also analyzed the expression of IGHV3–23 and IGHJ4 pair in AS and SM, and found ten (12, 13, 15–22 amino acids) and four (14, 16, 17 and 20 amino acids) types of amino acids lengths in AS and SM group, respectively (Fig. 3E, Supplementary Fig. S2G), suggesting that these CDR3 region structures had a high variability and SARS-CoV-2 specificity. Additional analysis coupled with the scRNA-seq data demonstrated that IGHV3–23 was differentially expressed in naive and memory B cells among four clusters (Supplementary Fig. S2K, Supplementary Table S8). Moreover, the skewed usage and the frequency of VH/VL combinations were detected among different groups. For instance, the IGHV4–44-IGLV1–44 combination was highly enriched in AS and SM group, the IGHV4–34-IGKV1D-39 combination had the highest frequencies in SM (Fig. 3F, Supplementary Fig. S2H), whereas IGLV3−9-IGHV6–1 and IGLV3−27-IGHV7−4−1 combinations were specifically enriched in AS (Supplementary Fig. S2I, 2 J, Supplementary Table S7), suggesting that the expanded BCR repertoire were associated with the viral infection states. Some of these findings are consistent with the published data (CoV-AbDab) [32], demonstrating the VJ gene usage and bias in different groups (Table 2 ).
Fig. 3.
BCR repertoire in asymptomatic and symptomatic COVID-19 patients.
(A) The skewed usage of IGKV genes in HC, AS and SM group, respectively. (B) The motif logo plot showing the CDR3 length distribution and amino acids frequency of IGKV1–9 in AS. The relative size of the letters indicated their frequency in the sequences and the total height of the letters depicts the information content of the position in bits. (C) The Heatmaps showing the VJ gene pairs of IGH, IGK and IGL in AS and SM. The color range indicating the usage counts of specific V-J gene pairs. (D) The Upset view showing the common and unique VJ pairs of IGH, IGK and IGL in HC, AS and SM group, respectively. (E). The motif logo plot showing the CDR3 length distribution and amino acids frequency of IGHV3–23-IGHJ4 in AS and SM. (F) The Heatmaps showing the V gene heavy and light chain combinations in AS. The color range indicating the usage counts. The error bars in Fig. 3A represent mean ± sd. p < 0.05 was considered significant. The samples include HC (n = 11), AS (n = 5) and SM (n = 35, total 33 patients and two patients were sampled twice at different disease stages).
Table 2.
BCR clonotypes identified in this study compared with the antibodies reported in the CoV-AbDab.
| ID | IG V gene | IG J gene | Binds | Protein & Epitope | Group | Oringin | Source |
|---|---|---|---|---|---|---|---|
| No.1 | IGKV2–24 | IGKJ2 | SARS-CoV2 | Unknow | AS | human | This study |
| NAa | |||||||
| No.2 | IGKV3–7 | IGKJ1 | SARS-CoV2 | Unknow | AS | human | This study |
| F26G3 | IGKV3–7 | IGKJ1 | SARS-CoV1 | Unknow | mouse | Michael Gubbins et al. [40] | |
| No.3 | IGHV3–23 | IGHJ4 | SARS-CoV2 | Unknow | SM, SDR | human | This study |
| CQTS127 | IGHV3–23 | IGHJ4 | SARS-CoV2 | S; RBD | human | [41] | |
| No.4 | IGKV3–20 | IGKJ1, 2 | SARS-CoV2 | Unknow | SM, SDR | human | This study |
| CQTS127 | IGKV3–20 | IGKJ2 | SARS-CoV2 | S; RBD | human | [42] | |
| No. 5 | IGKV1D-39 | IGKJ1 | SARS-CoV2 | Unknow | SM, SDR | human | This study |
| NA | |||||||
| No.6 | IGLV1–44 | IGLJ3 | SARS-CoV2 | Unknow | SM, SDR | human | This study |
| 12E; 15D et al. | IGLV1–44 | IGLJ3 | SARS-CoV1,2 | S; RBD | human | [43,44] | |
| No.7 | IGLV2–14 | IGLJ2 | SARS-CoV2 | Unknow | SM, SDR | human | This study |
| BLN2 | IGLV2–14 | IGLJ2 | SARS-CoV2 | S; NTD | Phage display | [45] |
Note: This Supplementary Table Shows the V J gene usage and bias in different groups. The clonotypes were compared with public data in CoV-AbDab [32]. NA: not available. HC: Healthy control; AS: Asymptomatic subjects; SM: Symptomatic patients, including the patients with severe disease (SD) and the patients recovered from the severe disease recovery (SDR).
4.4. The skewed usage of BCR repertoire in severe (SD) and recovery (SDR) COVID-19 patients
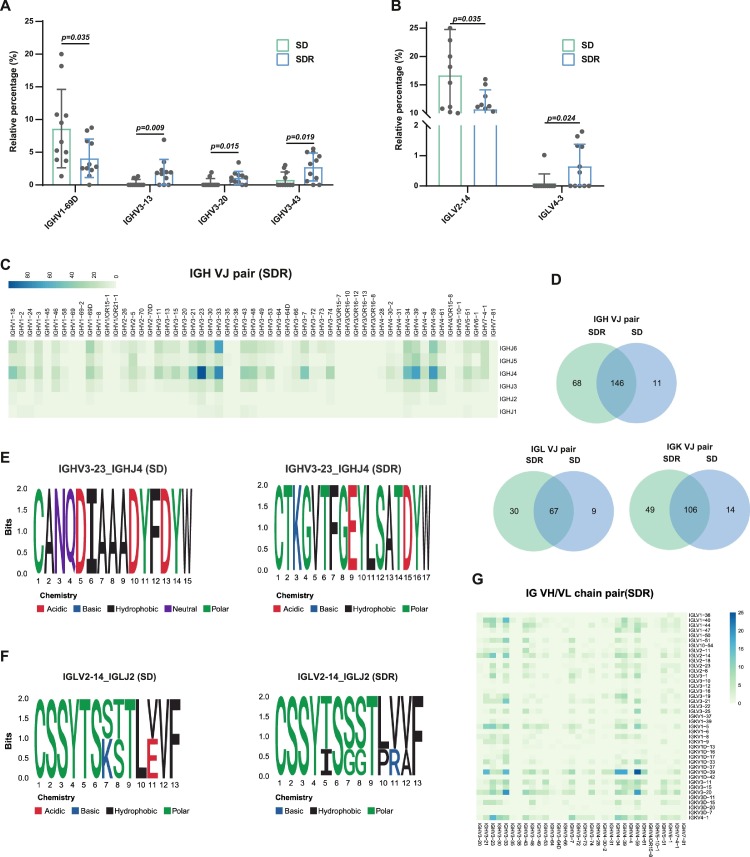
To study the biased V(D)J rearrangements of the BCR in the severe (SD) and recovery (SDR) COVID-19 patients, we compared the usage of V(D)J genes between these groups. IGLV2–14 and IGHV1–69D were dominant IGV genes in SD, whereas IGLV4–3 and several IGHV3 family genes (IGHV3–13, IGHV3–20, IGHV3–43) were enriched in SDR (Fig. 4 A, B, Supplementary Fig. S3A-C, Supplementary Table S3). We also performed dock analysis and showed that IGHV3–13 may bind to the S protein of SARS-CoV-2 (Supplementary Table S3). Moreover, we compared the common and unique VJ pairs between SD and SDR group, and found that the V-J pairs of IGKV1D39-IGKJ1, IGHV4–59-IGHJ4, IGHV4–39-IGHJ4 and IGHV3–23-IGHJ4 enriched in SDR, indicating that B cells might have undergone unique and specific V(D)J rearrangements in the recovery patients (Fig. 4C-D, Supplementary Fig. S3D, 3E, Supplementary Table S4–6). Furthermore, we identified the IGHV3–23-IGHJ4 and IGLV2–14-IGLJ2 expressed in the CDR3 of BCR in SD and SDR, and revealed the length distribution ranging from 8 to 23 amino acids (Fig. 4E, F, Supplementary Figs. S3F, 3G). Finally, we observed different IGHV–IGKV/LV combination preference in SD and SDR (Fig. 4G, Supplementary Figs. S3H-J). For instance, IGHV1–18-IGLV3–19, IGHV1–18-IGLV3–21 IGHV1–18-IGLV3–25, IGHV3–13–IGLV3–9, and IGHV6–1-IGLV4–60 specifically paired in SDR. Our results suggested that skewed usage of V(D)J genes and rearrangements may depend on the disease process and IGHV3–13 could be a potential candidate immunotherapy.
Fig. 4.
The skewed usage of BCRs in SD and SDR COVID-19 patients.
(A) The differential usage of IGH V genes in SD and SDR groups. (B) The differential usage of IGL V genes in SD and SDR groups. (C) The Heatmaps showing the VJ gene pairs of IGH, IGK and IGL in SDR. The color range indicating the usage counts of specific V-J gene pairs. (D) The Venn diagram showing the common and unique VJ pairs of IGH, IGK, and IGL in SD and SDR groups. (E) The motif logo plot showing the CDR3 length distribution and amino acids frequency of IGHV3–23_IGHJ4 in SD and SDR. (F) The motif logo plot showing the CDR3 length distribution and amino acids frequency of IGLV2–14_IGHJ2 in SD and SDR. (G) The Heatmaps showing the V gene heavy and light chain combinations in SDR. The color range indicating the usage counts. The error bars in Fig. 4A and B represent mean ± sd. p < 0.05 was considered significant. The samples include SD (n = 11, total 10 patients and one patient was sampled twice) and SDR (n = 11, total 10 patients and one sample was sampled twice).
5. Discussion
In this study, we identified a number of novel skewed usage of B-cell receptors in the COVID-19 patients with various clinical presentations using scBCR-seq coupled with the scRNA-seq data. In particular, we revealed that IGKV3–7 was highly enriched in asymptomatic subjects (AS). Interestingly, IGKV3–7 has been reported as the biased usage of VDJ genes associated with the immune response in kidney transplantation [33], suggesting that IGKV3–7 may play a role in discriminating self from non-self in humans, and consequently contribute to fighting the invading SASR-CoV-2 viruses and maintaining the symptom-free state of the AS individuals. Another significantly enriched gene in AS, IGKV2–24, has not been reported yet but warrants further studies (dock analysis, expression level test of the protein, flow cytometry-based competition assay, SARS-CoV-2 authentic viruses neutralization assay, functional repertoire analyses, etc.). On the other hand, IGHV3–13 was highly enriched in the recovery patients from the severe diseases (SDR). The dock analysis showed that IGHV3–13 could bind to the S protein of SARS-CoV-2. Interestingly, IGHV3–13 was reported to be associated with the protective antibodies against Ebola virus [34], suggesting that IGHV3–13 might be associated with the protective antibodies against SARS-CoV-2 infection.
We also examined and compared the usage of the V(D)J pairs and the variability of CDR3 region structure in the COVID-19 patients. Our finding that IGHV3–23-IGHJ4 was highly enriched in SM (compared with AS and HC) and SDR (compared with SD) is consistent with the result reported in a previous study [35,36]. In addition, our scRNA-seq data showed that IGHV3–23 was differentially expressed in four B cells clusters, suggesting that IGHV3–23-IGHJ4 might be involved in the humoral immune responses against SARS-CoV-2 infection.
Furthermore, we identified a number of VDJ gene and VH/VL chain combinational preference in SDR group, including IGHV3–23-IGHJ4, IGHV1–18-IGLV3–19, IGHV1–18-IGLV3–21, and IGHV1–1-IGLV3–25. Of note, a similar VDJ gene combinational preference IGKV3–20-IGKJ1 was reported in the massive bias in the immunoglobulin gene repertoire of primary vitreoretinal lymphoma [37]. In addition, IGHV1–18_IGLV3–20 has been reported as the anti-RBD antibody of SARS-COV-2 [38,39]. These results suggest that IGHV3–23-IGHJ4, IGHV1–18-IGLV3–19, IGHV1–18-IGLV3–21, and IGHV1–18-IGLV3–25 may have neutralizing activities against SASR-CoV-2 infection.
The limitations of this study include the relatively small sample size of the asymptomatic group, the variations in the time-points of sample collections due to the challenges from the disease outbreak, and lack of experimental validations on some of the novel skewed usage of B-cell receptors.
In summary, we have identified a number of novel skewed usage of B-cell receptors in COVID-19 patients with various clinical presentations. Our findings will not only advance the understanding of the B cell-mediated immune responses in COVID-19 patients, but may also help identify novel therapeutical antibodies against SARS-CoV-2 infection.
Funding
This study was supported by the Department of Science and Technology of Shaanxi Province (Grant No. 2020ZDXM2-SF-02) (CZ and BS) and the operational funds from The First Affiliated Hospital of Xi'an Jiaotong University (CZ and BS).
CRediT authorship contribution statement
Junpeng Ma: Methodology, Writing – original draft, Writing – review & editing. Han Bai: Methodology, Writing – original draft, Writing – review & editing. Tian Gong: Methodology, Writing – original draft, Writing – review & editing. Weikang Mao: Methodology, Writing – original draft. Yijun Nie: Methodology. Xuan Zhang: Methodology. Yanyan Da: Methodology. Xiaorui Wang: Methodology. Hongyu Qin: Methodology. Qiqi Zeng: Methodology. Fang Hu: Methodology. Xin Qi: Methodology. Bingyin Shi: Conceptualization, Resources, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition. Chengsheng Zhang: Conceptualization, Methodology, Resources, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition, Project administration.
Declarations of Competing Interest
The author Weikang Mao is employed by the LC-BIO TECHNOLOGIES (HANGZHOU) CO., LTD., China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are extremely grateful to all the patients and their families for participation of this study and providing the valuable information. We also thank our colleagues at The First Affiliated Hospital of Xi'an Jiaotong University and The Renmin Hospital of Wuhan University for their kind help and strong support throughout the course of this study. We would like to acknowledge all other supporting entities, including the FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858), the EOSC-hub (project# 777536), and the EGI-ACE (project# 101017567). In particular, the European e-Infrastructure projects for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MCC, INFN-PADOVA-STACK, INFN-LNL-2, NCG-INGRID-PT, TW-NCHC, CESGA, IFCA-LCG2, UA-BITP, SURFsara and NIKHEF.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.imlet.2022.08.006.
Appendix. Supplementary materials
References
- 1.WHO Coronavirus (COVID-19) Dashboard. https://www.who.int/.
- 2.Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 3.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Weekly epidemiological update on COVID-19: 11 May 2021. . Available online: https://www.who.int/publications/m/item/weekly-epidemiological -update-on-covid-19—11-may-2021.
- 6.Araf Y., Akter F., Tang Y.D., Fatemi R., Parvez S.A., Zheng C., Hossain G. Omicron variant of SARS-CoV-2: genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B1.617 by vaccine and convalescent serum. Cell. 2021;184(16) doi: 10.1016/j.cell.2021.06.020. 4220-4236.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen A.T.C., Coura-Filho G.B., Rehder M.H.H. Clinical Characteristics of Covid-19 in China. N. Engl. J. Med. 2020;382:1860. doi: 10.1056/NEJMc2005203. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5(49):eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McElvaney O.J., McEvoy N.L., McElvaney O.F., Carroll T.P., Murphy M.P., Dunlea D.M., Ni Choileain O., Clarke J., O'Connor E., Hogan G., et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020;202:812–821. doi: 10.1164/rccm.202005-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bui L.T., Winters N.I., Chung M.I., Joseph C., Gutierrez A.J., Habermann A.C., Adams T.S., Schupp J.C., Poli S., Peter L.M., et al. Single-cell RNA-sequencing reveals dysregulation of molecular programs associated with SARS-CoV-2 severity and outcomes in patients with chronic lung disease. bioRxiv. 2020 doi: 10.1101/2020.10.20.347187. [DOI] [Google Scholar]
- 15.Emery J.C., Russell T.W., Liu Y., Hellewell J., Pearson C.A., Group C.C.-W., Knight G.M., Eggo R.M., Kucharski A.J., Funk S., et al. The contribution of asymptomatic SARS-CoV-2 infections to transmission on the Diamond Princess Cruise Ship. elife. 2020;9:e58699. doi: 10.7554/eLife.58699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohn M.K., Hall A., Sepiashvili L., Jung B., Steele S., Adeli K. Pathophysiology of COVID-19: mechanisms underlying disease severity and progression. Physiology (Bethesda). 2020;35:288–301. doi: 10.1152/physiol.00019.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183. doi: 10.1016/j.cell.2020.11.029. e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei Q., Li Y., Hou H.Y., Wang F., Ouyang Z.Q., Zhang Y., Lai D.Y., Banga Ndzouboukou J.L., Xu Z.W., Zhang B., et al. Antibody dynamics to SARS-CoV-2 in asymptomatic COVID-19 infections. Allergy. 2021;76:551–561. doi: 10.1111/all.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achiron A., Gurevich M., Falb R., Dreyer-Alster S., Sonis P., Mandel M. SARS-CoV-2 antibody dynamics and B-cell memory response over time in COVID-19 convalescent subjects. Clin. Microbiol. Infect. 2021;27(9) doi: 10.1016/j.cmi.2021.05.008. 1349.e1-1349.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurevich M., Zilkha-Falb R., Sonis P., Magalashvili D., Menascu S., Flechter S., Dolev M., Mandel M., Achiron A. SARS-CoV-2 memory B and T cell profiles in mild COVID-19 convalescent patients. Int. J. Infect. Dis. 2022;115:208–214. doi: 10.1016/j.ijid.2021.12.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabets A., Bila G., Grytsko R., Samborskyy M., Rebets Y., Vari S.G., Pagneux Q., Barras A., Boukherroub R., Szunerits S., et al. The potential of developing pan-coronaviral antibodies to spike peptides in convalescent COVID-19 patients. Arch. Immunol. Ther. Exp. (Warsz.) 2021;69:5. doi: 10.1007/s00005-021-00607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wooding D.J., Bach H. Treatment of COVID-19 with convalescent plasma: lessons from past coronavirus outbreaks. Clin. Microbiol. Infect. 2020;26:1436–1446. doi: 10.1016/j.cmi.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueso T., Pouderoux C., Pere H., Beaumont A.L., Raillon L.A., Ader F., Chatenoud L., Eshagh D., Szwebel T.A., Martinot M., et al. Convalescent plasma therapy for B-cell-depleted patients with protracted COVID-19. Blood. 2020;136:2290–2295. doi: 10.1182/blood.2020008423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultheiss C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., et al. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53:442–455. doi: 10.1016/j.immuni.2020.06.024. e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Government of China . 8nd Ed. 2021. New Coronavirus Pneumonia Prevention and Control Program.http://www.nhc.gov.cn/jkj/s3577/202105/6f1e8ec6c4a540d99fafef52fc86d0f8/files/4a860a7e85d14d55a22fbab0bbe77cd9.pdf [Accessed May 14, 2021] [Google Scholar]
- 26.World Health Organization (2020): Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/health-topics/coronavirus/laboratory-diagnostics-for-novel-coronavirus. [Accessed March 19, 2020].
- 27.World Health Organization (2020): “COVID-19: surveillance, case investigation and epidemiological protocols; https://www.who.int/internal-publications-detail/considerations-in-the-investigation-of-cases-and-clusters-of-covid-19 [Accessed October 22, 2020].
- 28.Honorato R.V., Koukos P.I., Jimenez-Garcia B., Tsaregorodtsev A., Verlato M., Giachetti A., Rosato A., Bonvin A. Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.729513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Team, R.C. R Foundation for Statistical Computing; Vienna, Austria: 2008. R: A language and Environment For Statistical Computing.http://www.r-project.org Available online: [Google Scholar]
- 31.Wang X., Bai H., Ma J., Qin H., Zeng Q., Hu F., Jiang T., Mao W., Zhao Y., Chen X., et al. Identification of distinct immune cell subsets associated with asymptomatic infection, disease severity, and viral persistence in COVID-19 patients. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.812514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon H.J., Fang T., Lee J.G., Jang J.Y., Kim K., Choi S., Yan J.J., Ryu J.H., Koo T.Y., Ahn C., et al. VDJ gene usage of B Cell Receptors in peripheral blood of ABO-incompatible kidney transplantation patients. Transplant. Proc. 2018;50:1056–1062. doi: 10.1016/j.transproceed.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 34.Cagigi A., Misasi J., Ploquin A., Stanley D.A., Ambrozak D., Tsybovsky Y., Mason R.D., Roederer M., Sullivan N.J. Vaccine generation of protective ebola antibodies and identification of conserved B-cell signatures. J. Infect. Dis. 2018;218:S528–S536. doi: 10.1093/infdis/jiy333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.Y., Rustagi A., Rogers A.J., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28(4):516–525. doi: 10.1016/j.chom.2020.09.002. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belhouachi N., Xochelli A., Boudjoghra M., Lesty C., Cassoux N., Fardeau C., Tran T.H.C., Choquet S., Sarker B., Houillier C., et al. Primary vitreoretinal lymphomas display a remarkably restricted immunoglobulin gene repertoire. Blood Adv. 2020;4:1357–1366. doi: 10.1182/bloodadvances.2019000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 39.Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., Katsyv I., Rendeiro A.F., Amin A.D., Schapiro D., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gubbins M.J., Plummer F.A., Yuan X.Y., Johnstone D., Drebot M., Andonova M., Andonov A., Berry J.D. Molecular characterization of a panel of murine monoclonal antibodies specific for the SARS-coronavirus. Mol. Immunol. 2005;42:125–136. doi: 10.1016/j.molimm.2004.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.CN111909260A. Available online: https://patents.google.com/patent/CN111909260A/en.
- 42.WO2005060520. Available online: https://patentimages.storage.googleapis.com/5a/11/f4/7f766a82568126/WO2005060520A2.pdf.
- 43.Han X., Wang Y., Li S., Hu C., Li T., Gu C., Wang K., Shen M., Wang J., Hu J., et al. A rapid and efficient screening system for neutralizing antibodies and its application for the discovery of potent neutralizing antibodies to SARS-CoV-2 S-RBD. bioRxiv. 2020 doi: 10.1101/2020.08.19.253369. 2020.2008.2019.253369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noy-Porat T., Mechaly A., Levy Y., Makdasi E., Alcalay R., Gur D., Aftalion M., Falach R., Leviatan Ben-Arye S., Lazar S., et al. Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice. iScience. 2021;24 doi: 10.1016/j.isci.2021.102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in GEO, under accession code GSE165080 and GSE180118 are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080 [31] and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180118. Other supporting raw data are available from the corresponding author upon reasonable request. Source data are provided with this paper.