Abstract
Phenotypic plasticity and adaptive evolution enable population persistence in response to global change. However, there are few experiments that test how these processes interact within and across generations, especially in marine species with broad distributions experiencing spatially and temporally variable temperature and pCO2. We employed a quantitative genetics experiment with the purple sea urchin, Strongylocentrotus purpuratus, to decompose family-level variation in transgenerational and developmental plastic responses to ecologically relevant temperature and pCO2. Adults were conditioned to controlled non-upwelling (high temperature, low pCO2) or upwelling (low temperature, high pCO2) conditions. Embryos were reared in either the same conditions as their parents or the crossed environment, and morphological aspects of larval body size were quantified. We find evidence of family-level phenotypic plasticity in response to different developmental environments. Among developmental environments, there was substantial additive genetic variance for one body size metric when larvae developed under upwelling conditions, although this differed based on parental environment. Furthermore, cross-environment correlations indicate significant variance for genotype-by-environment interactive effects. Therefore, genetic variation for plasticity is evident in early stages of S. purpuratus, emphasizing the importance of adaptive evolution and phenotypic plasticity in organismal responses to global change.
Keywords: additive genetic variance, parental effects, plasticity, marine invertebrates, upwelling
1. Introduction
As phenotypic distributions of populations are being shaped by rapid environmental change, much attention has focused on individual species' ecological and evolutionary responses to the altered environments [1]. Processes of selection and phenotypic plasticity can occur simultaneously within a population, modifying demographic processes, and thereby linking ecological and adaptive evolutionary phenotypic change to population persistence [1,2]. Phenotypic plasticity is the main mechanism by which populations can respond to environmental change over the short term [3]. Adaptive plastic responses, defined as plasticity that shifts phenotypes towards trait values that maximize fitness, could occur across generations (parental or carry-over effects) or within generations (intra-generational plasticity). Adaptive parental effects are expected to occur when parental environments predict offspring environments, and when observed, have small but significant effects on offspring traits [4–6]. Alternatively, developmental plasticity is a type of intra-generational plasticity where environments experienced during early development affect later stage phenotypes. Both parental effects and developmental plasticity have the potential to shape population level responses to the environment and pinpointing when in the life cycle environmental change has the strongest effect is key for predicting organismal responses to change.
While phenotypic plasticity can facilitate population persistence, it has limited effectiveness during long-term environmental change. Phenotypic plasticity has developmental constraints that could limit organismal responses to directional increases in environmental change, such as temperature, and there may exist costs to maintaining plasticity [7,8]. Further, plasticity will only be advantageous as long as the range of phenotypes produced across environments by specific genotypes, or the reaction norms [9], continue to align with the phenotypic optima maximizing fitness in each environment [8]. When reaction norms are no longer adaptive across environments, evolutionary adaptation is the only way populations can persist [1,2]. Such microevolutionary responses can occur in population mean phenotype, or in the level of plasticity itself [1,10].
Adaptive evolutionary responses to changing or novel environments rely on the existence of additive genetic variance that aligns with the direction of selection on phenotypic variation [11,12]. Additive genetic variance can be environmentally dependent, thus should be estimated under a variety of scenarios representing predicted environmental changes [4]. Not only is the amount of adaptive variation of a trait dependent on the environment, but also the relative ranking among genotypes of additive genetic values can change across environments, signalling additive genetic variance for plasticity [10,13]. Evolutionary responses to selection, and hence population adaptation to change, relies on both environment-specific additive genetic variance in trait mean as well as the additive genetic variance in plasticity. To determine how populations will respond to global changes and persist, it is essential to simultaneously evaluate the separate contributions of plastic and evolutionary phenotypic shifts during population responses to environmental change.
Strongylocentrotus purpuratus are widely dispersed across coastal habitats along the California Current Large Marine Ecosystem. Throughout their range, extending from British Columbia in the north to Baja California in the south, S. purpuratus experience temperature and pH variation, mostly due to seasonal upwelling, which is expected to increase in frequency and intensity in the Anthropocene [14–17]. High pCO2 alters the carbonate chemistry in seawater, reduces pH, and directly impacts the ability of marine organisms to calcify, including early-stage sea urchins [18]. As S. purpuratus larvae are planktotrophs with long pelagic larval durations, body size and skeletal features are critical for the ability to capture food and can influence predation rates, swimming speeds and stability in flowing water [19–21]. Phenotypic plasticity in larval morphometrics has been observed before in S. purpuratus, both in response to high pCO2 alone [22–25] but also in upwelling conditions mirrored in this experiment [26–28]. In previous experiments, larval cultures were generated from pooled gametes of multiple adults, thus phenotypes represent treatment averages across multiple genotypes and lack resolution to separate the contributions of parental effects, developmental plasticity or genetic effects on variation in the measured traits [27–29]. Here, we used a quantitative genetic analysis to partition out the roles of the environment, genetics and parental effects on observed variation in phenotypic plasticity of larval body size morphometrics. Thus, our experimental design enabled us to further extend these studies by quantifying family-level variation in plastic responses to upwelling and non-upwelling conditions and compare evolvability to short-term plastic responses at ecological time scales, which together extend our knowledge of how marine organisms will respond to global change.
2. Material and methods
(a) . Collection and adult conditioning
Strongylocentrotus purpuratus is an external fertilizer that spawns large numbers of gametes between January and May. Adult urchins were collected by hand on SCUBA from two sites (25 km apart) with similar habitat quality [30], in August and September (site details in electronic supplementary material, S1). Urchins were placed in one of four 90 l glass tanks per treatment (10 urchins per tank, four tanks per treatment), while keeping track of site identity (details in electronic supplementary material). Adult conditioning was conducted over approximately four months under two regimes differing in temperature and pCO2: non-upwelling (N) (mean values 17°C and 596 µatm pCO2) and upwelling (U) (mean values 12.8°C and 1117 µatm pCO2; electronic supplementary material, S1 and table S1). Throughout this conditioning, urchins were fed Macrocystis pyrifera in excess once per week.
Temperature and pCO2 levels were maintained throughout the conditioning period using heat pumps regulated by Nema 4X digital temperature controllers and a flow-through CO2 mixing system, modified from Fangue et al. [31]. Treated seawater was evenly pumped from two reservoir tanks to conditioning tanks at a rate of 20 l h−1 and temperature, pH, salinity, total alkalinity and carbonate chemistry were monitored regularly (electronic supplementary material, S1).
(b) . Crossing design, spawning and larval culturing
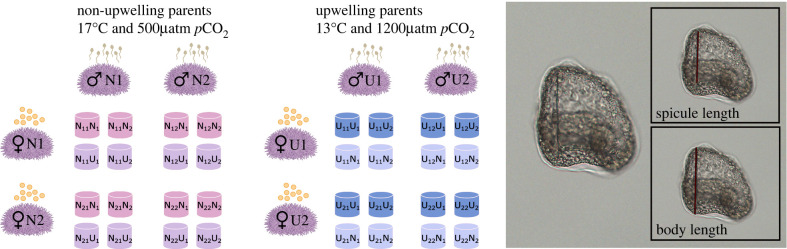
Owing to the large number of crosses necessary for this project, we employed a staggered cross-classified North Carolina II breeding design (figure 1). Spawning and generation of crosses began on 7 January 2019. Gametes from two males and two females conditioned in the N treatment were reciprocally crossed to yield four unique families. Each of these families was partitioned among four cultures, two reared in the N treatment (NN) and two reared in the U treatment (NU). The next day, the same crossing scheme was performed with parents from the U treatment, and families reared in either the U treatment (UU) or the N treatment (UN). The 16 cultures generated on a single day were designated as a block (1 parental treatment × 4 families × 4 cultures), and this block design was repeated 10 times in succession, alternating parental urchins from non-upwelling and upwelling, for a total of 160 cultures across 40 total families.
Figure 1.
Experimental crossing design. Adult urchins were conditioned for four months to either non-upwelling conditions (N) or upwelling conditions (U). Gametes from two males and two females from each condition were crossed reciprocally, generating four distinct crosses, each replicated four times. Two replicates from each cross were reared in the same condition as the parents (NN, UU) or the opposite condition (NU, UN). Two by two crosses for each parental condition were performed five times in succession for a total of 40 unique crosses. Spicule length and body length were measured in prism stage larvae, pictured. (Online version in colour.)
Fertilizations were performed in ambient seawater conditions and embryos were placed in rearing containers prior to the first cleavage, in either the same conditions as their parents or the reciprocal condition (figure 1). Larval cultures were set up in a flow-through seawater system with two reservoir tanks per treatment, as in the adult conditioning, feeding 6 l nested buckets (one bucket fitted with 30 µM mesh nested within another standard bucket) at a flow rate of 3 l h−1. Each pair of nested buckets formed one culture container. Temperature and pH of reservoir tanks were measured daily while salinity and pH of larval cultures was measured 24 h post fertilization (hpf) (electronic supplementary material, S1). Larval cultures were maintained at a concentration of 10 larvae ml−1 until the early echinopluteus stage, prism, defined by the beginning of tripartite gut differentiation, where the gut begins to form distinct sections (figure 1).
(c) . Morphometric measurements of eggs and larvae
Unfertilized egg and prism samples were preserved in 2% formalin buffered with 100 mM NaBO3 (pH 8.7) in FSW. Owing to differences in temperature-dependent developmental delay, prism larvae in N developmental treatments (17°C) were sampled between 45–46 hpf and prism larvae in U developmental treatments (13°C) were sampled between 55 and 56 hpf (figure 1). Photographs (n ≥ 30 eggs per dam; n ≥ 30 prism larvae per culture) were taken using a Motic 10MP digital camera fitted to an Olympus BX50 compound microscope and Motic Images Plus software. All measurements, calibrated using a stage micrometre, were obtained using ImageJ (https://imagej.nih.gov/ij/). For each unfertilized egg, three independent diameter measurements were averaged per egg to account for any potential irregularity in shape. For each prism, two measurements were taken, spicule length defined as the length from the tip of the body rod to the branching point of the postoral rod and body length. For each culture, the proportion of developmental abnormality (n ≥ 30 larvae per culture) was also scored. All measurements were taken by two researchers to minimize variation and bias, which was included in the models below.
(d) . Statistical analysis
Differences in egg diameter between treatments were quantified using a linear mixed model with a fixed effect of parental treatment (U or N) and random effects of dam identity and block using the lme4 package (v. 1.1–27.1) [32]. Relationships between egg diameter and prism morphometrics were assessed with a linear regression. Quantitative genetic linear mixed models employing a character state approach (where the expression of a single phenotype in a given environment defines a character state [10]) were used to decompose phenotypic variation in larval spicule and body lengths into contributions from plasticity, adaptive potential and parental effects. We fit separate, identical model structures for spicule and body length within a Bayesian framework and used a Markov chain Monte Carlo (MCMC) algorithm to sample posterior distributions as implemented in the package MCMCglmm (v. 2.29) [33]. All MCMCglmm models assumed Gaussian error distributions and response variables were multiplied by 100 before analyses to improve model convergence; results are reported for the scaled values of the response unless otherwise indicated.
For each larval trait, spicule length and body size, we modelled the interaction of each distinct parental conditioning environment (N and U) with the two rearing environments of their offspring (N and U). In the crossing design (figure 1), the gametes of parents were always crossed with gametes from parents of the same conditioning environment, meaning the data from N parents are independent of data from U parents. Thus, we modelled data from each parental conditioning environment in separate models. Using Bayesian inference allowed direct comparison of posterior probability distributions for model parameters of interest across different models [34].
For each larval trait and parental conditioning environment model (4 total), we fitted separate intercepts for each larval development environment (N and U) to estimate population mean larval plasticity across the two character states [10] and a measurer identity fixed effect (two-level continuous covariate with values –0.5 and 0.5) to control for an average difference between measurers. Random effects of dam and sire were fitted to estimate the variances in maternal or paternal effects, respectively. Random effects of block and culture identity were included to account for phenotypic similarity among larvae due to shared block effects or container environments, respectively. Preliminary models indicated homogeneity of variances between larval environments for the dam, sire, block and culture effects. Thus, a single, common variance across environments was fit for each of these random terms. We also fit separate larval environment residual variances, but the cross-environment covariance was fixed to zero as this is not estimable when individuals are only measured in a single environment.
Additive genetic (co)variances within and across larval rearing environments were estimated to evaluate the adaptive potential of larval morphological traits and to quantify variation in genotype-by-environment interactions. We fit random effects of individual identity and associated these with a generalized inverse of the numerator relatedness matrix [35,36] that was calculated from a pedigree constructed based on the breeding design using the nadiv package [37]. Cross-environment additive genetic covariances are estimable, unlike residual covariances, because related individuals in the two environments provide information about the cross-environment covariance of genetic effects [38,39]. To interpret our estimates of cross-environment additive genetic correlations, we ranked family mean additive genetic values for comparison between larval rearing environments (electronic supplementary material, S6).
Models employed diffuse normal prior distributions for all fixed effects (mean = 0, variance = 1010) and univariate parameter expanded prior distributions for rearing culture, block, dam and sire variance components with a scaling factor of 1000 to give scaled non-central F-distributions with one numerator and denominator degrees of freedom [33,40]. A multivariate parameter expanded prior was used for the additive genetic covariance matrix that gave a uniform marginal prior distribution for the correlation. A weak inverse Wishart prior was set for the matrix of residual variances (model details are provided within R code at https://github.com/qgevoeco/QGplasticity_S_purpuratus).
Models were run for an initial burn-in of 200 000 (spicule length) or 130 000 (body length) iterations, after which every 1000th iteration was retained in the posterior distribution to yield 2000 sample MCMC chains for each model that had absolute autocorrelation values <0.1. We report the marginal posterior mean, mode and 95% highest posterior density credible interval (95 %CI) and for key parameters plot full marginal posterior distributions alongside prior distributions to further facilitate interpretation (electronic supplementary material, S4).
Narrow-sense heritability was calculated as additive genetic variance (VA) divided by total phenotypic variance (VP), where VP = VA + Vdam + Vsire + Vculture + Vblock + Vresidual. Evolvability (IA) [41], which is a mean standardized additive genetic variance, was calculated as VA/INT2, where INT is the model intercept for a given developmental environment and represents the phenotypic mean, marginalizing over measurer effects. Heritability gives an absolute measure of expected evolutionary change, whereas evolvability expresses a proportional change and is therefore more suitable for comparative purposes [41]. Note, heritability and evolvability of the scaled response (i.e. spicule or body length × 100) are the same values for the response on the un-transformed scale. Posterior distributions were obtained for all summary statistics (e.g. heritability, IA, and differences between VAs as well as IAs) and differences between marginal posterior distributions by calculating desired values across each MCMC sample.
3. Results
(a) . Environmental conditioning reveals plasticity of larval traits
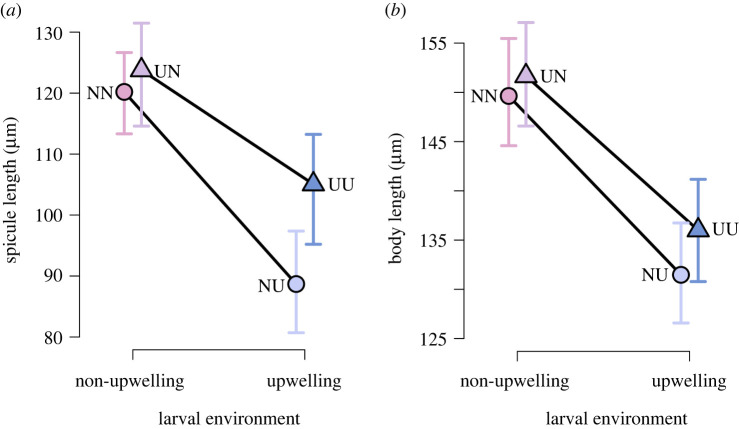
There was an effect of developmental environment on larval phenotypes: we observed a reduction in spicule length in larvae developed in U conditions compared to N (figure 2). However, spicule length in the U developmental environment depended on parental conditioning. Larvae where both the parents and embryonic development occurred in upwelling conditions (UU) had higher mean spicule length than larvae that developed in N conditions after parents were conditioned in U (NU) (figure 2a). For example, there is a 0.953 probability that UU spicule length is at least 7% larger than NU (or 6.2 µm larger) as calculated across the full probability distribution of differences (electronic supplementary material, S4). As with spicule length, embryonic development in U led to decreased mean body length (figure 2b), with the combined effect of parental and development U environments (UU) increasing mean body length from just the development U treatment (NU). After controlling for random effects of dam and block, there was no significant difference in egg diameter observed between the two parental treatments (p = 0.511). Further, egg size was not a good predictor of larval body size morphometrics (spicule length: R2 = −0.017, p = 0.5546, body size R2 = −0.025, p = 0.876) (electronic supplementary material, figure S1). The proportion of developmental abnormalities was scored among all crosses and was highest in larvae from parents conditioned to upwelling that experienced upwelling embryonic development as well (UU) (electronic supplementary material, figure S2).
Figure 2.
Strongylocentrotus purpuratus larvae exhibit phenotypic plasticity. Marginal posterior means and 95% credible intervals (error bars) of parameters estimated in linear mixed models for spicule (a) and body (b) length of larvae reared in either non-upwelling (N) or upwelling (U) developmental environments. Parents were either conditioned in the non-upwelling (circles) or upwelling (triangles) environments (black solid lines connect treatment means from the same parental environment). Plotted colours and letters refer to treatment combinations as detailed in figure 1. (Online version in colour.)
(b) . Components of variation in larval traits
To assess the potential evolutionary responses to abiotic conditions associated with upwelling, we quantified variance components of larval body size metrics. Additive genetic variance depended on developmental environment: additive genetic variances for spicule length are larger in the upwelling developmental environment (electronic supplementary material, tables S2 and figure S3a–d). The marginal posterior mean (95% CI) difference between additive genetic variance when larvae developed in U environments as opposed to N was 1.12 (−0.268 to 2.54) when parents were conditioned in N (i.e. NU–NN) and 0.407 (−0.854 to 1.51) when parents were conditioned in U (UU–UN). Though the credible intervals span zero for these differences, there is 0.945 and 0.770 probability that the estimates differ from one another (i.e. difference is greater than zero for the NU–NN and UU–UN differences, respectively). The similarity of the U developmental environment posterior means and modes as well as large differences between prior and posterior probability density curves indicate high posterior probability that is informed by the data and not the prior (electronic supplementary material, figure S3b,d). By contrast, body length shows much less additive genetic variance for all treatments (electronic supplementary material, S5 figure S6a–d). The posterior means for all treatments are less than approximately 0.25 and the lower credible interval limits all converge to zero indicating relatively high posterior probability at small values of effectively zero (electronic supplementary material, S4).
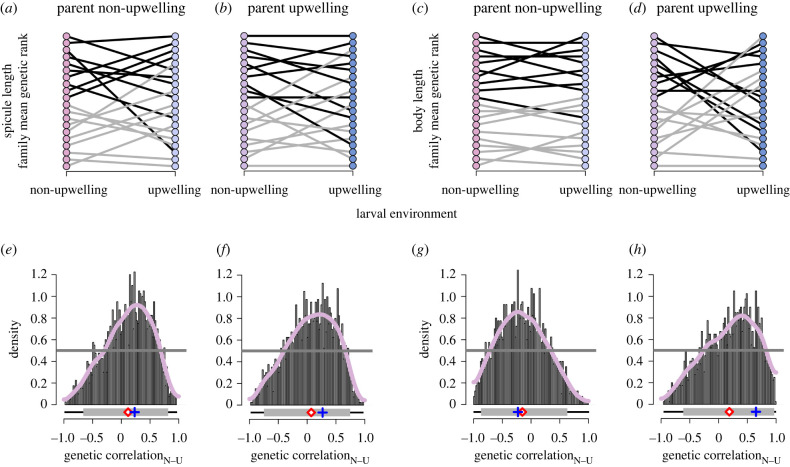
To determine the extent of among-genotype variability in the family-level plastic response, and hence genetic variation underlying phenotypic plasticity, we quantified cross-developmental environment genetic correlations. For spicule length, the marginal posterior distributions of the cross-development environment additive genetic correlations have posterior means and modes close to zero and are broad (figure 3e,f), with credible intervals that span most of the range of possible values, indicating some relative re-ranking of genotypes as they are expressed in the two development environments (figure 3a,b). The upper limits of these credible intervals are 0.81 or less (electronic supplementary material, table S2), excluding values near 1, hence indicative of significant variance for genotype-by-environment interactive effects. Similarly for body size, the marginal posterior distributions of the cross-developmental environment additive genetic correlations are broad (figure 3g,h), with means and modes close to zero and credible intervals that span most of the range of possible values (electronic supplementary material, table S3), indicating relative re-ranking of genotypes as they are expressed in the two development environments (figure 3c,d). For body size, there are differences in cross-development environmental genetic correlations depending on parental condition as there is approximately 0.63 posterior probability for a negative cross-environment genetic correlation among larvae of non-upwelling parents versus 0.67 posterior probability for a positive cross-environment genetic correlation among larvae of upwelling parents. This suggests varying magnitudes of variance in genotype-by-environment interactions (figure 3g,h), but uncertainty limits the importance of this conclusion.
Figure 3.
Variation in family-level genetic reaction norms and correlations. Ranked family mean additive genetic value for spicule length (a,b) and body length (c,d). Family mean additive genetic values were calculated across all posterior samples to produce a posterior distribution, from which the posterior mode was ranked for each larval environment. Black (top 10 ranked families in non-upwelling larval environment) and grey (bottom 10 ranked families in non-upwelling larval environment) lines connect family mean genetic value ranks across developmental environments and point colours refer to treatment combinations as detailed in figure 1. Cross-developmental environment additive genetic correlation of larval spicule length (e,f) and larval body size (g,h). Marginal posterior MCMC samples (histogram bars with the range of samples depicted underneath by the thin black line), kernel density estimate (pink line), posterior mean (red diamond) and mode (blue cross), 95% credible interval (grey bar) and prior density (grey line). (Online version in colour.)
For both spicule length and body size, dam, sire, block and culture variances that capture any remaining parental, non-additive genetic or environmental effects all independently contributed little to overall phenotypic variance (electronic supplementary material, tables S2, S3 and S5, and figures S4 and S7). Within each parental environment of both larval body size traits, sire and dam variances did not differ between development environments, and hence were constrained to be equal in the model (see Material and methods), indicating transgenerational parental effects did not vary based on offspring development environment. Residual variances were largely similar between development environments, both within and among parental treatments, and similar in magnitude to the additive genetic variance (electronic supplementary material, tables S2, S3 and S5, and figures S5 and S8).
(c) . Evolvability of larval body size and spicule length
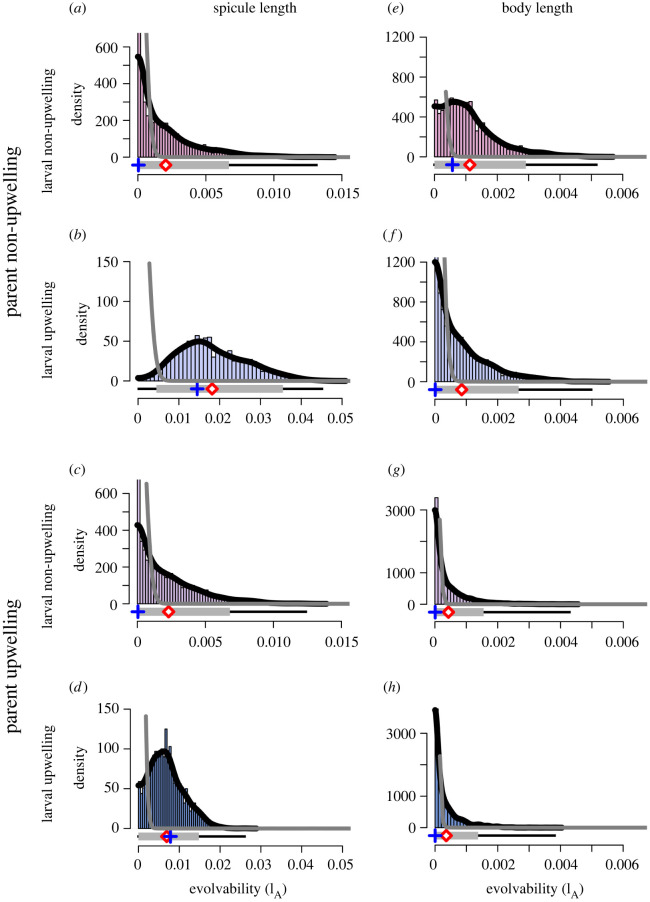
To quantify the potential for S. purpuratus evolutionary responses to U conditions simulated in the laboratory, we assessed potential differences in heritability (h2) and evolvability (IA) to allow comparisons across additive genetic variance estimates from different environments or even different traits. Similar to additive genetic variance, we observed relatively smaller values of heritability and evolvability in spicule length of larvae that developed in N environments (i.e. NN and UN; figure 4a–d; electronic supplementary material, table S2 and figure S3e–h). When larvae developed in U conditions, substantial levels of heritability and moderate evolvability were observed with differences in magnitude between the two larval U treatments depending on parental environment (figure 4b,d; electronic supplementary material, table S2 and figure S3f,h). The marginal posterior mean (95% CI) difference between evolvability when larvae developed in U environments as opposed to N was 0.0161 (0.000753–0.0344) when parents were conditioned in N (i.e. NU–NN) and 0.00462 (−0.00486 to 0.0146) when parents were conditioned in U (UU–UN). Though the credible interval spans zero for the difference between larval development environments when parents were conditioned in U (UU–UN), there is 0.676 probability that this difference is 0.0025 or greater. In contrast to spicule length, the heritability and evolvability values for body length were lower for both treatments when the parents were in N conditions (NN and NU) and decreased further when the parents were reared in the U environment (UN and UU; figure 4e–h; electronic supplementary material, table S3 and figure S6e–h).
Figure 4.
Evolvability. Strongylocentrotus purpuratus spicule (a–d) and body (e–h) length marginal posterior MCMC samples (histogram bars with sample range depicted underneath by the thin black line), kernel density estimate (black line), posterior mean (red diamond) and mode (blue cross), 95% credible interval (grey bar) and prior density (grey line) for the evolvability (IA). Colours refer to treatment combinations as detailed in figure 1. (Online version in colour.)
4. Discussion
(a) . The role of plasticity in shaping larval traits
We observed phenotypic plasticity in S. purpuratus larvae reared in different developmental environments, which has similarly been observed in other independent studies [27,28]. Both spicule length and larval body size were reduced when reared in upwelling conditions even after controlling for potentially confounding effects of developmental delay. We found evidence of genetic variation in phenotypic plasticity, or genotype-by-environment interactions (GxE), suggesting genotypes exhibit different plastic responses to an upwelling developmental environment (figure 3). Rankings of additive genetic values across families become reordered among full siblings exposed to different developmental conditions. Strongylocentrotus purpuratus habitats tend towards being highly heterogeneous, characterized by highly dynamic upwelling regimes that vary in time and space [29,42] which will probably grow in frequency and intensity in future years [15,16]. These heterogeneous environments appear to have favoured plasticity and maintenance of GxE in prism stage morphometrics, therefore slopes of reaction norms are not likely to be under strong directional selection. While it is known that larval body size morphometrics are important predictors of later stage survival and settlement, our results suggest prism stage morphometrics measured here are either under relaxed selection [43], or selection that maintains variation in GxE. While we measured morphometrics in early pre-feeding larvae, it is possible that later stage larval feeding morphometrics could be under stronger selection pressure, potentially contributing more to fitness, settlement, and survival. Ultimately, the temporal links between larval skeletal morphometrics and larval survival should be further investigated in each environment to discriminate between alternative explanations for the maintenance of variation in GxE. Nevertheless, the variation in additive genetic value between families in response to different developmental environments in our study, indicative of a genetic basis for phenotypic plasticity in early stages of S. purpuratus, has important implications for the ability of this ecologically important species to persist under future global change scenarios.
We investigated the role of parental effects, a form of phenotypic plasticity, on egg size and larval body morphometrics. Egg size, a fitness trait associated with fertilization success and postzygotic survival, is a direct result of maternal investment through provisioning of energy reserves [44–47]. We observed no differences in mean egg diameter between dams conditioned in the two treatments (electronic supplementary material, S3 figure S1) when controlling for random effect of dam, similarly to previous studies examining parental effects of upwelling stress [28,29]. Egg size has significant influences on larval survival and recruitment success for a diversity of broadcast spawning marine invertebrates; in echinoderms, egg volume and energetic content are highly correlated [48], however, egg size is not always a robust predictor of energetic content in planktotrophic species [45], including echinoderms [28,29,49,50]. Our study did not find egg size to predict larval size (electronic supplementary material, figure S1), although egg size was measured over a small range and energetic content was not quantified. However, parental effects on prism larvae morphometrics were observed (figure 2), suggesting parental conditioning induces latent effects that impact larval fitness while early development through gastrulation appears constrained and unaffected by the environment, effects that are similarly observed in a previous experiment in S. purpuratus [28]. This combined evidence of parental effects on larval morphometrics in S. purpuratus could be explained by parental investment in mRNAs critical for development, epigenetic processes or differential investment of key nutrients in the eggs [27,51,52]. Transgenerational plasticity is mostly likely to occur when parental environments are predictive of larval environments [53,54], which we observed: larvae developed in upwelling were larger when their parents were also conditioned in upwelling conditions (figure 2). This suggests that parental effects are a likely mechanism contributing to larval phenotypic change in response to environmental conditions in S. purpuratus. Predictable high magnitude variation in environmental parameters such as temperature and pH that occur throughout the life cycle of S. purpuratus is likely to favour the maintenance of phenotypic plasticity. If this predictability breaks down, broadcast spawning invertebrates such as S. purpuratus might be more likely to exhibit bet-hedging type strategies to maintain populations, although this strategy lacks empirical support in S. purpuratus populations studied to date [55].
(b) . The potential for adaptation to global change
Adaptation to global change relies on sufficient natural genetic variation and genetic correlations between selected traits. Larval body size is an important, often heritable, fitness trait among diverse marine invertebrates but can vary based on differences in environmental effects [25,56,57]. We observe higher additive genetic variance, heritability and evolvability for spicule length among larvae reared in upwelling conditions, compared to larvae reared in non-upwelling. This indicates more potential for adaptive responses to conditions expected to occur under anthropogenic change. Strongylocentrotus purpuratus spawning activity occurs seasonally between December and April, months characterized by upwelling episodes, which can last multiple days [29]; therefore, the conditions in our experiment are relevant to what larvae are likely to experience in the wild. Heritability values here are similar to previous estimates in S. purpuratus larval morphometric traits after exposure to high pCO2 [25]. Further, molecular experiments have shown that upwelling conditions induce a stress response in S. purpuratus larvae [51]. This indicates that we observe higher adaptive potential in larvae experiencing stressful environmental conditions, despite additive genetic variance observed to be lower in unfavourable conditions in most studies [58], including in sea urchins [57]. However, the majority of studies examining additive genetic variance under stressful conditions employ a novel stress, whereas the conditions in our experiment were chosen as endpoints of temperature and pH already occurring naturally in their environment.
Measures of evolvability allow us to quantify the relative extent to which phenotypes can evolve in response to selection. In particular, evolvability is better suited than heritability for comparing adaptive potential among environments, traits, or even species since evolvability expresses change in proportion to the current trait mean (heritability expresses potential absolute change) and heritability depends on the phenotypic variation in the population which itself can be affected by the selective environment independent of the amount of additive genetic variance [41]. We observe high evolvability in larvae reared under some conditions but not others, suggesting a strong role of the environment in the evolvability of larval fitness traits in S. purpuratus. For example, for larval spicule length, our posterior mean evolvability estimates those developing in non-upwelling conditions (0.00205 and 0.00230 for parental conditioning in non-upwelling and upwelling, respectively) are similar to the median evolvability of 0.001 for length measures from 1025 estimates compiled by Hansen & Pélabon [59]. However, our evolvability estimates for spicule length of larvae developing in upwelling conditions (0.0182 and 0.00692 for parental conditioning environments non-upwelling and upwelling, respectively) were well above the 75th percentile of 0.0047 from that same study. Minimal correlations among larval rearing environments suggest that the highly variable environment S. purpuratus experiences may limit the rate at which adaptation could occur. There is higher evolvability of spicule length in larvae produced from adults conditioned in non-upwelling but developed in upwelling (figure 4b, NU treatment) as opposed to those coming from parents conditioned to upwelling and during embryonic development (figure 4d, UU treatment). This difference in evolvability amounts to 0.0112 (posterior mean; 95 %CI: –0.00463 to 0.0301), which represents a potential evolutionary change in mean phenotype of approximately 1.1% more in the NU versus UU treatments over a single generation. This observation suggests a subtle effect of parental conditioning on the genetic contribution to phenotypic variance under upwelling conditions. We also find effects of parental conditioning on plasticity and genetic contributions to phenotypic variance. Plastic phenotypes, or those that shift in response to the environment, are biased toward traits that have high additive genetic variance [60], which we observe in larval responses to upwelling. These correlations can often be explained by developmental constraints that limit phenotypic change in a particular direction. This is likely true in S. purpuratus early stages– phenotypic change on spicule length appears to be less constrained, having both the potential to be phenotypically plastic and able to be acted on by selection, as opposed to body size which could be more constrained by development. However, spicule length plasticity is inherently limited by the body size of the larvae, so further study into the genetic correlations and covariances between these two traits would be insightful as to how these traits contribute to evolvability.
(c) . Trade-offs in larval fitness traits
Developmental environments have been shown to shape later stage phenotypes in a diversity of organisms and these changes in phenotypes can have trade-offs as well as latent effects on later stages [6,61,62]. We found subtle effects of parental environment on larval traits; of the larvae reared in the upwelling environment, having parents also conditioned to upwelling (UU) led to an increase in spicule length compared with larvae whose parents were conditioned in non-upwelling (NU). However, we observed a higher proportion of developmental abnormalities among UU crosses, characterized by embryos that failed to successfully gastrulate. At the time of adult collection, individuals were probably experiencing conditions more similar to non-upwelling, therefore higher abnormalities could be explained by a mismatch between wild and captive conditions for parental upwelling conditioned individuals. While upwelling parental exposure may confer some benefit to larvae developing in upwelling conditions, there is a compensatory trade-off in that many of the larvae derived from the UU crosses show higher mortality as evident by early developmental abnormalities. As only properly developed larvae were selected for morphometric analysis, this shows that the UU survivors were on average larger than UN individuals. There is a well-established trade-off between growth and sensitivity to high pCO2 in coastal marine invertebrates, where slowed growth or reallocation of energy in high pCO2 facilitates high tolerance [6,63]. In the tropical urchin Tripneustes gratilla, parental conditioning to high temperatures and high pCO2 led to more resilient larvae with a trade-off of reduced size [64]. While abnormality was high in UU crosses, the benefit of increased size relative to UN crosses suggests a complex role of parental effects on early life-history stages.
5. Conclusion
Climate models predict more frequent and severe incidences of upwelling in the future, which will directly impact calcifying organisms within the California Large Marine Ecosystem, such as S. purpuratus [15,16]. However, incorporating selection on larval body size into predictive models show that negative effects of OA are likely overestimated, as larval body size exhibits high heritability under these scenarios and S. purpuratus maintains large population sizes that will enable adaptive responses to selection [65]. Our data build upon this work to reveal that effects are maintained in more ecologically relevant upwelling conditions (high pCO2 and low temperature). Additionally, we report the influence of parental environment on estimates of adaptive genetic variation, which will alter how strong adaptation to increased upwelling may impact these populations. Further, we report genetic variation in phenotypic plasticity, or genotype-by-environment interactions, showing that phenotypic plasticity itself has potential to evolve in this population. Therefore, in considering future upwelling scenarios, it is likely that both phenotypic plasticity and adaptation will contribute to S. purpuratus population responses to stressful periods of upwelling.
Acknowledgements
We thank Clint Nelson for assistance with boating and sea urchin collection, and Christoph Pierre for kelp collections. We also thank Logan Kozal, Terence Leach, Jannine Chamorro, Juliet Wong, Sam Bogan, Maddie Housh and Avery DeSantis for invaluable assistance in data collection.
Ethics
Adult urchins were collected under the California Scientific Collecting Permit to G.E.H. (SC-1223).
Data accessibility
Data and all R code are freely available on GitHub: https://github.com/qgevoeco/QGplasticity_S_purpuratus.
The data are provided in electronic supplementary material [66].
Authors' contributions
M.E.S.: conceptualization, data curation, supervision, visualization, writing—original draft, writing—review and editing; M.E.W.: conceptualization, data curation, formal analysis, validation, visualization, writing—original draft, writing—review and editing; O.M.S.: data curation, writing—review and editing; G.E.H.: conceptualization, funding acquisition, project administration, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare no competing interests.
Funding
National Science Foundation award (grant no. IOS-1656262) to G.E.H., and diving and boating resources were provided through NSF award OCE-1232779 (director: Dr Robert Miller).
References
- 1.Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J. 2008. Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17, 167-178. ( 10.1111/j.1365-294X.2007.03413.x) [DOI] [PubMed] [Google Scholar]
- 2.Davis MB, Shaw RG, Etterson JR. 2005. Evolutionary responses to climate change. Ecology 86, 1704-1714. ( 10.1890/03-0788) [DOI] [Google Scholar]
- 3.Chevin LM, Collins S, Lefèvre F. 2013. Phenotypic plasticity and evolutionary demographic responses to climate change: taking theory out to the field. Funct. Ecol. 27, 967-979. ( 10.1111/j.1365-2435.2012.02043.x) [DOI] [Google Scholar]
- 4.Chirgwin E, Marshall DJ, Sgrò CM, Monro K. 2018. How does parental environment influence the potential for adaptation to global change? Proc. R. Soc. B 285, 20181374. ( 10.1098/rspb.2018.1374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uller T, Nakagawa S, English S. 2013. Weak evidence for anticipatory parental effects in plants and animals. J. Evol. Biol. 26, 2161-2170. ( 10.1111/jeb.12212) [DOI] [PubMed] [Google Scholar]
- 6.Swezey DS, Boles SE, Aquilino KM, Stott HK, Bush D. 2020. Evolved differences in energy metabolism and growth dictate the impacts of ocean acidification on abalone aquaculture. Proc. Natl Acad. Sci. USA 117, 1-7. ( 10.1073/pnas.2006910117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murren CJ, et al. 2015. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity 115, 293-301. ( 10.1038/hdy.2015.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendry AP. 2016. Key questions on the role of phenotypic plasticity in eco-evolutionary dynamics. J. Hered. 107, 25-41. ( 10.1093/jhered/esv060) [DOI] [PubMed] [Google Scholar]
- 9.Schmalhausen I. 1949. Factors of evolution: the theory of stabilizing selection. Philadelphia, PA: Blakiston. [Google Scholar]
- 10.Via S, Lande R. 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505-522. [DOI] [PubMed] [Google Scholar]
- 11.Lush J. 1943. Animal breeding plans. Ames, IA: Iowa State College Press. [Google Scholar]
- 12.Lande R. 1979. Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33, 402-416. ( 10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- 13.Falconer DS. 1989. Introduction to quantitative genetics, 3rd edn. New York, NY: John Wiley & Sons. [Google Scholar]
- 14.Feely RA, Sabine CL, Hernandez-Ayon JM, Ianson D, Hales B. 2008. Evidence for upwelling of corrosive ‘acidified’ water onto the continental shelf. Science 320, 1490-1492. [DOI] [PubMed] [Google Scholar]
- 15.Sydeman WJ, García-Reyes M, Schoeman DS, Rykaczewski RR, Thompson SA, Black BA, Bograd SJ. 2014. Climate change. Climate change and wind intensification in coastal upwelling ecosystems. Science 345, 77-80. ( 10.1126/science.1251635) [DOI] [PubMed] [Google Scholar]
- 16.Bakun A, Black BA, Bograd SJ, García-Reyes M, Miller AJ, Rykaczewski RR, Sydeman WJ. 2015. Anticipated effects of climate change on coastal upwelling ecosystems. Curr. Clim. Chang. Reports 1, 85-93. ( 10.1007/s40641-015-0008-4) [DOI] [Google Scholar]
- 17.Bednaršek N, Naish KA, Feely RA, Hauri C, Kimoto K, Hermann AJ, Michel C, Niemi A, Pilcher D. 2021. Integrated assessment of ocean acidification risks to pteropods in the Northern high latitudes: regional comparison of exposure, sensitivity and adaptive capacity. Front. Mar. Sci. 8, 1-23. ( 10.3389/fmars.2021.671497)35685121 [DOI] [Google Scholar]
- 18.Kroeker KJ, Kordas RL, Crim R, Hendriks IE, Ramajo L, Singh GS, Duarte CM, Gattuso J-P. 2013. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Chang. Biol. 19, 1884-1896. ( 10.1111/gcb.12179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen JD. 2008. Size-specific predation on marine invertebrate larvae. Biol. Bull. 214, 42-49. ( 10.2307/25066658) [DOI] [PubMed] [Google Scholar]
- 20.Chan KYK. 2012. Biomechanics of larval morphology affect swimming: Insights from the sand dollars Dendraster excentricus. Integr. Comp. Biol. 52, 458-469. ( 10.1093/icb/ics092) [DOI] [PubMed] [Google Scholar]
- 21.Strathmann RR. 1971. The feeding behavior of planktotrophic echinoderm larvae: mechanisms, regulation, and rates of suspensionfeeding. J. Exp. Mar. Bio. Ecol. 6, 109-160. ( 10.1016/0022-0981(71)90054-2) [DOI] [Google Scholar]
- 22.Hofmann GE, Barry JP, Edmunds PJ, Gates RD, Hutchins DA, Klinger T, Sewell MA. 2010. The effect of ocean acidification on calcifying organisms in marine ecosystems: an organism-to-ecosystem perspective. Annu. Rev. Ecol. Evol. Syst. 41, 127-147. ( 10.1146/annurev.ecolsys.110308.120227) [DOI] [Google Scholar]
- 23.Kroeker KJ, Kordas RL, Crim RN, Singh GG. 2010. Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol. Lett. 13, 1419-1434. ( 10.1111/j.1461-0248.2010.01518.x) [DOI] [PubMed] [Google Scholar]
- 24.Chan KYK, Tong CSD. 2020. Temporal variability modulates pH impact on larval sea urchin development. Conserv. Physiol. 8, 1-11. ( 10.1093/conphys/coaa008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly MW, Padilla-Gamiño JL, Hofmann GE. 2013. Natural variation and the capacity to adapt to ocean acidification in the keystone sea urchin Strongylocentrotus purpuratus. Glob. Chang. Biol. 19, 2536-2546. ( 10.1111/gcb.12251) [DOI] [PubMed] [Google Scholar]
- 26.Strader ME, Wong JM, Hofmann GE. 2020. Ocean acidification promotes broad transcriptomic responses in marine metazoans: a literature survey. Front. Zool. 17, 1-23. ( 10.1186/s12983-020-0350-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong JM, Johnson KM, Kelly MW, Hofmann GE. 2018. Transcriptomics reveal transgenerational effects in purple sea urchin embryos: adult acclimation to upwelling conditions alters the response of their progeny to differential p CO2 levels. Mol. Ecol. 27, 1120-1137. ( 10.1111/mec.14503) [DOI] [PubMed] [Google Scholar]
- 28.Wong JM, Kozal LC, Leach TS, Hoshijima U, Hofmann GE. 2019. Transgenerational effects in an ecological context: Conditioning of adult sea urchins to upwelling conditions alters maternal provisioning and progeny phenotype. J. Exp. Mar. Bio. Ecol. 517, 65-77. ( 10.1016/j.jembe.2019.04.006) [DOI] [Google Scholar]
- 29.Hoshijima U, Hofmann GE. 2019. Variability of seawater chemistry in a kelp forest environment is linked to in situ transgenerational effects in the purple sea urchin, Strongylocentrotus purpuratus. Front. Mar. Sci. 6, 1-18. ( 10.3389/fmars.2019.00062)36817748 [DOI] [Google Scholar]
- 30.Castorani MCN, Harrer SL, Miller RJ, Reed DC. 2021. Disturbance structures canopy and understory productivity along an environmental gradient. Ecol. Lett. 24, 2192-2206. ( 10.1111/ele.13849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fangue NA, O'Donnell MJ, Sewell MA, Matson PG, MacPherson AC, Hofmann GE. 2010. A laboratory-based, experimental system for the study of ocean acidification effects on marine invertebrate larvae. Limnol. Oceanogr. Methods 8, 441-452. ( 10.4319/lom.2010.8.441) [DOI] [Google Scholar]
- 32.Bates D, Mächler M, Bolker BM, Walker SC. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1-48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 33.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1-22. ( 10.1002/ana.22635)20808728 [DOI] [Google Scholar]
- 34.Morrissey MB, de Villemereuil P, Doligez B, Gimenez O. 2014. Bayesian approaches to the quantitative genetic analysis of natural populations. In Quantitative genetics in the wild (eds Charmantier A, Garant D, Kruuk LEB), pp. 228-253. Oxford, UK: Oxford University Press. [Google Scholar]
- 35.Henderson CR. 1973. Sire evaluation and genetic trends. J. Anim. Sci. 1973, 10-41. [Google Scholar]
- 36.Kruuk LEB. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. B 359, 873-890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolak ME. 2012. Nadiv: an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol. Evol. 3, 792-796. ( 10.1111/j.2041-210X.2012.00213.x) [DOI] [Google Scholar]
- 38.Mrode RA. 2015. Linear models for the prediction of animal breeding values, 2nd edn. Cambridge, MA: CABI Publishing. [Google Scholar]
- 39.Wolak ME, Roff DA, Fairbairn DJ. 2015. Are we underestimating the genetic variances of dimorphic traits? Ecol. Evol. 5, 590-597. ( 10.1002/ece3.1361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gelman A. 2006. Prior distributions for variance parameters in hierarchical models (Comment on Article by Browne and Draper). Bayesian Anal. 1, 515-534. ( 10.1214/06-BA117A) [DOI] [Google Scholar]
- 41.Houle D. 1992. Comparing evolvability and variability. Genetics 130, 195-204. ( 10.1093/genetics/130.1.195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapsenberg L, Hofmann GE. 2016. Ocean pH time-series and drivers of variability along the northern Channel Islands, California, USA. Limnol. Oceanogr. 61, 953-968. ( 10.1002/lno.10264) [DOI] [Google Scholar]
- 43.Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. 2009. Relaxed selection in the wild. Trends Ecol. Evol. 24, 487-496. ( 10.1016/j.tree.2009.03.010) [DOI] [PubMed] [Google Scholar]
- 44.Villinski JT, Villinski JC, Byrne M, Raff RA. 2002. Convergent maternal provisioning and life-history evolution in Echinoderms. Evolution 56, 1764-1775. ( 10.1111/j.0014-3820.2002.tb00190.x) [DOI] [PubMed] [Google Scholar]
- 45.Moran AL, McAlister JS. 2009. Egg size as a life history character of marine invertebrates: is it all it's cracked up to be? Biol. Bull. 216, 226-242. ( 10.2307/25548157) [DOI] [PubMed] [Google Scholar]
- 46.Falkner I, Sewell MA, Byrne M. 2015. Evolution of maternal provisioning in ophiuroid echinoderms: characterisation of egg composition in planktotrophic and lecithotrophic developers. Mar. Ecol. Prog. Ser. 525, 1-13. ( 10.3354/meps11217) [DOI] [Google Scholar]
- 47.Caballes CF, Pratchett MS, Kerr AM, Rivera-Posada JA. 2016. The role of maternal nutrition on oocyte size and quality, with respect to early larval development in the coral-eating starfish, Acanthaster planci. PLoS ONE 11, 1-21. ( 10.1371/journal.pone.0158007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran AL, McAlister JS, Whitehill EAG. 2013. Eggs as energy: revisiting the scaling of egg size and energetic content among echinoderms. Biol. Bull. 224, 184-191. ( 10.1086/BBLv224n3p184) [DOI] [PubMed] [Google Scholar]
- 49.McEdward LR. 1986. Comparative morphometrics of echinoderm larvae. I. Some relationships between egg size and initial larval form in echinoids. J. Exp. Mar. Bio. Ecol. 96, 251-265. ( 10.1016/0022-0981(86)90206-6) [DOI] [Google Scholar]
- 50.Mcedward LR, Janies DA. 1997. Relationships among development, ecology, and morphology in the evolution of echinoderm larvae and life cycles. Biol. J. Linn. Soc. 60, 381-400. ( 10.1006/bijl.1996.0107) [DOI] [Google Scholar]
- 51.Strader ME, Kozal LC, Leach TS, Wong JM, Chamorro JD, Housh MJ, Hofmann GE. 2020. Examining the role of DNA methylation in transcriptomic plasticity of early stage sea urchins: developmental and maternal effects in a kelp forest herbivore. Front. Mar. Sci. 7, 205. ( 10.3389/fmars.2020.00205) [DOI] [Google Scholar]
- 52.Eirin-Lopez JM, Putnam HM. 2019. Marine environmental epigenetics. Annu. Rev. Mar. Sci. 11, 335-368. ( 10.1146/annurev-marine-010318-095114) [DOI] [PubMed] [Google Scholar]
- 53.Donelan SC, Hellmann JK, Bell AM, Luttbeg B, Orrock JL, Sheriff MJ, Sih A. 2020. Transgenerational plasticity in human-altered environments. Trends Ecol. Evol. 35, 115-124. ( 10.1016/j.tree.2019.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall DJ, Uller T. 2007. When is a maternal effect adaptive? Oikos 116, 1957-1963. ( 10.1111/j.2007.0030-1299.16203.x) [DOI] [Google Scholar]
- 55.Flowers JM, Schroeter SC, Burton RS. 2002. The recruitment sweepstakes has many winners: genetic evidence from the sea urchin Strongylocentrotus purpuratus. Evolution 56, 1445-1453. ( 10.1111/j.0014-3820.2002.tb01456.x) [DOI] [PubMed] [Google Scholar]
- 56.Griffiths J, Johnson K, Sirovy K, Yeats M, Pan F, La Peyre J, Kelly M. 2021. Transgenerational plasticity and the capacity to adapt to low salinity in the eastern oyster, Crassostrea virginica. Proc. R. Soc. B 288, 20203118. ( 10.1098/rspb.2020.3118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunday JM, Crim RN, Harley CDG, Hart MW. 2011. Quantifying rates of evolutionary adaptation in response to ocean acidification. PLoS ONE 6, 1-8. ( 10.1371/journal.pone.0022881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charmantier A, Garant D. 2005. Environmental quality and evolutionary potential: lessons from wild populations. Proc. R. Soc. B 272, 1415-1425. ( 10.1098/rspb.2005.3117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen TF, Pélabon C. 2021. Evolvability: a quantitative-genetics perspective. Annu. Rev. Ecol. Evol. Syst. 52, 153-175. ( 10.1146/annurev-ecolsys-011121-021241) [DOI] [Google Scholar]
- 60.Noble DWA, Radersma R, Uller T. 2019. Plastic responses to novel environments are biased towards phenotype dimensions with high additive genetic variation. Proc. Natl Acad. Sci. USA 116, 13 452-13 461. ( 10.1073/pnas.1821066116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 62.Sasaki MC, Dam HG. 2020. Genetic differentiation underlies seasonal variation in thermal tolerance, body size, and plasticity in a short-lived copepod. Ecol. Evol. 10, 12 200-12 210. ( 10.1002/ece3.6851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan T-CF, Applebaum SL, Manahan DT. 2015. Experimental ocean acidification alters the allocation of metabolic energy. Proc. Natl Acad. Sci. USA 112, 4696-4701. ( 10.1073/pnas.1416967112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karelitz S, Lamare MD, Mos B, De Bari H, Dworjanyn SA, Byrne M. 2019. Impact of growing up in a warmer, lower pH future on offspring performance: transgenerational plasticity in a pan-tropical sea urchin. Coral Reefs 38, 1085-1095. ( 10.1007/s00338-019-01855-z) [DOI] [Google Scholar]
- 65.Kelly MW, Hofmann GE. 2013. Adaptation and the physiology of ocean acidification. Funct. Ecol. 27, 980-990. ( 10.1111/j.1365-2435.2012.02061.x) [DOI] [Google Scholar]
- 66.Strader ME, Wolak ME, Simon OM, Hofmann GE. 2022. Data from: Genetic variation underlies plastic responses to global change drivers in the purple sea urchin, Strongylocentrotus purpuratus. Figshare. ( 10.6084/m9.figshare.c.6146004) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Strader ME, Wolak ME, Simon OM, Hofmann GE. 2022. Data from: Genetic variation underlies plastic responses to global change drivers in the purple sea urchin, Strongylocentrotus purpuratus. Figshare. ( 10.6084/m9.figshare.c.6146004) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data and all R code are freely available on GitHub: https://github.com/qgevoeco/QGplasticity_S_purpuratus.
The data are provided in electronic supplementary material [66].