Abstract
Metabolic reprogramming is of great significance in the progression of various cancers and is critical for cancer progression, diagnosis, and treatment. Cellular metabolic pathways mainly include glycolysis, fat metabolism, glutamine decomposition, and oxidative phosphorylation. In cancer cells, reprogramming metabolic pathways is used to meet the massive energy requirement for tumorigenesis and development. Metabolisms are also altered in malignant osteosarcoma (OS) cells. Among reprogrammed metabolisms, alterations in aerobic glycolysis are key to the massive biosynthesis and energy demands of OS cells to sustain their growth and metastasis. Numerous studies have demonstrated that compared to normal cells, glycolysis in OS cells under aerobic conditions is substantially enhanced to promote malignant behaviors such as proliferation, invasion, metastasis, and drug resistance of OS. Glycolysis in OS is closely related to various oncogenes and tumor suppressor genes, and numerous signaling pathways have been reported to be involved in the regulation of glycolysis. In recent years, a vast number of inhibitors and natural products have been discovered to inhibit OS progression by targeting glycolysis-related proteins. These potential inhibitors and natural products may be ideal candidates for the treatment of osteosarcoma following hundreds of preclinical and clinical trials. In this article, we explore key pathways, glycolysis enzymes, non-coding RNAs, inhibitors, and natural products regulating aerobic glycolysis in OS cells to gain a deeper understanding of the relationship between glycolysis and the progression of OS and discover novel therapeutic approaches targeting glycolytic metabolism in OS.
Keywords: osteosarcoma, glycolysis, signaling pathway, Cancer Progression, key enzymes
1 Introduction
OS is a malignant bone tumor that primarily affects children and teenagers. It has a low incidence in the general population but is fatal (Davis et al., 2019; Ye et al., 2022). OS occurs in the metaphysis of long bones: the distal femur and proximal tibia (Saraf et al., 2018). One of the important features of OS is its heterogeneity. The complexity of the somatic genome of OS is the main cause of intratumor heterogeneity (Corre et al., 2020). Several genes (TP53, RB, MDM2, ATRX, and DLG2) have been found in OS repeated mutation (Chen et al., 2014). OS has a high proclivity for lung metastasis, contributing to its poor treatment and prognosis (Longhi et al., 2006). OS aggressiveness correlates with osteolysis makers, tumor-infiltrating osteoclasts are frequently found in OS, serum TRACP 5b and MMP9 levels are elevated (Avnet et al., 2008). Over the past few decades, the treatment options for OS have been continuously optimized, and the survival rate of OS patients has improved, but the mortality rate remains high. This necessitates a more comprehensive understanding of OS to identify the best treatment plan (Berner et al., 2015). Few cancer cells grow slowly, while osteosarcoma cells grow very fast and must synthesize large amounts of nutrients to survive.
For rapid growth, cancer cells enhance their metabolism by “metabolic reprogramming.” One of the most common metabolic changes is enhanced glycolysis, also known as the “Warburg effect,” manifested by increased glucose uptake and lactate production (Hsu and Sabatini, 2008). The Warburg effect was first described in the 1920s (Koppenol et al., 2011). Normal cells obtain energy by breaking down glucose through oxidative phosphorylation in the mitochondria. In contrast, cancer cells obtain energy by converting glucose into lactate via enhanced glycolysis, either in oxygen or hypoxia (Warburg, 1956). Glycolysis was previously thought to be typically characterized by increased glucose uptake and lactate production but inefficient adenosine triphosphate (ATP) production. This energy source accounts for more than half of the ATP supply of hypermetabolic tumor cells. Recent studies have found that tumor growth in vivo is related to the ratio of NAD+/NADH, and reducing the ratio of NAD+/NADH can inhibit cell proliferation (Gui et al., 2016).
When the demand for NAD exceeds the demand for ATP, cells undergo aerobic glycolysis (Luengo et al., 2021). Studies have shown that enhanced glycolysis is critical for cancer cells’ rapid growth and metastasis (Kroemer and Pouyssegur, 2008). Reliable and effective therapeutic drugs can be explored and developed by targeting the glycolytic pathway (Dang et al., 2009; Li et al., 2018). Over the past few decades, glycolysis has been extensively demonstrated for the growth, invasion, and treatment of various tumors, including OS. However, there is no systematic study and discussion on how glycolysis regulates and affects the progression and treatment of OS. This study summarizes the roles of key glycolysis enzymes, signaling pathways, non-coding RNAs, inhibitors, and natural products in OS and explores metabolic-targeted treatment strategies for OS.
2 The glycolysis in osteosarcoma
OS drugs, doxorubicin (ADR) or paclitaxel in combination with the glycolysis inhibitor 2-DG, are significantly more effective in reducing tumor growth than either drug alone (Maschek et al., 2004). This indicates that the glycolysis inhibitors combined with chemotherapeutic drugs improve the therapeutic effect in OS, so it is necessary to understand the mechanism of glycolysis in OS deeply. Designing small-molecule inhibitors target some key enzymes in the glycolytic pathway is a reliable solution for studying OS therapeutic strategies. For example, targeting glycolytic regulator (M2-PK) by synthetic peptide aptamers reduces the proliferation ability of OS cells (Spoden et al., 2008). According to many studies, enhanced glycolysis is the main metabolic alteration in OS, while many glycolytic enzymes have been shown to promote the tumorigenic activity of OS cells and are associated with poor prognosis in patients (Sottnik et al., 2011; Zhuo et al., 2015; Shen et al., 2019). Furthermore, this study reviewed some key glycolytic enzymes, signaling pathways, transcription factors, and non-coding RNAs in the glycolytic in OS cells.
2.1 Key enzymes of glycolysis in osteosarcoma
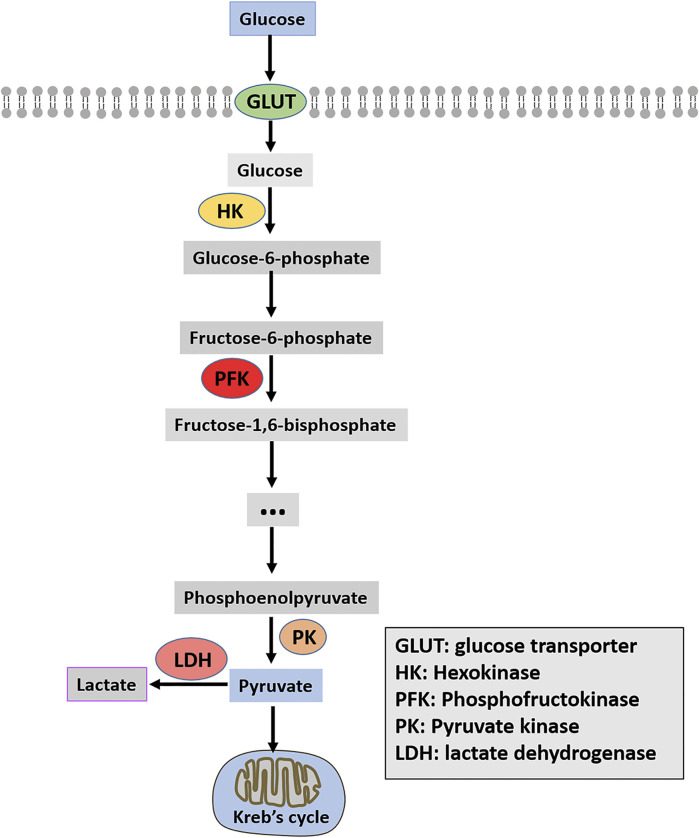
Enhancing glycolysis in cancer tissues and cells is closely related to the overexpression or enhanced activity of key glycolysis enzymes (Bian et al., 2022). Glycolysis-related enzymes include three rate-limiting enzymes: hexokinase (HK), phosphofructokinase (PFK), and pyruvate kinase (PK). Moreover, glucose transporter (GLUT) and lactate dehydrogenase (LDH) are also summarized in this paper and shown in Figure 1.
FIGURE 1.
Schematic diagram of glycolysis.
2.1.1 HK2
2.1.1.1 The function of HK2
HK2 is one of the subtypes of hexokinase, which functions as the first rate-limiting glycolysis enzyme in the glycolysis pathway and converts glucose to glucose-6-phosphate (G6P) (Wilson, 2003). HK2 is the most efficient of all subtypes of HK, and it has both an N-terminal active site domain and a C-terminal active site domain (Xu and Herschman, 2019). Glucose quickly enters the cell through the glucose transporter, where HK2 binds ATP and intracellular glucose to efficiently catalyze the production of glucose-6-phosphate (G6P), which promotes glycolysis (Mathupala et al., 2006). HK2 is also related to the progression of several cancers. HK2 is abnormally expressed in high amounts in many cancers, which is very important for tumor initiation, and maintenance and is closely related to cancer treatment and prognosis (Gong et al., 2012; Patra et al., 2013; Sato-Tadano et al., 2013).
2.1.1.2 The roles of HK2 in osteosarcoma
Significantly, HK2 is overexpressed in OS and is closely related to Ki67. Ki-67, also known as MKI-67, is a protein encoded by the MKI67 gene in humans. Expressed only in actively proliferating cells, is a proliferation-associated nuclear antigen. Clinically, ki67 is mainly used to label cells in the proliferative cycle and due to its overexpression in cancer cells, Ki-67 has been proposed as a prognostic biomarker for cancer (Li et al., 2015). The 5-year survival rate of patients with ki67 expression in osteosarcoma patients is extremely low (Zeng et al., 2021). The knockdown of HK2 causes a decrease in aerobic glycolysis, increasing cell apoptosis and decreasing cell colony formation ability. Furthermore, the effects mentioned above of HK2 can be blocked by the glycolysis inhibitor, 2-deoxy-d-glucose (2-DG), which further indicates that the effect of HK2 on the growth of OS is achieved by regulating the glycolysis pathway (Zhuo et al., 2015). Besides, HK2 is the direct transcriptional target of the classical signal pathway nuclear factor kappa B (NF-κB). HK2 overexpression can restore metabolic transfer induced by inhibiting NF-κB activity in OS cells and enhance the glycolysis pathway (Londhe et al., 2018). HK2 functions as a target for microRNAs (miRNAs) in OS cells (Song et al., 2017; Sun L. et al., 2020; Wan et al., 2021). Liu C. et al. (2019) suggested that low expression of miR-185 in OS negatively regulates the expression of HK2. Overexpression of miR-185 significantly suppressed the proliferation and migration of OS cells. However, HK2 overexpression could significantly weaken the inhibitory effect of miR-185 on glycolysis in OS cells. By inhibiting HK2, miR-185 inhibits glucose metabolism in OS cells, making it an excellent target for drugs. In addition, HK2 can be regulated by the ELK1-miR-134-PTBP1 signaling pathway, which affects the glycolysis and chemical resistance of OS cells (Zhang et al., 2021b). circRNA can also affect the progress of glycolysis and OS by regulating the expression of HK2 (Gu et al., 2020). Interestingly, HK2 can also be regulated by lncRNA. LncRNA-SARCC can increase the sensitivity of OS cells to cisplatin. The mechanism may be that miR-143 inhibits the expression of HK2 and reduces the glycolysis level of OS cells. The mechanism of lncRNA-SARCC-miR-143-HK2 provides a new therapeutic approach for studying cisplatin resistance in OS (Wen et al., 2020). A study illustrated that iron increased the expression of HK2 and GLUT1, and regulated the AMPK/mTORC1 signaling pathway (Ni et al., 2020). Overall, these results indicate that targeting the HK2 might be a potential therapeutically valuable strategy in OS.
2.1.2 Phosphofructokinase
2.1.2.1 The function of phosphofructokinase
The primary function of fructose-6-phosphate-1-kinase (PFK-1) is to catalyze the conversion of fructose-6-phosphate to fructose-6-diphosphate and adenosine diphosphate (ADP), thereby promoting glycolysis. As this process is irreversible, phosphofructokinase (PFK) becomes the second rate-limiting enzyme in glycolysis (Uyeda, 1979). The catalytic activity of PFK-1 is regulated by some metabolites, small molecule inhibitors, and intracellular proteins, such as adenosine triphosphate (ATP), F-6-P, and 2, 6-diphosphate fructose 6-diphosphate (F-2,6-BP), and adenosine diphosphate (ADP) (Sola-Penna et al., 2010; Bartrons et al., 2018). As a product of catalyzation by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK-2/FBPase-2, PFKFB), F-2,6-BP is one of the regulatory substances and the most potent positive allosteric effector of PFK-1 (Yi et al., 2019). Four different genes (PFKFB1, PFKFB2, PFKFB3, and PFKFB4) encode PFK-2/FBPase-2 (Ros and Schulze, 2013). These four genes are expressed differently in different human tissues and organs. PFKFB1 mainly exists in the liver and skeletal muscle, PFKFB2 is highly expressed in the myocardium, PFKFB3 is generally expressed and PFKFB4 is highly expressed in the testis (Yi et al., 2019). In mammals, three subtypes of PFK have been identified: PFK-M (muscle), PFK-P (plasma), and PFK-L (liver). The homology between PFK-M and PFK-L is about 68.6%, between PFK-M and PFK-P is about 70.3%, and between PFL-L and PFL-P is about 66.6% (Sola-Penna et al., 2010). The effect of PFK on tumor cells has been extensively researched. PFK is regulated by carcinogenic proteins (HIF-1α and c-myc) or tumor suppressor protein (p53) in many tumors, which affects tumor progression by altering the rate of glycolysis (Moreno-Sanchez et al., 2007; Yalcin et al., 2009).
2.1.2.2 The roles of phosphofructokinase in osteosarcoma
The role of PFKFB1 and PFKFB4 in OS has not been reported. PFKFB1 has four splice variants: PFKFB1-201, PFKFB1-202, PFKFB1-203 and PFKFB1-204 (Lee et al., 2003; Ros and Schulze, 2013). PFKFB-1 is lowly expressed in normal cells, but overexpressed in cancer cells (Minchenko et al., 2003; Kotowski et al., 2021). For example, the expression of PFKFB-1 is significantly correlated with the prognosis of bladder cancer patients (Zhang C. et al., 2020). PFKFB4 is overexpressed in multiple human cancers such as lung adenocarcinoma, supports tumor growth by synthesizing F-2,6-BP and is required for glycolysis in response to hypoxia and tumor growth (Chesney et al., 2014). Studies have demonstrated that PFKFB2, as the substrate of tumor inhibitor miR-1297, is negatively regulated by miR-1297 and affects the proliferation of OS by regulating glycolysis (Pan et al., 2020). SRC/ERK/C-MYC/PFKFB2 signal pathway in OS cells affects the glycolysis pathway by regulating PFKFB2 expression, affecting its proliferation (Zhao et al., 2018). Moreover, the tumor suppressor gene miR-26b is down-regulated in OS. It regulates the expression of the glycolysis-related protein (LDHA, GLUT1), invasion, and cell cycle markers (such as MMP-9, MMP-2, cyclin D1, and p27) by inhibiting the expression of mRNA and protein of PFKFB3, resulting in malignant progression of OS (Du et al., 2015). Recent studies have revealed that Rho-related coiled-coil kinase (ROCK) includes two members: ROCK1 and ROCK2. Dysregulated expression of ROCK is closely associated with enhanced metastatic ability and reduced patient survival in various cancers (de Sousa et al., 2020). Both ROCK2 and PFKFB3 proteins have significantly high expression in OS. Regarding mechanism, ROCK2 stabilizes PFKFB3 expression by blocking its ubiquitination degradation. Low-level ROCK2 knockouts can save OS cells from proliferation and metastatic loss by upregulating PFKFB3 (Deng et al., 2019). The ubiquitin-like protein FAT10 is highly expressed in various cancers (colorectal, liver, and gastric) and promotes cancer progression (Aichem and Groettrup, 2016). Similarly, ubiquitin-like protein FAT10 is highly expressed in OS tissues, while high levels of FAT10 are closely associated with tumor growth and survival time of OS patients. Experimental results showed that FAT10 upregulates the protein expression level of PFKFB3 by directly binding to EGFR and inhibiting its ubiquitination and degradation, thus inducing glycolysis (Deng et al., 2020). It can be observed that PFK, as a glycolytic enzyme, also affects malignant behaviors of OS, such as cell growth and metastasis.
2.1.3 Pyruvate kinase
2.1.3.1 The function of pyruvate kinase
Pyruvate kinase (PK) is the last rate-limiting enzyme in the glycolysis pathway. Its function is to catalyze the conversion of phosphoenolpyruvate (PEP) to pyruvate and convert ADP to ATP, thus promoting glycolysis (Harris et al., 2012). PK is encoded by two genes, PKLR and PKM, and it is divided into four subtypes: PKL (liver type competition), PKR (red blood cell competition), PKM1 (muscle competition isozyme M1), and PKM2 (muscle competition isozyme M1) (Dayton et al., 2016). The distribution of these four subtypes in the human body is diverse. PKL subtypes are highly expressed in kidney, liver and red blood cells; PKR mainly exists in red blood cells; PKM1 is enriched in the myocardium, skeletal muscle and brain tissues; PKM2 is mainly distributed in the brain and liver tissues (Israelsen et al., 2013). PKM2 is the most powerful and widely studied subtype and has abnormally high expression in many cancers, such as colorectal tumors, breast tumors, lung tumors, prostate tumors, and ovarian tumors, while the expression levels in the pharynx and testicular tumors are below or close to the critical value (Munoz-Colmenero et al., 2015). PKM2 can regulate the activity and localization of proteins through post-transcriptional modifications such as phosphorylation and acetylation, thus regulating cell proliferation ability (Israelsen and Vander Heiden, 2015). Lv et al. (2013) reported that the carcinogenic form of fibroblast growth factor receptor type 1 could directly phosphorylate PKM2 tyrosine residue 105 (Y105) to inhibit PKM2 and tumor growth. Furthermore, PKM2 was acetylated by p300 acetyltransferase at the K433 site, which interfered with the binding of FBP to block PKM2 activation. It promoted the nuclear accumulation and kinase activity of PKM2, thereby promoting the proliferation of cancer cells.
2.1.3.2 The roles of Pyruvate kinase in osteosarcoma
Of the four competing subtypes, only PKM2 is more studied in OS. Notably, PKM2 is expressed abnormally high in OS tissues and is more abundant than para cancerous tissues (60.2 vs. 26.1%, p < 0.001). In addition, high expression of PKM2 is positively correlated with kennel stage and distant metastasis. The higher the expression level of PKM in OS patients, the higher the overall survival (OS) time (Liu Z. X. et al., 2016). Moreover, circPVT1 binds to miR-423-5p and activates the Wnt5a/Ror2 signal through saponification of miR-423-5p, promoting OS glycolysis (upregulated by HK2, GLUT1, LDHA, and PKM2), proliferation and transfer (Wan et al., 2021). Pu et al. (2022) demonstrated that the interaction between lncRNA colon cancer-associated transcript-1 (lncCCAT1) and PKM2 promotes the phosphorylation of sterol regulatory element binding protein 2 (SREBP2) and enhances the Warburg effect, adipogenesis, and OS cell growth. As a direct target of miR-491-5p, PKM2 was negatively regulated by miR-491-5 in OS cells and PKM2 overexpression could rescue the inhibitory effect of miR-491-5p on the proliferation of OS cells (Chen T. et al., 2018). The knockdown of PKM in OS cells decreased the expression levels of cyclin D1 and Bcl-2 and increased the expression levels of Bax, caspase-3, and PARP. Moreover, knockdown of PKM inhibited the growth of OS cells and induced G1 cell cycle arrest and apoptosis in OS cells. In addition, the knockdown of PKM2 significantly inhibited the metastatic ability of OS cells. In vivo experiments displayed that the knockdown of PKM2 can reduce the growth rate of OS and induce the structural deterioration of tumor tissue (Yuan et al., 2018). Glucagon-like peptide 2 (GLP-2), a glucagon precursor-derived peptide, promotes the release of ingested nutrients from enteroendocrine cells (Suzuki et al., 2020). GLP2 inhibits the proliferation of OS cells both in vivo and in vitro. The specific mechanism is that GLP2 inhibits the expression of NF-κB expression in OS cells, resulting in decreased PKM2 and CyclinD1 (Lu et al., 2018). According to a case report, miR-1294 is down-expressed in OS cells. Overexpression of miR-1294 inhibits the growth and metastasis of OS cells, and induces G0/G1 stagnation and apoptosis. Besides, PKM2 is a direct target of miR-1294 in OS cells, PKM2 overexpression saves the decline of cell proliferation, migration, and invasion caused by miR-1294 overexpression (Yuan et al., 2020). IRF7 is a transcription factor and one of the interferon regulatory factors (IRFs). IRFs play important roles in cell differentiation, apoptosis, and cell cycle regulation in tumor development (Chen et al., 2017). PKM2 mediates the effect of IRF7 on glycolysis in OS cells. IRF7 inhibits PKM2 transcription, thus suppressing the glycolysis and resulting in a decline in the proliferation of OS cells (Li et al., 2022). Interestingly, PKM2 is closely related to chemotherapy drug resistance of OS stem cells. PKM2 overexpression can lead to the resistance of cisplatin by OS stem cells. Metformin treatment combined with PKM2 knockdown significantly reduces the semi-maximum inhibitory concentration (IC50) of cisplatin on HOS OS stem cells (Shang et al., 2017).
2.1.4 Glucose transporters
2.1.4.1 The function of glucose transporters
Human GLUTs are a 14-member family encoded by the SLC2 gene (Deng and Yan, 2016). Increased glucose uptake and metabolism are hallmarks of cancer cells, and the primary function of GLUTs is to facilitate glucose uptake across the plasma membrane (Barron et al., 2016). Generally, GLUT1 is expressed in almost all tissues but is abnormally expressed in high amounts in numerous cancers, including lung cancer, breast cancer, hepatocellular carcinoma, and kidney cancer (Barron et al., 2016). GLUT2 mainly transports glucose in hepatocytes and has a very high Km value and a high affinity for glucosamine (Deng and Yan, 2016). Several studies have shown that the high affinity and transportability of GLUT3 to glucose are prominent in neurons, and the expression of GLUT3 in gliomas and astrocytoma is significantly higher than that of GLUT1 (Barron et al., 2016). GLUT4 is mainly distributed in adipocytes, cardiomyocytes, and skeletal muscle cells in vivo and is essential for regulating systemic glucose homeostasis (Mueckler and Thorens, 2013).
2.1.4.2 The roles of glucose transporters in osteosarcoma
Numerous studies have reported that GLUT1 is a significant independent prognostic factor for many tumors, including OS. Kubo et al. (2015) immunohistochemically determined the expression of GLUT1 and the angiogenic activity of CD34 positive microvessels in patients with high-grade OS. Studies have suggested that glucose metabolism might negatively affect angiogenesis. GLUT1 is a potential marker of survival and drug therapy in patients with OS, with important implications for the prognosis of OS. GLUT1, as the downstream target gene of HIF1a, often affects the growth of OS and is the key protein related to angiogenesis and energy metabolism (Nepal et al., 2011). In addition, the protein level and mRNA level of GLUT1 are abnormally overexpressed in OS, and high levels of GLUT1 are significantly associated with lymph node metastasis, age and low survival rate (Fan et al., 2017). Knockdown of GLUT1 significantly slowed the proliferation of OS cells in vitro and in vivo (Fan et al., 2010; Jian et al., 2015). A study suggested that GLUT1 and GLUT4, downstream substrates of tumor suppressor p53, were negatively regulated by p53 protein to reduce glucose transport. The inhibitory effect of P53 on the transcriptional activity of the GLUT4 promoter was significantly greater than that on GLUT1 promoter. thereby degrading metabolic intensity and inhibiting tumor growth (Schwartzenberg-Bar-Yoseph et al., 2004). The main function of GLUT1 is to transport glucose, which is no different in OS. Studies have reported that GLUT1 is regulated by insulin in MG-63 cells of OS. Although insulin does not affect the expression of GLUT1, it can induce translocation to the plasma membrane (Cifuentes et al., 2011). Remarkably, GLUT1 can also be regulated by lncRNA. The expression level of HAND-AS1 was lower in OS than in normal tissues. Knockdown of HAND-AS1 promotes GLUT1 protein expression, suppressing OS cell proliferation. Non-coding RNA of HAND2-AS1 targets glucose metabolism and inhibits the proliferation of cancer cells in OS (Chen S. et al., 2019). In addition, miR-328-3p inhibited GLUT1 expression and glucose uptake rate in OS cells, and hBERA/miR-328 combined with OS chemotherapeutic drugs cisplatin or doxorubicin could significantly inhibit the proliferation of OS cells (Yi et al., 2020). OS is a highly invasive tumor, which is an important reason for its poor prognosis. As a tumor biological function regulator, LAIR-1 can inhibit the expression of GLUT1, thus reducing the expression of EMT-related molecules in OS cells and inhibiting the metastatic ability of OS cells (Zhang J. et al., 2020). Carcinogen STC2 enhances glycolysis and promotes the growth of OS cells by upregulating the expression of GLUT1 (Yu et al., 2021). The role of GLUT2 in OS has not been reported so far. A study demonstrated that ELK1 promotes the expression of GLUT3, HK2, PDK1 and PTBP1 by inhibiting miR-134 expression, while enhancing the chemical tolerance and aerobic glycolysis of OS cells (Zhang et al., 2021b). Altogether, glucose transporters (GLUTs), especially GLUT1, play a critical role in the glycolytic pathway and is a putative candidate target in OS treatment.
2.1.5 LDHA
2.1.5.1 The function of LDHA
Lactate dehydrogenase (LDH) catalyzes pyruvate to lactate and is a vital oxidoreductase in the glycolytic pathway. LDH comprises two subunits, LDH-A and LDH-B, assembled into five different isozymes (LDH1, LDH2, LDH3, LDH4, and LDH5). LDH-A, encoded by the LDHA gene, is the main skeletal muscle form and has a high affinity for pyruvate. LDH-B is encoded by the LDHB gene and exists primarily in the myocardium (Dawson et al., 1964; Ding et al., 2017). Due to the high affinity for pyruvate, LDHA prioritizes the conversion of pyruvate to lactic acid and NADH to NAD+. Simultaneously, LDHB has a high affinity for lactic acid under anaerobic conditions. When oxygen is sufficient, priority is given to converting lactic acid to pyruvate and NAD + to NADH (Urbanska and Orzechowski, 2019). Immunohistochemistry experiments indicated that LDHB expression in normal and cancer tissues had no significant difference. However, LDHA was found to be highly expressed in most cancer cells, which is the main reason why LDHA can be used as a biomarker of many malignant tumors, such as gastric cancer (Kolev et al., 2008), testicular germ cell tumor (Augoff and Grabowski, 2004), glioma (Hu S. et al., 2016) and melanoma (Brand et al., 2016). The function of LDHB in tumors has also been reported (Doherty and Cleveland, 2013). LDHB is expressed abnormally high in lung cancer cell lines and is closely associated with the survival rate of patients with lung adenocarcinoma (McCleland et al., 2013). As a key enzyme in the glycolysis pathway, LDHB is also an essential protein for triple-negative breast cancer. LDHB and other related glycolysis enzymes are significantly upregulated in triple-negative breast cancer. Knocking down LDHB suppresses the growth of breast cancer cells (McCleland et al., 2012).
2.1.5.2 The roles of LDHA in osteosarcoma
In OS, glycolytic enzymes such as LDHA, GLUT1, and HK1 can be negatively regulate the phenolic compound apigenin (4′,5,7-trihydroxyflavone), which attenuates the Warburg effect, mediated by PI3K/Akt/mTOR signal pathway dehydrogenase (Shi et al., 2022). In 2019, Wang et al. found three cell-active hLDHA inhibitors (38, 63, and 374) by targeting human lactate, and related experimental evidence that these inhibitors suppressed the proliferation of MG-63 cells. The IC50 values were 6.47 μM, 2.93 μM, and 6.10 μM, and hLDHA50 values were 3.03 μM, 0.63 μM, and 3.26 μM for inhibition by EC (Wang F. et al., 2020). Other studies have also designed a variety of inhibitors for LDHA and confirmed their inhibitory effect on LDHA by enzymatic assay in vitro and strongly inhibited the proliferation of OS cells by cytotoxicity assay (Cui et al., 2016; Fang et al., 2017). MicroRNAs (miRNAs) have been widely studied in OS growth, invasion, and migration. The miR-33b inhibits glycolysis by targeting the key enzyme lactate dehydrogenase A (LDHA), suppressing OS cells’ growth (Zheng et al., 2018). In 2016, compared with normal osteoblasts (hFOB1.19), LDHA was upregulated in OS cell lines. Treatment of OS cells with FX11, an LDHA inhibitor, down-regulated the activity of LDHA and suppressed the growth and metastasis abilities of OS cells in a dose-dependent manner. Moreover, the study also reported that 2-DG, a glycolysis inhibitor, completely blocked cell proliferation and invasion mediated by LDHA, suggesting that the carcinogenic activity of LDHA in OS may be dependent on glycolysis. Interestingly, inhibiting the pharmacological effects of c-myc or HIF-1α reduces the expression of LDHA (Gao et al., 2016). LDHA was also closely related to cisplatin resistance in OS. Bioinformatics analysis and luciferase assay demonstrated that glycolytic enzyme LDHA was the downstream substrate of miR-329-3p in OS cells. LDHA overexpression in miR-329-3p overexpressed cisplatin-resistant cells could restore glucose metabolism and increase cisplatin resistance in OS cells (Li G. et al., 2021). The miR-323a-3p negatively regulated mRNA and protein levels of LDHA in OS cells. The expression levels of LDHA and miR-323a-3p in OS tissues showed a negative correlation. LDHA promotes glycolysis in OS, while miR-323a-3p overexpression reduces the ability of OS cells to produce lactic acid (Chen H. et al., 2018). Circular RNAs (circRNAs) are critical in OS biology. For example, circATRNL1 is highly expressed in OS tissues and cells. CircATRNL1 regulates the expression of LDHA, promoting glycolysis and the progression of OS (Zhang et al., 2021a). Furthermore, LDHA is regulated by various circRNAs, which mediates glycolysis and the growth of OS (Wan et al., 2021; Jiang X. et al., 2022). A new study stated that abnormally high expression of KDM6B in OS promotes tumor metastasis in OS by positively regulating the expression level of LDHA, which raises the prospect of OS (Jiang Y. et al., 2021). There are also a few studies on the role of LDHB in OS. Remarkably, LDHB was highly expressed in OS cells, and the mRNA expression level of LDHB was significantly higher in metastatic OS. Furthermore, the poor prognosis of patients with OS frequently correlated with a high expression of LDHB. LDHB knockdown inhibits the growth and metastasis of OS cells. Statistical analysis of clinicopathological characteristics of LDHB disclosed that the expression levels of LDHB were related to the TNM stage of the tumor, recurrence, and survival in OS patients. Therefore, LDHB can be regarded as a prognostic marker of cancer recurrence and poor overall survival of OS (Li et al., 2016). Similarly, LDHB is highly expressed in OS tissues and cells, and the existence of the miR-141-3p/FUS/LDHB axis enhances the growth, migration, and invasion of OS cells (Wang, 2020).
2.2 Potential natural products of glycolysis in osteosarcoma
Many natural compounds and synthetic reagents targeting glycolytic enzymes have been developed, with related cell experiments and clinical trials proving that they can inhibit OS progression. For example, Icariside II, an active flavonoid extracted from Epimedium koreanum Nakai (Berberidaceae), has anticancer effects in various cancer cells (Kang et al., 2012; Wu et al., 2015) (Geng et al., 2014). It is a natural mTOR inhibitor that inhibits aberrant glycolysis in OS by inhibiting the cap-dependent translation of c-myc through mTORC1-4E-BP1 axis (Zhang et al., 2016). Bavachinin is a flavonate from the seeds of Proralea corylifolia L., native to eastern Asia, inducing HIF-α ubiquitin and proteasome-mediated degradation and inhibiting the synthesis of HIF-1 and the glucose metabolism of OS (Nepal et al., 2012). Resveratrol is a natural polyphenol extracted from grapes, mulberries, peanuts, and other plants. In OS, resveratrol upregulates Cx43 and E-cadherin and inhibits the Wnt/β-catenin signaling pathway, thereby inhibiting OS proliferation and glycolysis, inducing apoptosis, and reducing the invasiveness of U2-OS cells in vitro (Xie et al., 2017). Quercetin, an all-natural chemical found in a variety of plants and foods (Harborne and Williams, 2000), induces apoptosis in osteosarcoma cells through mitochondrial dysfunction and Akt dephosphorylation (Xie et al., 2011). The isoflavones formononetin and calycosin are the two main active components of traditional Chinese medicine Radix astragali. They are involved in the inactivation of ERK and Akt pathways, thereby inducing apoptosis of OS cells, and formononetin is more effective than calycosin (Yang et al., 2014). Oxymatrine and matrine are alkaloids extracted from the roots of Sophora flavescens and have various pharmacological activities. Oxymatrine induces human bone by inhibiting PI3K/Akt pathway mitochondrial-dependent apoptosis in sarcoma cells (Wei et al., 2014; Zhang et al., 2014). Selaginella tamariscina is a traditional medicinal plant for treating advanced cancer in the East and exerts an anti-metastatic effect on OS cells by inhibiting p38 and Akt signaling pathways (Yang et al., 2013). Tanshinone I is isolated from the Chinese herbal medicine Salvia miltiorrhiza and can significantly downregulate circ_0000376 expression in OS cells and inhibit the viability, migration, and invasion of OS cells through circ_0000376/miR-432-5p/BCL2 axis, glycolysis and trigger apoptosis of OS cells (Ye et al., 2022). Punicalagin is a polyphenolic compound extracted from pomegranate, which downregulates the expression of p65, survivin, XIAP, CIAP2 and other proteins, and inhibits the proliferation and metastasis of OS cells by inhibiting the NF-κB signaling pathway (Wang X. Z. et al., 2020). Chlorogenic acid (CGA) is a phenolic compound contained in plant-related products, although related levels are also found in other plant-related products such as herbs, fruits, and vegetables. Green coffee beans are the main source of CGA (Liang and Kitts, 2015). which activates ERK1/2 and inhibits the proliferation of OS cells (Sapio et al., 2020). Oridonin, a diterpenoid extracted from the herb Rubescens, inhibits the proliferation, migration, and invasion of OS cells by inhibiting the expression of matrix metalloproteinases and STAT3 signaling pathway (Du et al., 2019). Amentoflavone, a flavonoid extracted from Selaginella tamariscina, inhibits OS progression by inhibiting ERK/NF-κB signaling pathway (Lee et al., 2019). Oleanolic acid (OA), a naturally occurring triterpenoid, induces apoptosis in OS cells by inhibiting Notch signaling while inhibiting the proliferation and invasion of OS cells by suppressing SOX9/Wnt1 signaling (Xu et al., 2018) (Chen X. et al., 2021). The above potential natural products of glycolysis in osteosarcoma are displayed in Tables 1, 2.
TABLE 1.
List of glycolysis inhibitors that target OS cells.
| Inhibitors | Target glycolysis enzymes | References |
|---|---|---|
| Cytochalasin B | GLUT | Ogawa et al. (2021) |
| STF-31 | GLUT | Kraus et al. (2018) |
| GLU-MTX (glucose-methotrexate conjugate) | GLUT1 | Wozniak et al. (2021) |
| Insulin and parathyroid hormone (PTH) | GLUT1 | Thomas et al. (1995) |
| KRH102053 | HIF-1α | Nepal et al. (2011) |
| NSC-134754 and NSC-643735 | HIF-1α | Chau et al. (2005) |
| Bavachinin | HIF-1α | Nepal et al. (2012) |
| Icariside II | HIF-1α | Choi et al. (2008) |
| FX11 | LDHA | Gao et al. (2016) |
| Compound 11 | LDHA | Fang et al. (2017) |
| 2-((3-cyanopyridin-2-yl) thio) acetamide-containing compounds | LDHA | Cui et al. (2016) |
| Metformin | PKM2 | Shang et al. (2017) |
| 63 | LDHA | Fei Wang et al. (2020) |
| 12 | PDK2 | Cao et al. (2019) |
TABLE 2.
List of potential small molecule inhibitors and drugs for OS treatment in basic experimental or clinical trials.
2.3 Signaling pathways linking glycolysis and osteosarcoma
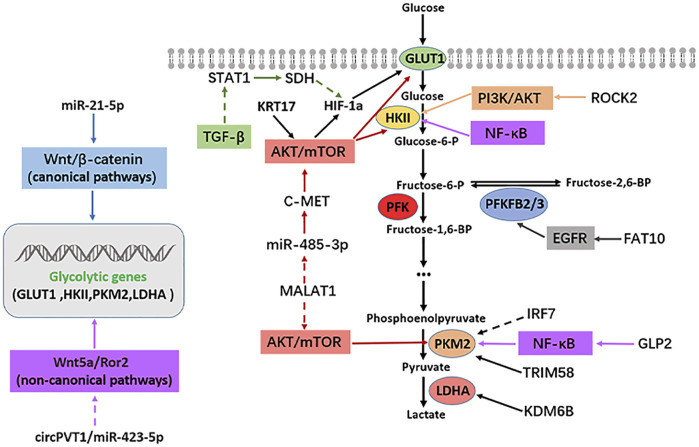
The rapid growth and metastasis of tumors are closely related to their enhanced glycolysis and the uptake rate. Glucose utilization efficiency in cancer cells is significantly increased (Hanahan and Weinberg, 2011; Jia et al., 2019). However, the interaction between enhanced metabolic pathways and a series of intracellular signal pathways, especially some carcinogenic signal pathways, is often an essential regulatory mechanism of tumor progression. Several signal pathways are involved in OS occurrence and development, such as PI3K/AKT, mTOR, Hippo, Notch, Wnt/β-catenin, VEGF, and TGF-β. This study summarized some classical signal pathways that participate in glycolysis and affect the progression of OS, such as PI3K/AKT, mTOR, EGFR, TGF-β, and NF-κB. The regulatory mechanism between the above signaling pathways, glycolytic enzymes, and OS is illustrated in Figure 2.
FIGURE2.
Signaling pathways regulating glycolysis in osteosarcoma. Solid arrows indicate facilitation, while dashed arrows indicate inhibition.
2.3.1 Phosphatidylinositol-3-kinase/AKT pathway
In terms of glycolysis, PI3K can be activated by numerous proteins, which can directly activate AKT. Activated AKT can regulate the expression of glycolysis-related genes directly or indirectly (Xie et al., 2019). As a result, the PI3K/AKT pathway is thought to be very crucial in inducing glycolysis. PI3Ks is a family of lipoprotein kinases. Phosphatidylinositol (PI) catalyzes 3-hydroxy phosphorylation to produce the second messenger, which plays a role in signal transduction (Engelman, 2009). PI3Ks are divided into three types, I-III, which has a conserved kinase domain (Engelman, 2009). PI3K/AKT pathway is a carcinogenic signal pathway in many tumors, including OS. PI3K/AKT pathway has been overactivated in OS and promotes its occurrence and development of OS. Interestingly, abnormal activation of the PI3K/AKT signaling pathway is involved in various steps of pathological development of OS, such as OS occurrence, proliferation, invasion, metastasis, angiogenesis, cell cycle, apoptosis and chemotherapy resistance (Zhang et al., 2015). PI3K/AKT pathway acts on several substrate proteins, and different regulatory mechanisms are produced according to different substrate proteins. PI3K/AKT pathway is critical in regulating the glycolysis level of OS. The activated PI3K/AKT pathway positively regulates the glycolysis-related enzyme HK2 in OS. HK2 is highly expressed in OS. Inhibition of HK2 gene expression reduces OS cell proliferation and increases apoptosis, while the activated PI3K/AKT signal pathway enhances aerobic glycolysis by promoting HK2 expression (Zhuo et al., 2015). In 2017, Ru et al. (2018) found that miR-564 was expressed abnormally low in OS, and miR-564 overexpression in OS significantly suppressed the mRNA and protein expression of AKT, and attenuated OS cell glycolysis and cell proliferation. ROCK2 is highly expressed in OS tissues compared to normal tissues. The mRNA and protein expression of glycolytic enzyme HK2 is regulated by ROCK2. The specific mechanism is that ROCK2 promotes the expression of HKII by activating PI3K/AKT signal pathway, and then induces glycolysis and proliferation of OS cells (Deng et al., 2021). In summary, OS cells are maintained and developed through the PI3K/AKT pathway, which mediates glycolysis and proliferation.
2.3.2 Rapamycin target protein pathway
Rapamycin target protein (mTOR) is a serine/threonine protein kinase classified as phosphatidylinositol-3-kinase (PI3K)-related protein family (Xie et al., 2019). The mTOR pathway regulates various cellular metabolism processes, such as glycolysis, amino acid metabolism, fatty acid metabolism, and lipid metabolism (Mossmann et al., 2018). Concerning glycolysis, the mTOR pathway is vital for regulating cell glycolysis. Cancer cells express glycolysis-related enzymes, which are regulated by the mTOR pathway, affecting the progression of tumors (Vijayakrishnapillai et al., 2018). The mTOR pathway has been extensively researched in tumors. The mTOR pathway is abnormally activated in many cancers, regulating the metabolism and the progression of cancer cells (Mossmann et al., 2018). In OS, the mTOR pathway is a key signal pathway that is overactivated and affects the OS progression. It is reported that multiple elements in the mTOR pathway are involved in the OS regulation (Hu K. et al., 2016). The mTOR is also often regulated by PI3K and AKT to form a PI3K/AKT/mTOR signal pathway. Interestingly, mTOR serves as an activator of AKT, so a positive feedback loop can be formed between AKT and mTOR, thus promoting the proliferation and migration of cancer cells (Zhang et al., 2015). In 2016, Wang et al. (2017) demonstrated that the mTORC1 pathway was highly activated in clinical specimens of OS. In OS, the mTORC1 regulates glycolysis, proliferation, and antioxidation of OS cells by mediating serine/glycine metabolism. Low expression of miR-485-3P is found in human OS tissues and cells with high expression of c-MET, AKT3, and MALAT1. Mechanistically, miR-485-3p binds to c-Met and AKT3 mRNAs and inhibits c-Met and AKT3/mTOR signaling pathways. Furthermore, MALAT1 interacts with miR-485-3p and inhibits c-Met and AKT3/mTOR signaling. The above regulatory mechanisms functionally inhibit glycolysis, growth, and metastasis in OS (Wang Q. et al., 2021). KRT17/AKT/mTOR/HIF-1α axis regulates glycolysis and proliferation in OS cells. KRT17 is highly expressed in OS tissues and cells, and KRT17 positively regulates the glycolysis and proliferation of OS cells. Specifically, KRT17 regulates the expression of p-AKT, p-mTOR, HIF-1a and GLUT1. The results of recovery experiment also showed that KRT17 controlled glycolysis and proliferation of OS by positively regulating AKT/mTOR/HIF-1α pathway (Yan et al., 2020). Additionally, S100A10 is highly expressed in OS tissues and cell lines, and S100 calcium-binding protein A10 (S100A10) regulates glycolytic metabolism by activating the AKT/mTOR pathway, promotes OS cell proliferation and metastasis, and inhibits apoptosis (Ling and Lu, 2022). Hence, the mTOR pathway plays a critical role in OS progression and the formulation of therapeutic strategies targeting this pathway is of utmost significance.
2.3.3 Epidermal growth factor receptor pathway
Epidermal growth factor receptor (EGFR) is a member of the ErbB receptor tyrosine kinase (RTK) family, which is vital to regulating epithelial cell physiology (Lemmon et al., 2014). EGFR also regulates autophagy and metabolism (Tan et al., 2016). Xanthohumol is a prebiotic flavonoid with anticancer activity isolated from Hops (Wei et al., 2018). Xanthohumol down-regulated EGFR expression, further inhibiting the activity of AKT1 and glycolysis while promoting apoptosis in colorectal cancer (Liu W. et al., 2019). A dual novel EGFR/HER2 inhibitor, KU004, can inhibit the expression of hexokinase II (HK2) through PI3K/AKT signaling pathway, thereby inhibiting glycolysis and tumor growth (Tian et al., 2017). EGFR is often widely studied as an oncogene in tumors. In OS, an earlier study detected the expression of EGFR by immunohistochemical method. The results indicated that EGFR protein expression level was detected in 57% of all cases analyzed in the form of strong membrane staining, and six of 12 OS cells showed moderate to high expression of EGFR, which is of paramount significance for the follow-up study of EGFR in OS (Wen et al., 2007). EGFR knockdown significantly suppressed the growth and migration of OS cells. On the contrary, EGFR overexpression promotes the proliferation and metastasis of OS cells. Based on its mechanism, EGFR positively regulates the expression of p-AKT and p-ERK and promotes OS progression. Besides, the level of p-EGFR increased in OS cells treated with gemcitabine, and the inhibition of proliferation and apoptosis rate increased more significantly in OS cells treated with gemcitabine combined with EGFR knockdown (Wang S. et al., 2021b). The regulation of glycolysis by EGFR has also been reported in OS. Ubiquitin-like protein FAT10 was significantly overexpressed in OS tissues and cells. The high expression level of FAT10 was closely related to tumor growth and the short survival time of OS patients. Regarding mechanism, FAT10 directly binds to EGFR, inhibits its ubiquitination and degradation, and positively regulates the expression of glycolysis protein PFKFB3. Regarding function, FAT10/EGFR/PFKFB3 axis promotes glycolysis and proliferation of OS cells (Deng et al., 2020).
2.3.4 Nuclear factor kappa B pathway
NF-κB is a family of dimer transcription factors involved in coordinating inflammatory response, innate and acquired immunity, differentiation, growth, and survival in almost all multicellular organisms (Mitchell et al., 2016). Hexokinase (HK) two is a direct transcriptional target of NF-κB. Canonical NF-κB signaling pathway inhibition reduced the expression level of HK2 in OS and inhibited OS aerobic glycolysis (Londhe et al., 2018). The activation of NF-κB leads to increased aerobic glycolysis rate and the up-regulation of GLUT3 through the loss of p53, indicating that NF-κB promotes the Warburg effect by upregulating glycolysis gene GLUT3 in cancer cells (Kawauchi et al., 2008). NF-κB can promote the Warburg effect by upregulating pyruvate kinase M2 (PKM2) (Yang et al., 2012). In OS, NF-κB inhibitors can induce apoptosis of ZOS and U2OS cells and inhibit OS’s growth. Furthermore, the authors have found that the NF-κB pathway may play a key role in OS cell survival, mainly mediated by GSK-3β and IKB kinase (IKK). When GSK-3β or IKK is blocked, IkBA is stabilized and trapped in the cytoplasm, damaging the cell survival pathway and promoting apoptosis of OS cells (Tang et al., 2012). In OS, NF-κB inhibitors can induce apoptosis of OS cells and suppress their progression. In addition, the NF-κB pathway may take part in OS cell survival, mediated by GSK-3β and IKB kinase (IKK). When GSK-3β or IKK is blocked, IkBA is stabilized and trapped in the cytoplasm, directly damaging cell survival and promoting apoptosis of OS cells (Londhe et al., 2018).
2.3.5 Transforming growth factor β pathway
Transforming growth factor β (TGF-β) is the prototype of the TGF-β family of growth and differentiation factors. TGF-β is a bifunctional regulator which suppresses or promotes cell growth. TGF-β regulates numerous events in normal physiological processes, and the interference of TGF-β signals is related to the pathoge nesis of connective tissue disorder, fibrosis, and tumor (Morikawa et al., 2016). The role of the TGF-β signaling pathway in glycolysis has also been extensively studied. It was found that TGF-β1 induces PFKFB3 expression by activating p38/MAPK and PI3K/AKT signaling pathways in glioma cells, resulting in an increase in fructose 2,6-diphosphate concentration, glucose uptake, glycolysis flux, and lactic acid production (Rodriguez-Garcia et al., 2017). In lung cancer cells, TGF-β negatively regulates PDK4 expression and the downregulation of PDK4 leads to the transition from glycolysis to oxidative phosphorylation (OXPHOS) (Hua et al., 2020). It is also important to note that, TGF-β is highly expressed in OS cells and tissues. It promotes the progression of OS by activating PI3K/AKT pathway (Ma K. et al., 2020). The regulation of glycolysis by TGF-β has also been reported in OS. TGF-β weakens succinate dehydrogenase (SDH) expression by reducing the transcription factor STAT1, which leads to the accumulation of succinate and the increase of HIF-1α in OS cells, then changes the glycolysis state and drug resistance of OS cells (Xu et al., 2021). Moreover, TGF-β inhibitor RepSox inhibits OS proliferation and EMT and promotes apoptosis by inhibiting the JNK/Smad3 signal pathway (He et al., 2021).
2.3.6 Wnt pathway
The Wnt signaling pathway is vital in cancer and has classical and non-classical types. It is crucial in the occurrence and development of cancer (Zhan et al., 2017). The role of the Wnt pathway in regulating glycolysis has been widely studied. In 2014, Pate et al. established that blocking Wnt signal transduction in colon cancer cells significantly reduced glycolysis metabolism and tumor growth (Pate et al., 2014). The Wnt signaling pathway was widely studied in epithelial tumors as a carcinogenic pathway. The role of the Wnt signaling pathway in mesenchymal tumors such as OS mains undefined and controversial (Singla et al., 2020). The Wnt signal pathway is involved in regulating the glycolysis of OS. Furthermore, miR-21-5p is overexpressed in OS, promoting the expression of glycolysis genes (GLUT-1, LDHA, HK2, and PKM2) by promoting Wnt/β-catenin pathway activity, thereby promoting cell proliferation and inducing apoptosis in OS (Wu Z. et al., 2021). Another study proved that the non-classical Wnt signaling pathway might also be involved in regulating glycolysis in OS. In another research, miR-423-5p was expressed in low levels in OS tissues and cells, whereas Wnt5a/Ror2 and circPVT1 were highly expressed. The circPVT1 interacts with miR-423-5p and sponges miR-423-5p, then activates Wnt5a/Ror2 signal. Functionally, circPVT1 promotes the growth and metastasis of OS through the circPVT1/miR-423-5p/Wnt5a/Ror2OS axis (Wan et al., 2021).
In summary, multiple carcinogenic pathways regulate OS glycolysis. For decades, researchers have been looking for alternatives to chemotherapy for OS, eager to find drugs with fewer side effects and higher efficacy. In OS, a considerable number of targeted signal pathway inhibitors have been shown to slow down the progression of OS significantly. The inhibitors of signaling pathways involved in glycolysis regulation in OS are summarized in Table 2.
2.4 Glycolysis-associated transcription factors in osteosarcoma
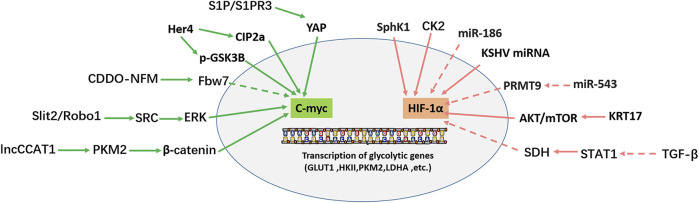
The glycolysis pathway is regulated by a series of transcription factors, such as HIF-1α, p53, and c-myc. c-myc is linked with the cancer progression as an oncogene in most cancers and increases aerobic glycolysis by activating related genes. Under the anoxic condition, glucose metabolism affects the expression of HIF-1α, which further induces the expression of aerobic glycolysis-related enzymes. Figure 3 shows that c-myc and HIF-1α regulate glycolysis in OS.
FIGURE 3.
Transcription factors (C-myc, HIF-1α) regulate the transcription of glycolytic genes. Solid arrows indicate facilitation, while dashed arrows indicate inhibition.
2.4.1 C-myc
The MYC family comprises three members: c-myc, N-myc, and L-myc, which are involved in various cellular activities: cell metabolism, differentiation, division, and cell death (Yoshida, 2018). In most tumors, MYC is a transcription factor; as an oncogene, MYC promotes tumor cell proliferation and metastasis. Dysfunctional MYC leads to abnormal transcription and affects cell fate by changing cell metabolism (Dang, 2016; Carroll et al., 2018; Beaulieu et al., 2020). C-myc is the most extensively studied and deregulated member of the MYC family in cancer (Duffy et al., 2021). C-myc positively regulates the glycolysis pathway via various mechanisms. As an important intracellular transcription factor, c-myc promotes glycolysis by transactivating and promoting the expression of genes related to glycolysis (Gruning et al., 2010; Han et al., 2012). Furthermore, as the target of several oncogenes or tumor suppressor genes, c-myc participates in regulating many signal pathways, such as MEK-ERK, Wnt/β-catenin, PI3K/AKT/mTOR, and STAT3/c-myc (Han et al., 2012; Li K. et al., 2021; Tang et al., 2022). In 2012, Han et al. (2012) found that c-myc overexpression in OS significantly enhanced the invasive ability of OS cells by promoting the MEK-ERK pathway. Moreover, c-myc is involved in cell proliferation, metastasis, apoptosis, cell cycle, and other cellular activities in OS (Wu et al., 2012; Gao J. et al., 2020; Fan et al., 2020).
C-myc is also essential for glycolysis regulation in OS. HER4 overexpression in OS enhances CIP2a-mediated c-myc S62 phosphorylation (c-myc stable) and reduces GSK3β-mediated c-myc T58 phosphorylation (c-myc unstable). Two pathways maintain c-myc stability by reducing their degradation of c-myc (Han et al., 2021). Therefore, HER4 promotes glycolysis and tumor growth in OS by upregulating c-myc positive regulation of glycolysis-related genes (GLUT1, HK2, KFPFB3, and PKM2) (Han et al., 2021). N-formyl morpholine substituent of CDDO (CDDO-NFM), a novel synthesized triterpenoid, can effectively suppress the proliferation of OS cells. CDDO-NFM promotes the degradation of c-myc by increasing the expression of Fbw7 protein, resulting in a significant decrease in the expression of glycolysis-related genes (GLUT1, PKM2, HK2, and LDHA) and blocking the glycolysis pathway (Gao et al., 2019). In 2019, Shen et al. (2019) found that S1P/S1PR3/YAP/c-myc/PGAM1 axis in OS enhances aerobic glycolysis. Sphingosine 1-phosphate receptor 3 (S1PR3) is a member of the G protein-coupled receptor (GPCRs) family and is critical in tumor imaging and targeted therapy. S1PR3 and its specific ligand S1P are highly expressed in OS. S1P/S1PR3 inhibits the phosphorylation of YAP and promotes the entry of YAP into the nucleus. YAP entering the nucleus forms a YAP/c-myc complex with c-myc. C-myc enhances glycolysis by promoting the transcription of the glycolytic enzyme PGAM1. TY52156 (S1PR3 antagonist) and methotrexate showed a synergistic effect on inhibiting OS cell growth in vitro and in vivo (Shen et al., 2019). Zhao et al. (2018) found that the high expression of slit guidance ligand-2 (SLIT2) and roundabout guidance receptor-1 (ROBO1) in OS is closely related to the decrease in overall survival rate in OS patients. SLIT2/ROBO1 axis enhances the expression of glycolysis-related genes by activating SRC/ERK/c-myc/PFKFB2 signal pathway. Regarding function, the SLIT2/ROBO1 axis promotes glycolysis and proliferation of OS cells, inhibits apoptosis, and promotes OS. The lncRNA colon cancer-associated transcript-1 (lncCCAT1) interacts with PKM2. PKM2 further promotes the phosphorylation of sterol regulatory element-binding protein 2 (SREBP2) and activates MYC transcriptional activity, thereby promoting glycolysis and adipogenesis. In addition, CDC25A dephosphorylates PKM2 and promotes the transactivation of PKM2, which in turn enhances the transcriptional expression of c-myc and upregulates the expression of glycolysis genes (PKM2, LDHA and GLUT1). The above mechanisms promote the Warburg effect, adipogenesis, and OS cell growth (Pu et al., 2022).
2.4.2 HIF-1α
Hypoxia-inducible factor-1α (HIF-1α) is a significant transcription factor that improves cell viability in a hypoxic environment, which is precisely regulated by hypoxia and hyperglycemia. HIF-1α and glucose interact with each other. HIF-1α induces the expression of glycolytic enzymes, while glucose metabolism affects the accumulation of HIF-1α in some cells (Xiao et al., 2013). In addition, HIF-1α is one of the critical transcription factors in tumor progression, and cancer targeted therapy. The effect of HIF-1α varies with or without oxygen. In an oxygen immersion environment, HIF-1α is completely inactivated and destroyed by the ubiquitin-proteasome pathway (UPP). However, in an anaerobic environment, it escapes damage, enters the nucleus, and then upregulates many genes involved in cancer progression (Rashid et al., 2021). HIF-1α activates the transcription of numerous genes related to tumor biology, including apoptosis, cell proliferation, angiogenesis, and glucose metabolism. Hypoxia and genetic changes in tumors can lead to HIF-1α overexpression. Compared with normal tissues, HIF-1α is highly expressed in different kinds of tumors, such as colon cancer, gastric cancer, lung cancer, skin cancer, pancreatic cancer, and breast cancer, including human OS, which is significantly associated with metastasis (Zhong et al., 1999). HIF-1α is strongly expressed in OS tissues compared with para cancerous tissues, and is associated with poor prognosis in patients with OS (Yang et al., 2007). In 2015, Guan et al. suggested that hypoxia promotes the metastasis of OS cells by activating HIF-1α/CXCR4 pathway (Guan et al., 2015). Furthermore, hypoxia and increased HIF-1α protein expression regulate the growth and metastasis of OS cells by upregulating VEGF expression (Liu M. et al., 2016). Simultaneously, HIF-1α overexpression upregulated the level of NDUFA4L2, thus promoting the metastasis and epithelial-mesenchymal transformation of OS cells by inhibiting ROS production (Xu et al., 2020). Yan et al. (2020) reported that KRT17 is highly expressed in osteosarcoma tissues and osteosarcoma cell lines, and promotes the proliferation and glycolysis of osteosarcoma cells by activating the AKT/mTOR/HIF1α pathway. TGF-β negative transcription factor STAT1 further inhibited the expression of succinate dehydrogenase (SDH). The decrease of SDH leads to HIF-1α upregulation, which changes the metabolic level of glucose and the drug resistance of OS cells (Xu et al., 2021). The miR-186 inhibits the proliferation, invasion, and glycolysis of OS cells by targeting HIF-1α and pituitary tumor transforming gene 1 (PTTG1) (Xiao et al., 2018). Interestingly, there is a negative correlation between the expression of miR-543 and protein arginine methyltransferase 9 (PRMT9) in OS samples and cell lines. There is a positive regulation of glycolysis by the miR-543/PRMT9/HIF-1α axis, thus promoting OS progression (Zhang H. et al., 2017a). Additionally, in the OS U2OS cell line, a Ser/Thr protein kinase CK2, which is highly expressed in many cancers, enhances glycolysis by upregulating HIF-1α expression (Zonta et al., 2021). Another study found that sphingosine kinase 1 (SphK1) increases the glycolysis level of OS cells by over-regulating HIF-1α expression and participates in the progression of doxorubicin resistance in OS cells (Ren and Su, 2020). Furthermore, Kaposi’s sarcoma herpesvirus (KSHV) miRNA promotes HIF-1α expression in OS to enhance glycolysis (Yogev et al., 2014).
2.5 Glycolysis-associated non-coding RNAs in osteosarcoma
Transcriptional regulation in mammalian cells is a complex regulatory process vital for cell life activities, such as differentiation, development, and metabolism (Carninci et al., 2005; Gustincich et al., 2006). There are numerous types of non-coding RNAs (ncRNAs), such as miRNAs, piRNAs, snoRNAs, lncRNAs and circRNA (Esteller, 2011). Some of these ncRNAs, such as miRNA, lncRNA and circRNA, have been researched extensively. Some scholars believe these ncRNAs are epigenetic regulators that affect various biological processes, including bone metabolism (Yang et al., 2020). This study reviewed the effects of three kinds of miRNA (Table 3), lncRNA (Table 4), and circRNA (Table 5) on glucose metabolism in OS cells. The ncRNAs can directly or indirectly regulate the expression of glycolysis-related enzymes to affect cell glycolysis and then affect OS proliferation, invasion, migration, and other malignant behavior.
TABLE 3.
miRNAs regulate glycolysis enzymes in osteosarcoma.
| miRNAs | Target glycolysis enzymes | Involvement of factors | Function | References |
|---|---|---|---|---|
| miR-34c-5p | ALDOA | miR-34c-5p/ALDOA | Down-regulate | Shen et al. (2020) |
| miR-132 | Glut1 | miR-132/Glut1 | Down-regulate | Rafat et al. (2021) |
| miR-150 | GLUT1 | miR-150/GLUT1 | Down-regulate | Yuan et al. (2017) |
| miR-522-3p | GLUT1 | miR-522-3p/GLUT1 | Up-regulate | Rong Chen et al. (2019) |
| miR-328-3p | GLUT1 | miR-328-3p/LAT1/SLC7A5/GLUT1 | Down-regulate | Yi et al. (2020) |
| miR-485-3p | GLUT1, HK2, PKM2 | c-MET, AKT3/mTOR/GLUT1 | Down-regulate | Qing Wang et al. (2021) |
| miR-21-5p | GLUT1, LDHA, HK2, PKM2 | miR-21-5p/Wnt/β-catenin/GLUT | Up-regulate | Ziyan Wu et al. (2021) |
| miR-186 | HIF-1α | miR-186/HIF-1 | Down-regulate | Xiao et al. (2018) |
| miR-543 | HIF-1α | miR-543/PRMT9/HIF-1α | Up-regulate | Heng Zhang et al. (2017a) |
| miR-1225-3p | HK2 | miR-1225-3p/KCNH1 | Down-regulate | Zhengmao Li et al. (2021) |
| miR-125b | HK2 | miR-125b/HK2 | Down-regulate | Wu et al. (2017) |
| miR-134 | HK2 | ELK1/miR-134/PTBP1 | Down-regulate | Zhang et al. (2021b) |
| miR-143 | HK2 | lncRNA-SARCC/miR-143/HK2 | Down-regulate | Wen et al. (2020) |
| miR-144 | HK2 | miR-144/CXCR4 | Down-regulate | Jun Liu et al. (2020b) |
| miR-181b | HK2 | miR-181b/HK2 | Down-regulate | Gu et al. (2020) |
| miR-335-5p | HK2, PKM2 | miR-335-5p/Myh9/HK2, PKM2 | Down-regulate | Cao et al. (2021) |
| miR-372-3p | HK2 | miR-372-3p/MAPK7/HK2 | Down-regulate | Yuanpeng Gao et al. (2020) |
| miR-423-5p | HK2, PKM2, GLUT1, LDHA | miR-423-5p-Wnt5a/Ror2/HK2 | Down-regulate | Wan et al. (2021) |
| miR-615 | HK2 | PI3K/AKT/HK2 | Down-regulate | Limin Sun et al. (2020) |
| miR1244 | HK2, LDHA | miR1244/TRIM44 | Down-regulate | Huo and Dou, (2021) |
| miR-497 | HK2 | miR-497/HK2 | Down-regulate | Song et al. (2017) |
| miR-185 | HK2 | miR-185/HK2 | Down-regulate | Chaojian Liu et al. (2019) |
| miR-323a-3p | LDHA | miR-323a-3p/LDHA | Down-regulate | Hanwen Chen et al. (2018) |
| miR-329-3p | LDHA | miR-329-3p/LDHA | Down-regulate | Gang Li et al. (2021) |
| miR-33b | LDHA | miR-33b/LDHA | Down-regulate | Zheng et al. (2018) |
| miR-409-3p | LDHA | miR-409-3p/LDHA | Down-regulate | Zhang et al. (2021a) |
| miR-15b-5p | PDK4 | miR-15b-5p/PDK4 | Down-regulate | Weng et al. (2018) |
| miR-1297 | PFKFB2 | miR-1297/PFKFB2 | Down-regulate | Pan et al. (2020) |
| miR-26b | PFKFB3 | miR-26b, PFKFB3 | Down-regulate | Zheng et al. (2015) |
| miR-26b | PFKFB3, LDHA, GLUT-1 | miR-26b/PFKFB3 | Down-regulate | Du et al. (2015) |
| miR-542-3p | PFKM | miR-542-3p/PFKM | Down-regulate | Jia et al. (2021) |
| miR-23b-3p | PGC1α | miR-23b-3p/PGC1α | Down-regulate | Zhu et al. (2019) |
| miR-365a-3p | PGK1 | miR-365a-3p/PGK1 | Down-regulate | Pan et al. (2022) |
| miR-1294 | PKM2 | miR-1294/PKM2 | Down-regulate | Yuan et al. (2020) |
| miR-491-5p | PKM2 | miR-491-5p/PKM2 | Down-regulate | Ting Chen et al. (2018) |
| miR-198 | PFKFB4 | miR-198/E2F2 | Down-regulate | Liu et al. (2022) |
| miR-578 | LDHA, PDK1 | miR-578-LDHA/PDK1 | Down-regulate | Hu et al. (2021) |
TABLE 4.
lncRNAs regulate glycolysis enzymes in osteosarcoma.
| lncRNA | Target glycolysis enzymes | Involvement of factors | Function | References |
|---|---|---|---|---|
| lncRNA HAND2-AS1 | GLUT1 | lncRNA HAND2-AS1/GLUT1 | Down-regulate | Shunguang Chen et al. (2019) |
| lncRNA PVT1 | HK2 | lncRNA PVT1/miR-497/HK2 | Up-regulate | Song et al. (2017) |
| lncRNA FEZF1-AS1 | HK2 | lncRNA FEZF1-AS1/miR-144/CXCR4 | Up-regulate | Jun Liu et al. (2020b) |
| lncRNA SARCC | HK2 | lncRNA SARCC/miR-143/HK2 | Down-regulate | Wen et al. (2020) |
| lncRNA TUG1 | HK2 | lncRNA TUG1/HK2 | Up-regulate | Han et al. (2018) |
| lncRNA CCAT1 | PKM2 | lncRNA CCAT1/PKM2/SREBP2 | Up-regulate | Pu et al. (2022) |
| lncRNA HCG18 | PGK1 | lncRNA HCG18/miR-365a-3p/PGK1 | Up-regulate | Pu et al. (2022) |
| lncRNA XLOC_005950 | PFKM | lncRNA XLOC_005950/miR-542-3p/PFKM | Up-regulate | Jia et al. (2021) |
| lncRNA KCNQ1OT1 | ALDOA | lncRNA KCNQ1OT1/miR-34c-5p/ALDOA | Up-regulate | Shen et al. (2020) |
TABLE 5.
cicrRNAs regulate glycolysis enzymes in osteosarcoma.
| circRNA | Target glycolysis enzymes | Involvement of factors | Function | References |
|---|---|---|---|---|
| CircRNA PVT1 | HK2, PKM2, GLUT1, LDHA | CircRNA PVT1/miR-423-5p/Wnt5a/Ror2/HK2 | Up-regulate | Wan et al. (2021) |
| CircRNA FAT1(e2) | HK2 | CircRNA FAT1(e2)/miR-181b/HK2 | Up-regulate | Gu et al. (2020) |
| CircRNA 0056285 | HK2, LDHA | CircRNA 0056285/miR-1244/TRIM44/HK2 | Up-regulate | Huo and Dou, (2021) |
| CircRNA 0016347 | HK2 | CircRNA 0016347/miR-1225-3p/KCNH1/HK2 | Up-regulate | Zhengmao Li et al. (2021) |
| CircRNA Circ_0001721 | HK2 | CircRNA Circ_0001721/miR-372-3p/MAPK7/HK2 | Up-regulate | Yuanpeng Gao et al. (2020) |
| CircRNA CNST | LDHA, PDK1 | CircRNA CNSTmiR-578/LDHA, PDK1 | Up-regulate | Hu et al. (2021) |
| CircRNA ATRNL1 | LDHA | CircRNA ATRNL1/miR-409-3p/LDHA | Up-regulate | Zhang et al. (2021a) |
| CircRNA 1244 | HK2, LDHA | CircRNA 1244/miR1244/HK2, LDHA | Up-regulate | Huo and Dou, (2021) |
| CircRNA TP2A2 | HK2, PKM2 | CircRNA TP2A2/miR-335-5p/Myh9/HK2, PKM2 | Up-regulate | Cao et al. (2021) |
2.5.1 miRNAs
miRNA is a highly conserved endogenous non-protein-coding RNA about 19–25 nucleotides in length, whose transcription process is independent of other genes. miRNAs degrade or induce translational silencing by binding to the 3′ untranslated regions (3′-UTRs) of target mRNAs, affecting cell proliferation, differentiation, apoptosis, and ontogeny (Jackson and Standart, 2007). As revealed in Table 3, most miRNAs inhibit the glycolysis and progression of OS cells. Chen et al. reported that miR-522-3p and GLUT1 are highly expressed and positively correlated with OS and that high levels of miR-522-3p are associated with low survival rates. Regarding the mechanism, miR-522-3p increases GLUT1 expression and glucose uptake, thus promoting the proliferation of OS cells (Chen R. et al., 2019). In addition, miR-543 is highly expressed in OS tissues and cells and positively regulates glycolysis through miR-543/PRMT9/HIF-1α axis to promote OS progression (Zhang H. et al., 2017a). miR-328-3p regulates GLUT1-mediated glucose uptake and metabolism in OS cells, and in addition, hBERA/miR-328 combined treatment with cisplatin or doxorubicin demonstrated strong synergy in inhibiting OS cell proliferation (Yi et al., 2020). Also, miRNAs can regulate glycolysis in OS cells by acting on signaling pathways. This study demonstrated that miR-485-3p was decreased and c-MET, AKT3, and MALAT1 were increased in human OS tissues and cells. miR-485-3p directly binds to c-MET and AKT3 mRNA and inhibits OS cell glycolysis, proliferation, migration, and invasion by reducing glycolysis-related proteins and migration-related proteins by inhibiting c-MET and AKT3/mTOR pathways (Wang Q. et al., 2021). Whereas miR-21-5p is overexpressed in OS. miR-21-5p upregulates Warburg effector proteins (GLUT1, LDHA, HK2, and PKM2) by activating Wnt/β-catenin signaling, promoting OS cell proliferation and metastasis (Wu Z. et al., 2021).
2.5.2 lncRNAs
LncRNAs are non-coding RNA with a length of more than 200 nucleotides, but due to the lack of open reading frames, they cannot encode proteins (Prensner and Chinnaiyan, 2011). However, recent studies have stated that lncRNAs not only directly regulate gene expression but also interact with miRNAs as competing endogenous RNAs (Zheng et al., 2019). As illustrated in Table 4, most lncRNAs positively regulate glycolysis, while a few lncRNAs inhibit the glycolysis pathway of OS cells. For example, lncRNAHAND2-AS1 inhibits glycolysis through negative regulation of GLUT1 (Chen S. et al., 2019). And lncRNA SARCC inhibits glycolysis through negative regulation of HK2 (Wen et al., 2020). In addition, lncRNA SNHG4 is highly expressed in OS tissues and cell lines, miR-377-3p is downregulated in OS, and SNHG4 acts as a competing endogenous RNA with sponge miR-377-3p to promote OS tumor proliferation and migration (Huang Y. F. et al., 2020). LncRNA FTX expression has also upregulated in OS, and lncRNA FTX inhibition suppressed the proliferation and migration of OS cells by regulating miR-320a/TXNRD1 (Huang S. et al., 2020). Similarly, lncRNA ROR1-AS1 is upregulated in OS tissues, ROR1-AS1 acts as a sponge for miR-504, and ROR1-AS1 overexpression suppressed miR-504 expression in miR-63 cells. LncRNA ROR1-AS1 accelerates OS invasion and proliferation by regulating miR-504 (Wu et al., 2020). LncRNAs can also regulate the proliferation or apoptosis of OS cells through signaling pathways. For instance, lncRNA MEG3 overexpression in OS cells can inhibit their proliferation and promote their apoptosis by inhibiting the Notch signaling pathway (Chen et al., 2020). Furthermore, lncRNA GAS5 is downregulated in human OS tissues and cell lines, and GAS5 acts as a competing endogenous RNA regulating the PI3K/AKT pathway by sponging miR-23a-3p, promoting PTEN expression and inhibiting PTEN expression in OS cell growth and invasion (Liu J. et al., 2020a).
2.5.3 Circular RNAs
Circular RNAs (circRNAs) are endogenous non-coding RNAs widely present in eukaryotic cells. Recently, circRNAs have been demonstrated to act as competing endogenous RNAs, namely miRNA sponges, to regulate the expression of target genes, as well as act as transcriptional regulators or RNA-binding proteins to indirectly regulate genes at the post-transcriptional level (Jeck et al., 2013; Du et al., 2016). CircRNAs have also been reported to promote OS glycolysis, as illustrated in Table 5. For example, circTADA2A is highly expressed in OS tissues and cell lines, and circTADA2A can easily sponge miR-203a-3p to upregulate CREB3 expression, which was identified as a driver gene in OS that promotes progression and metastasis (Wu et al., 2019) circCAMSAP1, also known as hsa_circ_0004338, is significantly upregulated in human OS tissues and cell lines and promotes OS progression and metastasis by sponging miR-145-5p and regulating FLI1 expression (Chen Z. et al., 2021). In addition, OS circ_001422 expression was significantly increased in cell lines and tissues, and the competitive interaction between circ_001422 and miR-195-5p increased FGF2 expression and initiated PI3K/Akt signaling, i.e., circ_001422 via regulation of miR-195-5p/FGF2/PI3K/Akt axis accelerates OS tumorigenesis and metastasis (Yang et al., 2021). Similarly, circRNA_103801 can accelerate the proliferation of OS cells by sponging miR-338-3p and regulating HIF-1/Rap1/PI3K-Akt pathway (Li Z. Q. et al., 2021).
3 Conclusion and future perspectives
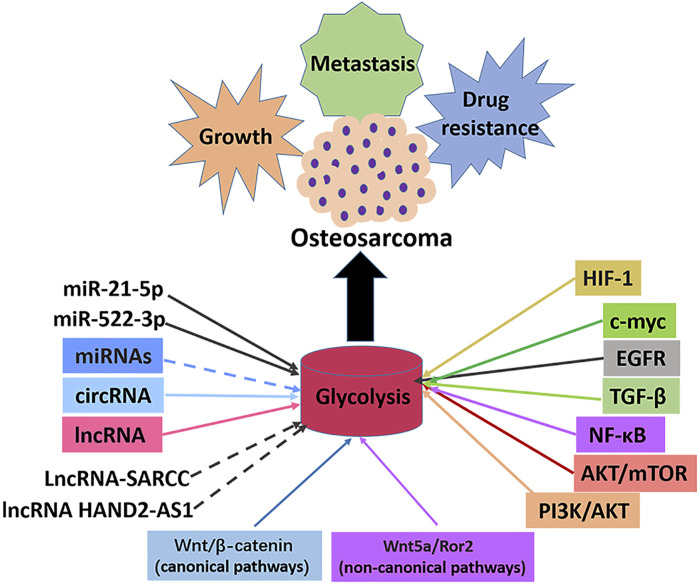
The abnormal activity of glycolysis is a sign of metabolic reprogramming of cancer cells and is very important for developing tumors. Similarly, hyperactive glycolysis in OS, a highly invasive tumor, maintains and promotes various malignant behaviors of OS cells. A series of enzymes mainly catalyzes the glycolysis pathway to promote the transformation of energy substances. In cancer cells, oncogenes and tumor suppressor genes change the level of glycolysis by directly or indirectly regulating the expression and enzyme activity of glycolysis-related genes. Numerous studies have shown that the classical signaling and glycolysis pathways in OS overlap and influence each other. This study reviewed several key enzymes involved in glycolysis, classical and non-classical signal pathways involved in glycolysis, and the role of intracellular transcription factors in the progression and treatment of OS (Figure 4). As a result of the complexity of the glycolysis pathway in OS, no single inhibitor of glycolysis can be effective in treating OS. It is imperative to find more key genes in the glycolysis pathway and OS occurrence and development to help find more effective metabolic drug targets. A summary of some inhibitors involved in the glycolysis pathway of OS is presented in this article. There is no reliable evidence that these inhibitors inhibit cellular glycolysis. However, the combined use of signal pathways targeting small molecules and glycolysis targeted inhibitors provides new insights for treating OS. Since most current research results were from OS cell experiments, the real prospect of clinical application remains unknown, so developing glycolysis therapy for OS is a challenge. In conclusion, the co-development of the glycolysis pathway, conducting basic research on OS progression, and providing corresponding clinical treatment will benefit OS patients.
FIGURE 4.
Signal pathways, transcription factors and ncRNAs that regulate glycolysis in osteosarcoma. The enhancement of glycolysis in osteosarcoma promotes the malignant behaviors such as proliferation, metastasis and drug resistance of osteosarcoma cells. Most miRNAs inhibit the progression of osteosarcoma by inhibiting glycolysis, with the exception of miR-21-5p and miR-522-3p. Most lncRNAs promotes the progression of osteosarcoma by promoting glycolysis, with the exception of lncRNA-SARCC and lncRNAHAND2-AS1. All circRNAs promote osteosarcoma progression by promoting glycolysis. Transcription factors c-myc and HIF-1α promote glycolysis and osteosarcoma progression. Signal pathways PI3K/AKT, AKT/mTOR, NF-κB, TGF-β, EGFR, Wnt/β-catenin, Wnt5a/Ror2 promote glycolysis pathway and osteosarcoma progression.
Acknowledgments
The authors acknowledge the use of Science Slides that is used to create Figures.
Author contributions
ZF and YO designed the research study and performed the research. YO provided help and advice on the production of pictures. ZF wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by grants from National Natural Science Foundation of China (82060492), Natural Science Foundation of Jiangxi province (S2021QNZD2L0110).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adamski J., Price A., Dive C., Makin G. (2013). Hypoxia-induced cytotoxic drug resistance in osteosarcoma is independent of HIF-1Alpha. PLoS One 8, e65304. 10.1371/journal.pone.0065304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichem A., Groettrup M. (2016). The ubiquitin-like modifier FAT10 in cancer development. Int. J. Biochem. Cell. Biol. 79, 451–461. 10.1016/j.biocel.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Augoff K., Grabowski K. (2004). Significance of lactate dehydrogenase measurements in diagnosis of malignancies. Pol. Merkur. Lek. 17, 644–647. [PubMed] [Google Scholar]
- Avnet S., Longhi A., Salerno M., Halleen J. M., Perut F., Granchi D., et al. (2008). Increased osteoclast activity is associated with aggressiveness of osteosarcoma. Int. J. Oncol. 33, 1231–1238. [PubMed] [Google Scholar]
- Bagatell R., Norris R., Ingle A. M., Ahern C., Voss S., Fox E., et al. (2014). Phase 1 trial of temsirolimus in combination with irinotecan and temozolomide in children, adolescents and young adults with relapsed or refractory solid tumors: A children's oncology group study. Pediatr. Blood Cancer 61, 833–839. 10.1002/pbc.24874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron C. C., Bilan P. J., Tsakiridis T., Tsiani E. (2016). Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism. 65, 124–139. 10.1016/j.metabol.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Bartrons R., Simon-Molas H., Rodriguez-Garcia A., Castano E., Navarro-Sabate A., Manzano A., et al. (2018). Fructose 2, 6-bisphosphate in cancer cell metabolism. Front. Oncol. 8, 331. 10.3389/fonc.2018.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavelloni A., Focaccia E., Piazzi M., Orsini A., Ramazzotti G., Cocco L., et al. (2019). Therapeutic potential of nvp-bkm120 in human osteosarcomas cells. J. Cell. Physiol. 234, 10907–10917. 10.1002/jcp.27911 [DOI] [PubMed] [Google Scholar]
- Beaulieu M. E., Castillo F., Soucek L. (2020)., 9. Cells, E1038. 10.3390/cells9041038 Structural and biophysical insights into the function of the intrinsically disordered myc oncoprotein Cells [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner K., Johannesen T. B., Berner A., Haugland H. K., Bjerkehagen B., Bohler P. J., et al. (2015). Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 54, 25–33. 10.3109/0284186X.2014.923934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X., Jiang H., Meng Y., Li Y. P., Fang J., Lu Z. (2022). Regulation of gene expression by glycolytic and gluconeogenic enzymes. Trends Cell. Biol. S0962-8924 (22), 00036–00038. 10.1016/j.tcb.2022.02.003 [DOI] [PubMed] [Google Scholar]
- Brand A., Singer K., Koehl G. E., Kolitzus M., Schoenhammer G., Thiel A., et al. (2016). LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell. Metab. 24, 657–671. 10.1016/j.cmet.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Cao W., Wang Z., Han X., Liu J., Wang W. (2019). In vitro cytotoxicity screening to identify novel anti-osteosarcoma therapeutics targeting pyruvate dehydrogenase kinase 2. Bioorg. Med. Chem. Lett. 29, 126665. 10.1016/j.bmcl.2019.126665 [DOI] [PubMed] [Google Scholar]
- Cao X., Meng X., Fu P., Wu L., Yang Z., Chen H. (2021). circATP2A2 promotes osteosarcoma progression by upregulating MYH9. Open Med. 16, 1749–1761. 10.1515/med-2021-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N. (2018). The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425. 10.1007/s11684-018-0650-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Gamero A. M., Borges K. S., Da Silva Silveira V., Lira R. C., de Paula Gomes Queiroz R., Valera F. C., et al. (2012). Inhibition of nuclear factor-κB by dehydroxymethylepoxyquinomicin induces schedule-dependent chemosensitivity to anticancer drugs and enhances chemoinduced apoptosis in osteosarcoma cells. Anticancer. Drugs 23, 638–650. 10.1097/CAD.0b013e328350e835 [DOI] [PubMed] [Google Scholar]
- Chau N. M., Rogers P., Aherne W., Carroll V., Collins I., Mcdonald E., et al. (2005). Identification of novel small molecule inhibitors of hypoxia-inducible factor-1 that differentially block hypoxia-inducible factor-1 activity and hypoxia-inducible factor-1alpha induction in response to hypoxic stress and growth factors. Cancer Res. 65, 4918–4928. 10.1158/0008-5472.CAN-04-4453 [DOI] [PubMed] [Google Scholar]
- Chawla S. P., Staddon A. P., Baker L. H., Schuetze S. M., Tolcher A. W., D'Amato G. Z., et al. (2012). Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J. Clin. Oncol. 30, 78–84. 10.1200/JCO.2011.35.6329 [DOI] [PubMed] [Google Scholar]
- Chen H., Gao S., Cheng C. (2018). MiR-323a-3p suppressed the glycolysis of osteosarcoma via targeting LDHA. Hum. Cell. 31, 300–309. 10.1007/s13577-018-0215-0 [DOI] [PubMed] [Google Scholar]
- Chen L., Wang J., Li J. W., Zhao X. W., Tian L. F. (2020). LncRNA MEG3 inhibits proliferation and promotes apoptosis of osteosarcoma cells through regulating Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 581–590. 10.26355/eurrev_202001_20034 [DOI] [PubMed] [Google Scholar]
- Chen R., Lin J., Yan W., Chen D. (2019). miR-522-3p promotes osteosarcoma cell growth by regulating glucose uptake and GLUT1 expression. Onco. Targets. Ther. 12, 9053–9058. 10.2147/OTT.S217324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Xu X., Lu S., Hu B. (2019). Long non-coding RNA HAND2-AS1 targets glucose metabolism and inhibits cancer cell proliferation in osteosarcoma. Oncol. Lett. 18, 1323–1329. 10.3892/ol.2019.10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Li Y., Cao W., Liu Y. (2018). miR-491-5p inhibits osteosarcoma cell proliferation by targeting PKM2. Oncol. Lett. 16, 6472–6478. 10.3892/ol.2018.9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Bahrami A., Pappo A., Easton J., Dalton J., Hedlund E., et al. (2014). Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell. Rep. 7, 104–112. 10.1016/j.celrep.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang Y., Zhang S., Wang A., Du Q., Wang Z. (2021). Oleanolic acid inhibits osteosarcoma cell proliferation and invasion by suppressing the SOX9/Wnt1 signaling pathway. Exp. Ther. Med. 21, 443. 10.3892/etm.2021.9883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J., Li J., Lu N., Shen X. Z. (2017). Interferon regulatory factors: A key to tumour immunity. Int. Immunopharmacol. 49, 1–5. 10.1016/j.intimp.2017.05.010 [DOI] [PubMed] [Google Scholar]
- Chen Z., Xu W., Zhang D., Chu J., Shen S., Ma Y., et al. (2021). circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression. Mol. Ther. Nucleic Acids 23, 1120–1135. 10.1016/j.omtn.2020.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney J., Clark J., Klarer A. C., Imbert-Fernandez Y., Lane A. N., Telang S. (2014). Fructose-2, 6-bisphosphate synthesis by 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 5, 6670–6686. 10.18632/oncotarget.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. J., Eun J. S., Kim D. K., Li R. H., Shin T. Y., Park H., et al. (2008). Icariside II from Epimedium koreanum inhibits hypoxia-inducible factor-1alpha in human osteosarcoma cells. Eur. J. Pharmacol. 579, 58–65. 10.1016/j.ejphar.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Cifuentes M., Garcia M. A., Arrabal P. M., Martinez F., Yanez M. J., Jara N., et al. (2011). Insulin regulates GLUT1-mediated glucose transport in MG-63 human osteosarcoma cells. J. Cell. Physiol. 226, 1425–1432. 10.1002/jcp.22668 [DOI] [PubMed] [Google Scholar]
- Corre I., Verrecchia F., Crenn V., Redini F., Trichet V. (2020). The osteosarcoma microenvironment: A complex but targetable ecosystem. Cells 9, E976. 10.3390/cells9040976 e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Lv W., Qu Y., Ma R., Wang Y. W., Xu Y. J., et al. (2016). Discovery of 2-((3-cyanopyridin-2-yl)thio)acetamides as human lactate dehydrogenase A inhibitors to reduce the growth of MG-63 osteosarcoma cells: Virtual screening and biological validation. Bioorg. Med. Chem. Lett. 26, 3984–3987. 10.1016/j.bmcl.2016.06.083 [DOI] [PubMed] [Google Scholar]
- Dai B., Yan T., Zhang A. (2017). ROR2 receptor promotes the migration of osteosarcoma cells in response to Wnt5a. Cancer Cell. Int. 17, 112. 10.1186/s12935-017-0482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V. (2016). A time for MYC: Metabolism and therapy. Cold Spring Harb. Symp. Quant. Biol. 81, 79–83. 10.1101/sqb.2016.81.031153 [DOI] [PubMed] [Google Scholar]
- Dang C. V., Le A., Gao P. (2009). MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479–6483. 10.1158/1078-0432.CCR-09-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. E., Bolejack V., Ryan C. W., Ganjoo K. N., Loggers E. T., Chawla S., et al. (2019). Randomized double-blind phase II study of regorafenib in patients with metastatic osteosarcoma. J. Clin. Oncol. 37, 1424–1431. 10.1200/JCO.18.02374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D. M., Goodfriend T. L., Kaplan N. O. (1964). Lactic dehydrogenases: Functions of the two types rates of synthesis of the two major forms can Be correlated with metabolic differentiation. Science 143, 929–933. 10.1126/science.143.3609.929 [DOI] [PubMed] [Google Scholar]
- Dayton T. L., Jacks T., Vander Heiden M. G. (2016). PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730. 10.15252/embr.201643300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa G. R., Vieira G. M., Das Chagas P. F., Pezuk J. A., Brassesco M. S. (2020). Should we keep rocking? Portraits from targeting Rho kinases in cancer. Pharmacol. Res. 160, 105093. 10.1016/j.phrs.2020.105093 [DOI] [PubMed] [Google Scholar]
- Deng B., Deng J., Yi X., Zou Y., Li C. (2021). ROCK2 promotes osteosarcoma growth and glycolysis by up-regulating HKII via phospho-PI3K/AKT signalling. Cancer Manag. Res. 13, 449–462. 10.2147/CMAR.S279496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D., Yan N. (2016). GLUT, SGLT, and SWEET: Structural and mechanistic investigations of the glucose transporters. Protein Sci. 25, 546–558. 10.1002/pro.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Deng J., Yi X., Zou Y., Liu H., Li C., et al. (2020). Ubiquitin-like protein FAT10 promotes osteosarcoma glycolysis and growth by upregulating PFKFB3 via stabilization of EGFR. Am. J. Cancer Res. 10, 2066–2082. [PMC free article] [PubMed] [Google Scholar]
- Deng X., Yi X., Deng J., Zou Y., Wang S., Shan W., et al. (2019). ROCK2 promotes osteosarcoma growth and metastasis by modifying PFKFB3 ubiquitination and degradation. Exp. Cell. Res. 385, 111689. 10.1016/j.yexcr.2019.111689 [DOI] [PubMed] [Google Scholar]
- Ding J., Karp J. E., Emadi A. (2017). Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: Interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. 19, 353–363. 10.3233/CBM-160336 [DOI] [PubMed] [Google Scholar]
- Doherty J. R., Cleveland J. L. (2013). Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692. 10.1172/JCI69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J. Y., Wang L. F., Wang Q., Yu L. D. (2015). miR-26b inhibits proliferation, migration, invasion and apoptosis induction via the downregulation of 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3 driven glycolysis in osteosarcoma cells. Oncol. Rep. 33, 1890–1898. 10.3892/or.2015.3797 [DOI] [PubMed] [Google Scholar]
- Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zhang J., Yan S., Tao Z., Wang C., Huang M., et al. (2019). Oridonin inhibits the proliferation, migration and invasion of human osteosarcoma cells via suppression of matrix metalloproteinase expression and STAT3 signalling pathway. J. BUON 24, 1175–1180. [PubMed] [Google Scholar]
- Duffy M. J., O'Grady S., Tang M., Crown J. (2021). MYC as a target for cancer treatment. Cancer Treat. Rev. 94, 102154. 10.1016/j.ctrv.2021.102154 [DOI] [PubMed] [Google Scholar]
- Engelman J. A. (2009). Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat. Rev. Cancer 9, 550–562. 10.1038/nrc2664 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Fan J., Mei J., Zhang M. Z., Yuan F., Li S. Z., Yu G. R., et al. (2017). Clinicopathological significance of glucose transporter protein-1 overexpression in human osteosarcoma. Oncol. Lett. 14, 2439–2445. 10.3892/ol.2017.6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhou J. Q., Yu G. R., Lu D. D. (2010). Glucose transporter protein 1-targeted RNA interference inhibits growth and invasion of the osteosarcoma cell line MG63 in vitro . Cancer biother. Radiopharm. 25, 521–527. 10.1089/cbr.2010.0784 [DOI] [PubMed] [Google Scholar]
- Fan Y., Jia X., Xie T., Zhu L., He F. (2020). Radiosensitizing effects of cmyc gene knockdowninduced G2/M phase arrest by intrinsic stimuli via the mitochondrial signaling pathway. Oncol. Rep. 44, 2669–2677. 10.3892/or.2020.7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang A., Zhang Q., Fan H., Zhou Y., Yao Y., Zhang Y., et al. (2017). Discovery of human lactate dehydrogenase A (LDHA) inhibitors as anticancer agents to inhibit the proliferation of MG-63 osteosarcoma cells. Medchemcomm 8, 1720–1726. 10.1039/c7md00222j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Zuo Q., Jiang T., Song H., Zhou J. (2019). A newly synthesized oleanolic acid derivative inhibits the growth of osteosarcoma cells in vitro and in vivo by decreasing c-MYC-dependent glycolysis. J. Cell. Biochem. 120, 9264–9276. 10.1002/jcb.28202 [DOI] [PubMed] [Google Scholar]
- Gao J., Ma S., Yang F., Chen X., Wang W., Zhang J., et al. (2020). miR193b exhibits mutual interaction with MYC, and suppresses growth and metastasis of osteosarcoma. Oncol. Rep. 44, 139–155. 10.3892/or.2020.7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Tu D. N., Li H., Jiang J. X., Cao X., You J. B., et al. (2016). Pharmacological or genetic inhibition of LDHA reverses tumor progression of pediatric osteosarcoma. Biomed. Pharmacother. 81, 388–393. 10.1016/j.biopha.2016.04.029 [DOI] [PubMed] [Google Scholar]
- Gao Y., Ma H., Gao Y., Tao K., Fu L., Ren R., et al. (2020). CircRNA Circ_0001721 promotes the progression of osteosarcoma through miR-372-3p/MAPK7 Axis. Cancer Manag. Res. 12, 8287–8302. 10.2147/CMAR.S244527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y. D., Yang L., Zhang C., Kong L. Y. (2014). Blockade of epidermal growth factor receptor/mammalian target of rapamycin pathway by Icariside II results in reduced cell proliferation of osteosarcoma cells. Food Chem. Toxicol. 73, 7–16. 10.1016/j.fct.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Gobin B., Huin M. B., Lamoureux F., Ory B., Charrier C., Lanel R., et al. (2015). BYL719, a new alpha-specific PI3K inhibitor: Single administration and in combination with conventional chemotherapy for the treatment of osteosarcoma. Int. J. Cancer 136, 784–796. 10.1002/ijc.29040 [DOI] [PubMed] [Google Scholar]
- Gong L., Cui Z., Chen P., Han H., Peng J., Leng X. (2012). Reduced survival of patients with hepatocellular carcinoma expressing hexokinase II. Med. Oncol. 29, 909–914. 10.1007/s12032-011-9841-z [DOI] [PubMed] [Google Scholar]
- Gruning N. M., Lehrach H., Ralser M. (2010). Regulatory crosstalk of the metabolic network. Trends biochem. Sci. 35, 220–227. 10.1016/j.tibs.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Gu H., Cheng X., Xu J., Zhou K., Bian C., Chen G., et al. (2020). Circular RNA circFAT1(e2) Promotes Osteosarcoma Progression and Metastasis by Sponging miR-181b and Regulating HK2 Expression. Biomed. Res. Int. 2020, 3589871. 10.1155/2020/3589871 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guan G., Zhang Y., Lu Y., Liu L., Shi D., Wen Y., et al. (2015). The HIF-1α/CXCR4 pathway supports hypoxia-induced metastasis of human osteosarcoma cells. Cancer Lett. 357, 254–264. 10.1016/j.canlet.2014.11.034 [DOI] [PubMed] [Google Scholar]
- Gui D. Y., Sullivan L. B., Luengo A., Hosios A. M., Bush L. N., Gitego N., et al. (2016). Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity to metformin. Cell. Metab. 24, 716–727. 10.1016/j.cmet.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustincich S., Sandelin A., Plessy C., Katayama S., Simone R., Lazarevic D., et al. (2006). The complexity of the mammalian transcriptome. J. Physiol. 575, 321–332. 10.1113/jphysiol.2006.115568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Wang Y., Bi W. (2012). C-Myc overexpression promotes osteosarcoma cell invasion via activation of MEK-ERK pathway. Oncol. Res. 20, 149–156. 10.3727/096504012x13522227232237 [DOI] [PubMed] [Google Scholar]
- Han J., Zhang Y., Xu J., Zhang T., Wang H., Wang Z., et al. (2021). Her4 promotes cancer metabolic reprogramming via the c-Myc-dependent signaling axis. Cancer Lett. 496, 57–71. 10.1016/j.canlet.2020.10.008 [DOI] [PubMed] [Google Scholar]
- Han X., Yang Y., Sun Y., Qin L., Yang Y. (2018). LncRNA TUG1 affects cell viability by regulating glycolysis in osteosarcoma cells. Gene 674, 87–92. 10.1016/j.gene.2018.06.085 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: The next generation. Cell. 144, 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harborne J. B., Williams C. A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55, 481–504. 10.1016/s0031-9422(00)00235-1 [DOI] [PubMed] [Google Scholar]
- Harris I., Mccracken S., Mak T. W. (2012). PKM2: A gatekeeper between growth and survival. Cell. Res. 22, 447–449. 10.1038/cr.2011.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Gao J., Zheng L., Liu S., Ye L., Lai H., et al. (2021). TGF‑β inhibitor RepSox suppresses osteosarcoma via the JNK/Smad3 signaling pathway. Int. J. Oncol. 59, 84. 10.3892/ijo.2021.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang B. H., Kubo T., Healey J. H., Yang R., Nathan S. S., Kolb E. A., et al. (2004). Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res. 64, 2734–2739. 10.1158/0008-5472.can-03-1952 [DOI] [PubMed] [Google Scholar]
- Hsu P. P., Sabatini D. M. (2008). Cancer cell metabolism: Warburg and beyond. Cell. 134, 703–707. 10.1016/j.cell.2008.08.021 [DOI] [PubMed] [Google Scholar]
- Hu K., Dai H. B., Qiu Z. L. (2016). mTOR signaling in osteosarcoma: Oncogenesis and therapeutic aspects (Review). Oncol. Rep. 36, 1219–1225. 10.3892/or.2016.4922 [DOI] [PubMed] [Google Scholar]
- Hu R., Chen S., Yan J. (2021). Blocking circ-CNST suppresses malignant behaviors of osteosarcoma cells and inhibits glycolysis through circ-CNST-miR-578-LDHA/PDK1 ceRNA networks. J. Orthop. Surg. Res. 16, 300. 10.1186/s13018-021-02427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Jiang Q., Luo D., Zhao L., Fu X., Chen Y., et al. (2016). miR-200b is a key regulator of tumor progression and metabolism targeting lactate dehydrogenase A in human malignant glioma. Oncotarget 7, 48423–48431. 10.18632/oncotarget.10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Wang Z., Chen M., Chen X., Liang W. (2018). The anti-osteosarcoma cell activity by a mTORC1/2 dual inhibitor RES-529. Biochem. Biophys. Res. Commun. 497, 499–505. 10.1016/j.bbrc.2018.02.050 [DOI] [PubMed] [Google Scholar]
- Hua W., Ten Dijke P., Kostidis S., Giera M., Hornsveld M. (2020). TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell. Mol. Life Sci. 77, 2103–2123. 10.1007/s00018-019-03398-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhu X., Ke Y., Xiao D., Liang C., Chen J., et al. (2020). LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. Cancer Biol. Ther. 21, 379–387. 10.1080/15384047.2019.1702405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. F., Lu L., Shen H. L., Lu X. X. (2020). LncRNA SNHG4 promotes osteosarcoma proliferation and migration by sponging miR-377-3p. Mol. Genet. Genomic Med. 8, e1349. 10.1002/mgg3.1349 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang Y., Xu Y. Q., Feng S. Y., Zhang X., Ni J. D. (2020). LncRNA TDRG1 promotes proliferation, invasion and epithelial-mesenchymal transformation of osteosarcoma through PI3K/AKT signal pathway. Cancer Manag. Res. 12, 4531–4540. 10.2147/CMAR.S248964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo S., Dou D. (2021). Circ_0056285 regulates proliferation, apoptosis and glycolysis of osteosarcoma cells via miR-1244/TRIM44 Axis. Cancer Manag. Res. 13, 1257–1270. 10.2147/CMAR.S290645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen W. J., Dayton T. L., Davidson S. M., Fiske B. P., Hosios A. M., Bellinger G., et al. (2013). PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 155, 397–409. 10.1016/j.cell.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen W. J., Vander Heiden M. G. (2015). Pyruvate kinase: Function, regulation and role in cancer. Semin. Cell. Dev. Biol. 43, 43–51. 10.1016/j.semcdb.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Standart N. (2007). How do microRNAs regulate gene expression? Sci. STKE. 2007, re1. 10.1126/stke.3672007re1 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Lu M., Jung K. H., Park J. H., Yu L., Onuchic J. N., et al. (2019). Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 116, 3909–3918. 10.1073/pnas.1816391116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Wang Y., Sun X., Zhao X., Zhang Y., Xu S., et al. (2021). Effect of lncRNA XLOC_005950 knockout by CRISPR/Cas9 gene editing on energy metabolism and proliferation in osteosarcoma MG63 cells mediated by hsa-miR-542-3p. Oncol. Lett. 22, 669. 10.3892/ol.2021.12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian F., Yuan F., Jiong M., Zhu X. Z., Yu G. R., Lu D. D. (2015). Silencing of glucose transporter protein-1 by RNA interference inhibits human osteosarcoma Mg63 cells growth in vivo . Technol. Cancer Res. Treat. 14, 243–248. 10.7785/tcrt.2012.500412 [DOI] [PubMed] [Google Scholar]
- Jiang H., Zeng Z. (2015). Dual mTORC1/2 inhibition by INK-128 results in antitumor activity in preclinical models of osteosarcoma. Biochem. Biophys. Res. Commun. 468, 255–261. 10.1016/j.bbrc.2015.10.119 [DOI] [PubMed] [Google Scholar]
- Jiang J., Wang W., Xiang W., Jiang L., Zhou Q. (2021). The phosphoinositide 3-kinase inhibitor ZSTK474 increases the susceptibility of osteosarcoma cells to oncolytic vesicular stomatitis virus VSVΔ51 via aggravating endoplasmic reticulum stress. Bioengineered 12, 11847–11857. 10.1080/21655979.2021.1999372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Zhang Q., Fan Y., Li J., Zhang J., Wang W., et al. (2022). MYC inhibition reprograms tumor immune microenvironment by recruiting T lymphocytes and activating the CD40/CD40L system in osteosarcoma. Cell. Death Discov. 8, 117. 10.1038/s41420-022-00923-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Guo S., Wang S., Zhang Y., Chen H., Wang Y., et al. (2022). EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 82, 831–845. 10.1158/0008-5472.CAN-21-2988 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Li F., Gao B., Ma M., Chen M., Wu Y., et al. (2021). KDM6B-mediated histone demethylation of LDHA promotes lung metastasis of osteosarcoma. Theranostics 11, 3868–3881. 10.7150/thno.53347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. H., Jeong S. J., Kim S. H., Kim J. H., Jung J. H., Koh W., et al. (2012). Icariside II induces apoptosis in U937 acute myeloid leukemia cells: Role of inactivation of STAT3-related signaling. PLoS One 7, e28706. 10.1371/journal.pone.0028706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi K., Araki K., Tobiume K., Tanaka N. (2008). p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell. Biol. 10, 611–618. 10.1038/ncb1724 [DOI] [PubMed] [Google Scholar]
- Kolev Y., Uetake H., Takagi Y., Sugihara K. (2008). Lactate dehydrogenase-5 (LDH-5) expression in human gastric cancer: Association with hypoxia-inducible factor (HIF-1alpha) pathway, angiogenic factors production and poor prognosis. Ann. Surg. Oncol. 15, 2336–2344. 10.1245/s10434-008-9955-5 [DOI] [PubMed] [Google Scholar]
- Koppenol W. H., Bounds P. L., Dang C. V. (2011). Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337. 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- Kotowski K., Rosik J., Machaj F., Supplitt S., Wiczew D., Jablonska K., et al. (2021)., 13. Cancers (Basel), 909. 10.3390/cancers13040909 Role of PFKFB3 and PFKFB4 in cancer: Genetic basis, impact on disease development/progression, and potential as therapeutic targets Cancers [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D., Reckenbeil J., Veit N., Kuerpig S., Meisenheimer M., Beier I., et al. (2018). Targeting glucose transport and the NAD pathway in tumor cells with STF-31: A re-evaluation. Cell. Oncol. 41, 485–494. 10.1007/s13402-018-0385-5 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Pouyssegur J. (2008). Tumor cell metabolism: cancer's achilles' heel. Cancer Cell. 13, 472–482. 10.1016/j.ccr.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Kubo T., Shimose S., Fujimori J., Furuta T., Arihiro K., Ochi M. (2015). Does expression of glucose transporter protein-1 relate to prognosis and angiogenesis in osteosarcoma? Clin. Orthop. Relat. Res. 473, 305–310. 10.1007/s11999-014-3910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijjer M. L., van Den Akker B. E., Hilhorst R., Mommersteeg M., Buddingh E. P., Serra M., et al. (2014). Kinome and mRNA expression profiling of high-grade osteosarcoma cell lines implies Akt signaling as possible target for therapy. BMC Med. Genomics 7, 4. 10.1186/1755-8794-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Kim K. S., An H. K., Kim C. H., Moon H. I., Lee Y. C. (2013). Dendropanoxide induces autophagy through ERK1/2 activation in MG-63 human osteosarcoma cells and autophagy inhibition enhances dendropanoxide-induced apoptosis. PLoS One 8, e83611. 10.1371/journal.pone.0083611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Li Y., Uyeda K., Hasemann C. A. (2003). Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase. J. Biol. Chem. 278, 523–530. 10.1074/jbc.M209105200 [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Chung J. G., Chien Y. T., Lin S. S., Hsu F. T. (2019). Suppression of ERK/NF-κB activation is associated with amentoflavone-inhibited osteosarcoma progression in vivo . Anticancer Res. 39, 3669–3675. 10.21873/anticanres.13515 [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Schlessinger J., Ferguson K. M. (2014). The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 6, a020768. 10.1101/cshperspect.a020768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow P. C., Tian Q., Ong Z. Y., Yang Z., Ee P. L. (2010). Antitumor activity of natural compounds, curcumin and PKF118-310, as Wnt/β-catenin antagonists against human osteosarcoma cells. Invest. New Drugs 28, 766–782. 10.1007/s10637-009-9311-z [DOI] [PubMed] [Google Scholar]
- Li C., Chen Y., Bai P., Wang J., Liu Z., Wang T., et al. (2016). LDHB may be a significant predictor of poor prognosis in osteosarcoma. Am. J. Transl. Res. 8, 4831–4843. [PMC free article] [PubMed] [Google Scholar]
- Li G., Li Y., Wang D. Y. (2021). Overexpression of miR-329-3p sensitizes osteosarcoma cells to cisplatin through suppression of glucose metabolism by targeting LDHA. Cell. Biol. Int. 45, 766–774. 10.1002/cbin.11476 [DOI] [PubMed] [Google Scholar]
- Li K., Yu H., Zhao C., Li J., Tan R., Chen L. (2021). Down-regulation of PRR11 affects the proliferation, migration and invasion of osteosarcoma by inhibiting the Wnt/β-catenin pathway. J. Cancer 12, 6656–6664. 10.7150/jca.62491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., et al. (2018). Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 33, 368–385. 10.1016/j.ccell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Li L. T., Jiang G., Chen Q., Zheng J. N. (2015). Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 11, 1566–1572. 10.3892/mmr.2014.2914 [DOI] [PubMed] [Google Scholar]
- Li X., Liu Y., Liao S., Lin C., Moro A., Liu J., et al. (2021). Polyphyllin VII induces apoptosis and autophagy via mediating H2O2 levels and the JNK pathway in human osteosarcoma U2OS cells. Oncol. Rep. 45, 180–190. 10.3892/or.2020.7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Song X., Liu Z., Li Q., Huang M., Su B., et al. (2019). Upregulation of miR-214 induced radioresistance of osteosarcoma by targeting PHLDA2 via PI3K/Akt signaling. Front. Oncol. 9, 298. 10.3389/fonc.2019.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Fu Y., Ouyang W., He M., Wang Y., Wang X., et al. (2021). Circ_0016347 promotes osteosarcoma progression by regulating miR-1225-3p/KCNH1 Axis. Cancer Biother Radiopharm.. Mary Ann Liebert, Inc. 10.1089/cbr.2019.3349 [DOI] [PubMed] [Google Scholar]
- Li Z., Geng M., Ye X., Ji Y., Li Y., Zhang X., et al. (2022). IRF7 inhibits the Warburg effect via transcriptional suppression of PKM2 in osteosarcoma. Int. J. Biol. Sci. 18, 30–42. 10.7150/ijbs.65255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Q., Wang Z., Zhang Y., Lu C., Ding Q. L., Ren R., et al. (2021). CircRNA_103801 accelerates proliferation of osteosarcoma cells by sponging miR-338-3p and regulating HIF-1/Rap1/PI3K-Akt pathway. J. Biol. Regul. Homeost. Agents 35, 1021–1028. 10.23812/20-725-A [DOI] [PubMed] [Google Scholar]
- Lian Z., Han J., Huang L., Wei C., Fan Y., Xu J., et al. (2019). Retraction of: A005, a novel inhibitor of phosphatidylinositol 3-kinase/mammalian target of rapamycin, prevents osteosarcoma-induced osteolysis. Carcinogenesis 40, e1–e13. 10.1093/carcin/bgy036 [DOI] [PubMed] [Google Scholar]
- Liang C., Li H., Shen C., Lai J., Shi Z., Liu B., et al. (2012). Genistein potentiates the anti-cancer effects of gemcitabine in human osteosarcoma via the downregulation of Akt and nuclear factor-κB pathway. Anticancer. Agents Med. Chem. 12, 554–563. 10.2174/187152012800617867 [DOI] [PubMed] [Google Scholar]
- Liang N., Kitts D. D. (2015). Role of chlorogenic acids in controlling oxidative and inflammatory stress conditions. Nutrients 8, E16. 10.3390/nu8010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F., Lu Q. (2022). S100 calcium-binding protein A10 contributes to malignant traits in osteosarcoma cells by regulating glycolytic metabolism via the AKT/mTOR pathway. Bioengineered 13, 12298–12308. 10.1080/21655979.2022.2071022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Wu Y., Zhou Y., Peng D. (2014). Endothelin A receptor antagonism enhances inhibitory effects of anti-ganglioside GD2 monoclonal antibody on invasiveness and viability of human osteosarcoma cells. PLoS One 9, e93576. 10.1371/journal.pone.0093576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Cai L., Li H. (2019). miR185 regulates the growth of osteosarcoma cells via targeting Hexokinase 2. Mol. Med. Rep. 20, 2774–2782. 10.3892/mmr.2019.10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chen M., Ma L., Dang X., Du G. (2020a). LncRNA GAS5 suppresses the proliferation and invasion of osteosarcoma cells via the miR-23a-3p/PTEN/PI3K/AKT pathway. Cell. Transpl. 29, 963689720953093. 10.1177/0963689720953093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Feng G., Li Z., Li R., Xia P. (2020b). Long non-coding RNA FEZF1-AS1 modulates CXCR4 to promote cell proliferation, Warburg effect and suppress cell apoptosis in osteosarcoma by sponging miR-144. Onco. Targets. Ther. 13, 2899–2910. 10.2147/OTT.S235970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Luo B., Zhao M. (2017). Bmi‑1‑‑targeting suppresses osteosarcoma aggressiveness through the NF‑κB signaling pathway. Mol. Med. Rep. 16, 7949–7958. 10.3892/mmr.2017.7660 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu J., Wu J., Zhou L., Pan C., Zhou Y., Du W., et al. (2015). ZD6474, a new treatment strategy for human osteosarcoma, and its potential synergistic effect with celecoxib. Oncotarget 6, 21341–21352. 10.18632/oncotarget.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhao J., Feng G., Li R., Jiao J. (2022). Silencing of circ-CDK14 suppresses osteosarcoma progression through the miR-198/E2F2 axis. Exp. Cell. Res. 414, 113082. 10.1016/j.yexcr.2022.113082 [DOI] [PubMed] [Google Scholar]
- Liu M., Wang D., Li N. (2016). MicroRNA-20b downregulates HIF-1α and inhibits the proliferation and invasion of osteosarcoma cells. Oncol. Res. 23, 257–266. 10.3727/096504016X14562725373752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li W., Liu H., Yu X. (2019). Xanthohumol inhibits colorectal cancer cells via downregulation of Hexokinases II-mediated glycolysis. Int. J. Biol. Sci. 15, 2497–2508. 10.7150/ijbs.37481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. C., Tsai J. J., Weng Y. S., Hsu F. T. (2020). Regorafenib suppresses epidermal growth factor receptor signaling-modulated progression of colorectal cancer. Biomed. Pharmacother. 128, 110319. 10.1016/j.biopha.2020.110319 [DOI] [PubMed] [Google Scholar]
- Liu Y., He J., Chen X., Li J., Shen M., Yu W., et al. (2014). The proapoptotic effect of formononetin in human osteosarcoma cells: Involvement of inactivation of ERK and Akt pathways. Cell. Physiol. biochem. 34, 637–645. 10.1159/000363029 [DOI] [PubMed] [Google Scholar]
- Liu Z. X., Hong L., Fang S. Q., Tan G. H., Huang P. G., Zeng Z., et al. (2016). Overexpression of pyruvate kinase M2 predicts a poor prognosis for patients with osteosarcoma. Tumour Biol. 37, 14923–14928. 10.1007/s13277-016-5401-7 [DOI] [PubMed] [Google Scholar]
- Londhe P., Yu P. Y., Ijiri Y., Ladner K. J., Fenger J. M., London C., et al. (2018). Classical NF-κB metabolically reprograms sarcoma cells through regulation of hexokinase 2. Front. Oncol. 8, 104. 10.3389/fonc.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi A., Errani C., de Paolis M., Mercuri M., Bacci G. (2006). Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat. Rev. 32, 423–436. 10.1016/j.ctrv.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Lu Y., Lu D., Hu Y. (2018). GLP2 promotes directed differentiation from osteosarcoma cells to osteoblasts and inhibits growth of osteosarcoma cells. Mol. Ther. Nucleic Acids 10, 292–303. 10.1016/j.omtn.2017.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luengo A., Li Z., Gui D. Y., Sullivan L. B., Zagorulya M., Do B. T., et al. (2021). Increased demand for NAD(+) relative to ATP drives aerobic glycolysis. Mol. Cell. 81, 691–707 e6. 10.1016/j.molcel.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Xu Y. P., Zhao D., Li F. L., Wang W., Sasaki N., et al. (2013). Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol. Cell. 52, 340–352. 10.1016/j.molcel.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Zhang C., Li W. (2020). TGF-beta is associated with poor prognosis and promotes osteosarcoma progression via PI3K/Akt pathway activation. Cell. Cycle 19, 2327–2339. 10.1080/15384101.2020.1805552 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma L., Xu Y., Xu X. (2020). Targeted MEK inhibition by cobimetinib enhances doxorubicin's efficacy in osteosarcoma models. Biochem. Biophys. Res. Commun. 529, 622–628. 10.1016/j.bbrc.2020.06.082 [DOI] [PubMed] [Google Scholar]
- Maloney C., Kallis M. P., Edelman M., Tzanavaris C., Lesser M., Soffer S. Z., et al. (2020). Gefitinib inhibits invasion and metastasis of osteosarcoma via inhibition of macrophage receptor interacting serine-threonine kinase 2. Mol. Cancer Ther. 19, 1340–1350. 10.1158/1535-7163.MCT-19-0903 [DOI] [PubMed] [Google Scholar]
- Manara M. C., Nicoletti G., Zambelli D., Ventura S., Guerzoni C., Landuzzi L., et al. (2010). NVP-BEZ235 as a new therapeutic option for sarcomas. Clin. Cancer Res. 16, 530–540. 10.1158/1078-0432.CCR-09-0816 [DOI] [PubMed] [Google Scholar]
- Martins-Neves S. R., Paiva-Oliveira D. I., Fontes-Ribeiro C., Bovee J., Cleton-Jansen A. M., Gomes C. M. F. (2018). IWR-1, a tankyrase inhibitor, attenuates Wnt/β-catenin signaling in cancer stem-like cells and inhibits in vivo the growth of a subcutaneous human osteosarcoma xenograft. Cancer Lett. 414, 1–15. 10.1016/j.canlet.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Maschek G., Savaraj N., Priebe W., Braunschweiger P., Hamilton K., Tidmarsh G. F., et al. (2004). 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo . Cancer Res. 64, 31–34. 10.1158/0008-5472.can-03-3294 [DOI] [PubMed] [Google Scholar]
- Mathupala S. P., Ko Y. H., Pedersen P. L. (2006). Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 25, 4777–4786. 10.1038/sj.onc.1209603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M. L., Adler A. S., Deming L., Cosino E., Lee L., Blackwood E. M., et al. (2013). Lactate dehydrogenase B is required for the growth of KRAS-dependent lung adenocarcinomas. Clin. Cancer Res. 19, 773–784. 10.1158/1078-0432.CCR-12-2638 [DOI] [PubMed] [Google Scholar]
- McCleland M. L., Adler A. S., Shang Y., Hunsaker T., Truong T., Peterson D., et al. (2012). An integrated genomic screen identifies LDHB as an essential gene for triple-negative breast cancer. Cancer Res. 72, 5812–5823. 10.1158/0008-5472.CAN-12-1098 [DOI] [PubMed] [Google Scholar]
- Minchenko O., Opentanova I., Caro J. (2003). Hypoxic regulation of the 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase gene family (PFKFB-1-4) expression in vivo . FEBS Lett. 554, 264–270. 10.1016/s0014-5793(03)01179-7 [DOI] [PubMed] [Google Scholar]
- Mitchell S., Vargas J., Hoffmann A. (2016). Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 8, 227–241. 10.1002/wsbm.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa S., Sugimoto N., Yamamoto N., Shirai T., Nishida H., Hayashi K., et al. (2012). Caffeine induces apoptosis of osteosarcoma cells by inhibiting AKT/mTOR/S6K, NF-κB and MAPK pathways. Anticancer Res. 32, 3643–3649. [PubMed] [Google Scholar]
- Moreno-Sanchez R., Rodriguez-Enriquez S., Marin-Hernandez A., Saavedra E. (2007). Energy metabolism in tumor cells. FEBS J. 274, 1393–1418. 10.1111/j.1742-4658.2007.05686.x [DOI] [PubMed] [Google Scholar]
- Moriceau G., Ory B., Mitrofan L., Riganti C., Blanchard F., Brion R., et al. (2010). Zoledronic acid potentiates mTOR inhibition and abolishes the resistance of osteosarcoma cells to RAD001 (everolimus): Pivotal role of the prenylation process. Cancer Res. 70, 10329–10339. 10.1158/0008-5472.CAN-10-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M., Derynck R., Miyazono K. (2016). TGF-Beta and the TGF-beta family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8, a021873. 10.1101/cshperspect.a021873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann D., Park S., Hall M. N. (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757. 10.1038/s41568-018-0074-8 [DOI] [PubMed] [Google Scholar]
- Mueckler M., Thorens B. (2013). The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 34, 121–138. 10.1016/j.mam.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Colmenero A., Fernandez-Suarez A., Fatela-Cantillo D., Ocana-Perez E., Dominguez-Jimenez J. L., Diaz-Iglesias J. M. (2015). Plasma tumor M2-pyruvate kinase levels in different cancer types. Anticancer Res. 35, 4271–4276. [PubMed] [Google Scholar]
- Nepal M., Choi H. J., Choi B. Y., Kim S. L., Ryu J. H., Kim D. H., et al. (2012). Anti-angiogenic and anti-tumor activity of Bavachinin by targeting hypoxia-inducible factor-1α. Eur. J. Pharmacol. 691, 28–37. 10.1016/j.ejphar.2012.06.028 [DOI] [PubMed] [Google Scholar]
- Nepal M., Gong Y. D., Park Y. R., Soh Y. (2011). An activator of PHD2, KRH102140, decreases angiogenesis via inhibition of HIF-1α. Cell. biochem. Funct. 29, 126–134. 10.1002/cbf.1732 [DOI] [PubMed] [Google Scholar]
- Ni S., Kuang Y., Yuan Y., Yu B. (2020). Mitochondrion-mediated iron accumulation promotes carcinogenesis and Warburg effect through reactive oxygen species in osteosarcoma. Cancer Cell. Int. 20, 399. 10.1186/s12935-020-01494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Rainusso N., Lee Y. C., Dawson B., Coarfa C., Han R., et al. (2019). Tegavivint and the beta-catenin/ALDH Axis in chemotherapy-resistant and metastatic osteosarcoma. J. Natl. Cancer Inst. 111, 1216–1227. 10.1093/jnci/djz026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Sasaki A., Ono K., Ohshika S., Ishibashi Y., Yamada K. (2021). Uptake of fluorescent D- and L-glucose analogues, 2-NBDG and 2-NBDLG, into human osteosarcoma U2OS cells in a phloretin-inhibitable manner. Hum. Cell. 34, 634–643. 10.1007/s13577-020-00483-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P. J., Liu Y. C., Hsu F. T. (2019). Protein kinase B and extracellular signal-regulated kinase inactivation is associated with regorafenib-induced inhibition of osteosarcoma progression in vitro and in vivo . J. Clin. Med. 8, E900. 10.3390/jcm8060900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P. J., Tsai J. J., Liu Y. C. (2017). Amentoflavone inhibits metastatic potential through suppression of ERK/NF-κB activation in osteosarcoma U2OS cells. Anticancer Res. 37, 4911–4918. 10.21873/anticanres.11900 [DOI] [PubMed] [Google Scholar]
- Pan X., Guo J., Liu C., Pan Z., Yang Z., Yao X., et al. (2022). LncRNA HCG18 promotes osteosarcoma growth by enhanced aerobic glycolysis via the miR-365a-3p/PGK1 axis. Cell. Mol. Biol. Lett. 27, 5. 10.1186/s11658-021-00304-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Li H., Tan J., Weng X., Zhou L., Weng Y., et al. (2020). miR-1297 suppresses osteosarcoma proliferation and aerobic glycolysis by regulating PFKFB2. Onco. Targets. Ther. 13, 11265–11275. 10.2147/OTT.S274744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate K. T., Stringari C., Sprowl-Tanio S., Wang K., Teslaa T., Hoverter N. P., et al. (2014). Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 33, 1454–1473. 10.15252/embj.201488598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra K. C., Wang Q., Bhaskar P. T., Miller L., Wang Z., Wheaton W., et al. (2013). Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 24, 213–228. 10.1016/j.ccr.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S. F., Lee C. Y., Hour M. J., Tsai S. C., Kuo D. H., Chen F. A., et al. (2014). Curcumin-loaded nanoparticles enhance apoptotic cell death of U2OS human osteosarcoma cells through the Akt-Bad signaling pathway. Int. J. Oncol. 44, 238–246. 10.3892/ijo.2013.2175 [DOI] [PubMed] [Google Scholar]
- Pignochino Y., Dell'Aglio C., Basirico M., Capozzi F., Soster M., Marchio S., et al. (2013). The Combination of Sorafenib and Everolimus Abrogates mTORC1 and mTORC2 upregulation in osteosarcoma preclinical models. Clin. Cancer Res. 19, 2117–2131. 10.1158/1078-0432.CCR-12-2293 [DOI] [PubMed] [Google Scholar]
- Prensner J. R., Chinnaiyan A. M. (2011). The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407. 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu F., Liu J., Jing D., Chen F., Huang X., Shi D., et al. (2022). LncCCAT1 interaction protein PKM2 upregulates SREBP2 phosphorylation to promote osteosarcoma tumorigenesis by enhancing the Warburg effect and lipogenesis. Int. J. Oncol. 60, 44. 10.3892/ijo.2022.5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Huang T., Chen Y., Li X., Gong W., Jiang G., et al. (2018). X-linked inhibitor of apoptosis protein inhibitor Embelin induces apoptosis via PI3K/Akt pathway and inhibits invasion in osteosarcoma cells. J. Cancer Res. Ther. 14, S648–S655. 10.4103/0973-1482.203599 [DOI] [PubMed] [Google Scholar]
- Rafat M., Moraghebi M., Afsa M., Malekzadeh K. (2021). The outstanding role of miR-132-3p in carcinogenesis of solid tumors. Hum. Cell. 34, 1051–1065. 10.1007/s13577-021-00544-w [DOI] [PubMed] [Google Scholar]
- Rashid M., Zadeh L. R., Baradaran B., Molavi O., Ghesmati Z., Sabzichi M., et al. (2021). Up-down regulation of HIF-1α in cancer progression. Gene 798, 145796. 10.1016/j.gene.2021.145796 [DOI] [PubMed] [Google Scholar]
- Ren X. F., Mu L. P., Jiang Y. S., Wang L., Ma J. F. (2015). LY2109761 inhibits metastasis and enhances chemosensitivity in osteosarcoma MG-63 cells. Eur. Rev. Med. Pharmacol. Sci. 19, 1182–1190. [PubMed] [Google Scholar]
- Ren X., Su C. (2020). Sphingosine kinase 1 contributes to doxorubicin resistance and glycolysis in osteosarcoma. Mol. Med. Rep. 22, 2183–2190. 10.3892/mmr.2020.11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RodriguezGarcia A., Samso P., Fontova P., Simon-Molas H., Manzano A., Castano E., et al. (2017). TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 284, 3437–3454. 10.1111/febs.14201 [DOI] [PubMed] [Google Scholar]
- Ros S., Schulze A. (2013). Balancing glycolytic flux: The role of 6-phosphofructo-2-kinase/fructose 2, 6-bisphosphatases in cancer metabolism. Cancer Metab. 1, 8. 10.1186/2049-3002-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru N., Zhang F., Liang J., Du Y., Wu W., Wang F., et al. (2018). MiR-564 is down-regulated in osteosarcoma and inhibits the proliferation of osteosarcoma cells via targeting Akt. Gene 645, 163–169. 10.1016/j.gene.2017.12.028 [DOI] [PubMed] [Google Scholar]
- Sapio L., Salzillo A., Illiano M., Ragone A., Spina A., Chiosi E., et al. (2020). Chlorogenic acid activates ERK1/2 and inhibits proliferation of osteosarcoma cells. J. Cell. Physiol. 235, 3741–3752. 10.1002/jcp.29269 [DOI] [PubMed] [Google Scholar]
- Saraf A. J., Fenger J. M., Roberts R. D. (2018). Osteosarcoma: Accelerating progress makes for a hopeful future. Front. Oncol. 8, 4. 10.3389/fonc.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Tadano A., Suzuki T., Amari M., Takagi K., Miki Y., Tamaki K., et al. (2013). Hexokinase II in breast carcinoma: A potent prognostic factor associated with hypoxia-inducible factor-1α and ki-67. Cancer Sci. 104, 1380–1388. 10.1111/cas.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg-BarYoseph F., Armoni M., Karnieli E. (2004). The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 64, 2627–2633. 10.1158/0008-5472.can-03-0846 [DOI] [PubMed] [Google Scholar]
- Shang D., Wu J., Guo L., Xu Y., Liu L., Lu J. (2017). Metformin increases sensitivity of osteosarcoma stem cells to cisplatin by inhibiting expression of PKM2. Int. J. Oncol. 50, 1848–1856. 10.3892/ijo.2017.3950 [DOI] [PubMed] [Google Scholar]
- Shen Y., Xu J., Pan X., Zhang Y., Weng Y., Zhou D., et al. (2020). LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell. Death Dis. 11, 278. 10.1038/s41419-020-2485-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhao S., Wang S., Pan X., Zhang Y., Xu J., et al. (2019). S1P/S1PR3 axis promotes aerobic glycolysis by YAP/c-MYC/PGAM1 axis in osteosarcoma. EBioMedicine 40, 210–223. 10.1016/j.ebiom.2018.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lian K., Jia J. (2022). Apigenin suppresses the Warburg effect and stem-like properties in SOSP-9607 cells by inactivating the PI3K/Akt/mTOR signaling pathway. Evid. Based. Complement. Altern. Med. 2022, 3983637. 10.1155/2022/3983637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla A., Wang J., Yang R., Geller D. S., Loeb D. M., Hoang B. H. (2020). Wnt signaling in osteosarcoma. Adv. Exp. Med. Biol. 1258, 125–139. 10.1007/978-3-030-43085-6_8 [DOI] [PubMed] [Google Scholar]
- Sola-Penna M., Da Silva D., Coelho W. S., Marinho-Carvalho M. M., Zancan P. (2010). Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life 62, 791–796. 10.1002/iub.393 [DOI] [PubMed] [Google Scholar]
- Song J., Wu X., Liu F., Li M., Sun Y., Wang Y., et al. (2017). Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 490, 217–224. 10.1016/j.bbrc.2017.06.024 [DOI] [PubMed] [Google Scholar]
- Sottnik J. L., Lori J. C., Rose B. J., Thamm D. H. (2011). Glycolysis inhibition by 2-deoxy-D-glucose reverts the metastatic phenotype in vitro and in vivo . Clin. Exp. Metastasis 28, 865–875. 10.1007/s10585-011-9417-5 [DOI] [PubMed] [Google Scholar]
- Spoden G. A., Mazurek S., Morandell D., Bacher N., Ausserlechner M. J., Jansen-Durr P., et al. (2008). Isotype-specific inhibitors of the glycolytic key regulator pyruvate kinase subtype M2 moderately decelerate tumor cell proliferation. Int. J. Cancer 123, 312–321. 10.1002/ijc.23512 [DOI] [PubMed] [Google Scholar]
- Sun J., Wu W., Tang X., Zhang F., Ju C., Liu R., et al. (2021). HDAC6 inhibitor WT161 performs anti-tumor effect on osteosarcoma and synergistically interacts with 5-FU. Biosci. Rep. 41, BSR20203905. 10.1042/BSR20203905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. Y., Hou Y. J., Yin Y. B., Wang F. Z., Yang M. F., Zhang Y. Y., et al. (2020). CCT128930 induces G1-phase arrest and apoptosis and synergistically enhances the anticancer efficiency of VS5584 in human osteosarcoma cells. Biomed. Pharmacother. 130, 110544. 10.1016/j.biopha.2020.110544 [DOI] [PubMed] [Google Scholar]
- Sun L., Wang P., Zhang Z., Zhang K., Xu Z., Li S., et al. (2020). MicroRNA-615 functions as a tumor suppressor in osteosarcoma through the suppression of HK2. Oncol. Lett. 20, 226. 10.3892/ol.2020.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Shan H. J., Yin G., Zhang X. Y., Huang Y. M., Li H. J. (2022). The anti-osteosarcoma cell activity by the sphingosine kinase 1 inhibitor SKI-V. Cell. Death Discov. 8, 48. 10.1038/s41420-022-00838-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Lu Y., Zhu J., Yu L., Ma B., et al. (2018). Oridonin prevents epithelial-mesenchymal transition and TGF-β1-induced epithelial-mesenchymal transition by inhibiting TGF-β1/Smad2/3 in osteosarcoma. Chem. Biol. Interact. 296, 57–64. 10.1016/j.cbi.2018.09.013 [DOI] [PubMed] [Google Scholar]
- Suzuki R., Brown G. A., Christopher J. A., Scully C. C. G., Congreve M. (2020). Recent developments in therapeutic peptides for the glucagon-like peptide 1 and 2 receptors. J. Med. Chem. 63, 905–927. 10.1021/acs.jmedchem.9b00835 [DOI] [PubMed] [Google Scholar]
- Tan X., Lambert P. F., Rapraeger A. C., Anderson R. A. (2016). Stress-induced EGFR trafficking: Mechanisms, functions, and therapeutic implications. Trends Cell. Biol. 26, 352–366. 10.1016/j.tcb.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Zhang Z., Qian H., Chen J., Wang Y., Chen X., et al. (2015). (-)-Epigallocatechin-3-gallate inhibits osteosarcoma cell invasiveness by inhibiting the MEK/ERK signaling pathway in human osteosarcoma cells. J. Environ. Pathol. Toxicol. Oncol. 34, 85–93. 10.1615/jenvironpatholtoxicoloncol.2015011925 [DOI] [PubMed] [Google Scholar]
- Tang H. Y., Guo J. Q., Sang B. T., Cheng J. N., Wu X. M. (2022). PDGFRβ modulates aerobic glycolysis in osteosarcoma HOS cells via the PI3K/AKT/mTOR/c-Myc pathway. Biochem. Cell. Biol. 100, 75–84. 10.1139/bcb-2021-0305 [DOI] [PubMed] [Google Scholar]
- Tang Q. L., Xie X. B., Wang J., Chen Q., Han A. J., Zou C. Y., et al. (2012). Glycogen synthase kinase-3β, NF-κB signaling, and tumorigenesis of human osteosarcoma. J. Natl. Cancer Inst. 104, 749–763. 10.1093/jnci/djs210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. M., Rogers S. D., Sleeman M. W., Pasquini G. M., Bringhurst F. R., Ng K. W., et al. (1995). Modulation of glucose transport by parathyroid hormone and insulin in UMR 106-01, a clonal rat osteogenic sarcoma cell line. J. Mol. Endocrinol. 14, 263–275. 10.1677/jme.0.0140263 [DOI] [PubMed] [Google Scholar]
- Tian C., Yuan Z., Xu D., Ding P., Wang T., Zhang L., et al. (2017). Inhibition of glycolysis by a novel EGFR/HER2 inhibitor KU004 suppresses the growth of HER2+ cancer. Exp. Cell. Res. 357, 211–221. 10.1016/j.yexcr.2017.05.019 [DOI] [PubMed] [Google Scholar]
- Tingting R., Wei G., Changliang P., Xinchang L., Yi Y. (2010). Arsenic trioxide inhibits osteosarcoma cell invasiveness via MAPK signaling pathway. Cancer Biol. Ther. 10, 251–257. 10.4161/cbt.10.3.12349 [DOI] [PubMed] [Google Scholar]
- Urbanska K., Orzechowski A. (2019). Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. Int. J. Mol. Sci. 20, E2085. 10.3390/ijms20092085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda K. (1979). Phosphofructokinase. Adv. Enzymol. Relat. Areas Mol. Biol. 48, 193–244. 10.1002/9780470122938.ch4 [DOI] [PubMed] [Google Scholar]
- van Cruijsen H., Voest E. E., Punt C. J., Hoekman K., Witteveen P. O., Meijerink M. R., et al. (2010). Phase I evaluation of cediranib, a selective VEGFR signalling inhibitor, in combination with gefitinib in patients with advanced tumours. Eur. J. Cancer 46, 901–911. 10.1016/j.ejca.2009.12.023 [DOI] [PubMed] [Google Scholar]
- Vijayakrishnapillai L. M. K., Desmarais J. S., Groeschen M. N., Perlin M. H. (2018). Deletion of ptn1, a PTEN/TEP1 orthologue, in Ustilago maydis reduces pathogenicity and teliospore development. J. Fungi (Basel) 5, 1. 10.3390/jof5010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Liu Y., Long F., Tian J., Zhang C. (2021). circPVT1 promotes osteosarcoma glycolysis and metastasis by sponging miR-423-5p to activate Wnt5a/Ror2 signaling. Cancer Sci. 112, 1707–1722. 10.1111/cas.14787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. W., Wu L., Cao Y., Yang L., Liu W., E X. Q., et al. (2017). A novel mechanism of mTORC1-mediated serine/glycine metabolism in osteosarcoma development. Cell. Signal. 29, 107–114. 10.1016/j.cellsig.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Wang F., Zhao Q., Liu J., Wang Z., Kong D. (2020). Identification of human lactate dehydrogenase A inhibitors with anti-osteosarcoma activity through cell-based phenotypic screening. Bioorg. Med. Chem. Lett. 30, 126909. 10.1016/j.bmcl.2019.126909 [DOI] [PubMed] [Google Scholar]
- Wang L. (2020). MiR-141-3p overexpression suppresses the malignancy of osteosarcoma by targeting FUS to degrade LDHB. Biosci. Rep. 40, BSR20193404. 10.1042/BSR20193404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu M. J., Bu J., Deng J. L., Jiang B. Y., Jiang L. D., et al. (2021). miR-485-3p regulated by MALAT1 inhibits osteosarcoma glycolysis and metastasis by directly suppressing c-MET and AKT3/mTOR signalling. Life Sci. 268, 118925. 10.1016/j.lfs.2020.118925 [DOI] [PubMed] [Google Scholar]
- Wang S., Wang Y., Huang Z., Wei H., Wang X., Shen R., et al. (2021a). Stattic sensitizes osteosarcoma cells to epidermal growth factor receptor inhibitors via blocking the interleukin 6-induced STAT3 pathway. Acta Biochim. Biophys. Sin. 53, 1670–1680. 10.1093/abbs/gmab146 [DOI] [PubMed] [Google Scholar]
- Wang S., Wei H., Huang Z., Wang X., Shen R., Wu Z., et al. (2021b). Epidermal growth factor receptor promotes tumor progression and contributes to gemcitabine resistance in osteosarcoma. Acta Biochim. Biophys. Sin. 53, 317–324. 10.1093/abbs/gmaa177 [DOI] [PubMed] [Google Scholar]
- Wang X., Lai P., Zhang Z., Huang M., Wang L., Yin M., et al. (2014). Targeted inhibition of mTORC2 prevents osteosarcoma cell migration and promotes apoptosis. Oncol. Rep. 32, 382–388. 10.3892/or.2014.3182 [DOI] [PubMed] [Google Scholar]
- Wang X. Z., Zhang S. F., Yang Z. H., Ye Z. W., Liu J. (2020). Punicalagin suppresses osteosarcoma growth and metastasis by regulating NF-κB signaling. J. Biol. Regul. Homeost. Agents 34, 1699–1708. 10.23812/20-23-A [DOI] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123, 309–314. 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- Wei D., Zhu X., Li S., Liu G., Wang Y., Wang W., et al. (2021). Tideglusib suppresses stem-cell-like features and progression of osteosarcoma by inhibiting GSK-3β/NOTCH1 signaling. Biochem. Biophys. Res. Commun. 554, 206–213. 10.1016/j.bbrc.2020.12.055 [DOI] [PubMed] [Google Scholar]
- Wei J., Zhu Y., Xu G., Yang F., Guan Z., Wang M., et al. (2014). Oxymatrine extracted from Sophora flavescens inhibited cell growth and induced apoptosis in human osteosarcoma MG-63 cells in vitro . Cell. biochem. Biophys. 70, 1439–1444. 10.1007/s12013-014-0078-2 [DOI] [PubMed] [Google Scholar]
- Wei S., Sun T., Du J., Zhang B., Xiang D., Li W. (2018). Xanthohumol, a prenylated flavonoid from Hops, exerts anticancer effects against gastric cancer in vitro . Oncol. Rep. 40, 3213–3222. 10.3892/or.2018.6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J. F., Jiang Y. Q., Li C., Dai X. K., Wu T., Yin W. Z. (2020). LncRNA-SARCC sensitizes osteosarcoma to cisplatin through the miR-143-mediated glycolysis inhibition by targeting Hexokinase 2. Cancer Biomark. 28, 231–246. 10.3233/CBM-191181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y. H., Koeppen H., Garcia R., Chiriboga L., Tarlow B. D., Peters B. A., et al. (2007). Epidermal growth factor receptor in osteosarcoma: Expression and mutational analysis. Hum. Pathol. 38, 1184–1191. 10.1016/j.humpath.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Weng Y., Shen Y., He Y., Pan X., Xu J., Jiang Y., et al. (2018). The miR-15b-5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect. Biochem. Biophys. Res. Commun. 503, 2749–2757. 10.1016/j.bbrc.2018.08.035 [DOI] [PubMed] [Google Scholar]
- Wilson J. E. (2003). Isozymes of mammalian hexokinase: Structure, subcellular localization and metabolic function. J. Exp. Biol. 206, 2049–2057. 10.1242/jeb.00241 [DOI] [PubMed] [Google Scholar]
- Wozniak M., Pastuch-Gawolek G., Makuch S., Wisniewski J., Krenacs T., Hamar P., et al. (2021). In vitro and in vivo efficacy of a novel glucose-methotrexate conjugate in targeted cancer treatment. Int. J. Mol. Sci. 22, 1748. 10.3390/ijms22041748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H., Lin K. H., Fu B. S., Hsu F. T., Tsai J. J., Weng M. C., et al. (2021). Sorafenib induces apoptosis and inhibits NF-κB-mediated anti-apoptotic and metastatic potential in osteosarcoma cells. Anticancer Res. 41, 1251–1259. 10.21873/anticanres.14882 [DOI] [PubMed] [Google Scholar]
- Wu J., Song T., Liu S., Li X., Li G., Xu J. (2015). Icariside II inhibits cell proliferation and induces cell cycle arrest through the ROS-p38-p53 signaling pathway in A375 human melanoma cells. Mol. Med. Rep. 11, 410–416. 10.3892/mmr.2014.2701 [DOI] [PubMed] [Google Scholar]
- Wu X., Cai Z. D., Lou L. M., Zhu Y. B. (2012). Expressions of p53, c-MYC, BCL-2 and apoptotic index in human osteosarcoma and their correlations with prognosis of patients. Cancer Epidemiol. 36, 212–216. 10.1016/j.canep.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Wu X., Yan L., Liu Y., Shang L. (2020). LncRNA ROR1-AS1 accelerates osteosarcoma invasion and proliferation through modulating miR-504. Aging (Albany NY) 13, 219–227. 10.18632/aging.103498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., He H., Wu B., Wen J., Guo Z., Luo Y., et al. (2017). miR-125b suppresses the aerobic glycolysis of osteosarcoma HOS cells by downregulating the expression of hexokinase-2. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 33, 1365–1370. [PubMed] [Google Scholar]
- Wu Y., Xie Z., Chen J., Chen J., Ni W., Ma Y., et al. (2019). Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol. Cancer 18, 73. 10.1186/s12943-019-1007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zhou Z., Zhang W., Yu Y. (2021). MiR-21-5p inhibition attenuates Warburg effect and stemness maintenance in osteosarcoma cells via inactivation of Wnt/β-catenin signaling. Acta Biochim. Pol. 68, 725–732. 10.18388/abp.2020_5631 [DOI] [PubMed] [Google Scholar]
- Xiao H., Gu Z., Wang G., Zhao T. (2013). The possible mechanisms underlying the impairment of HIF-1α pathway signaling in hyperglycemia and the beneficial effects of certain therapies. Int. J. Med. Sci. 10, 1412–1421. 10.7150/ijms.5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Wei Z., Li Y., Zhou X., Chen J., Wang T., et al. (2018). miR186 functions as a tumor suppressor in osteosarcoma cells by suppressing the malignant phenotype and aerobic glycolysis. Oncol. Rep. 39, 2703–2710. 10.3892/or.2018.6394 [DOI] [PubMed] [Google Scholar]
- Xie D., Zheng G. Z., Xie P., Zhang Q. H., Lin F. X., Chang B., et al. (2017). Antitumor activity of resveratrol against human osteosarcoma cells: A key role of Cx43 and wnt/β-catenin signaling pathway. Oncotarget 8, 111419–111432. 10.18632/oncotarget.22810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Yin J., Jia Q., Wang J., Zou C., Brewer K. J., et al. (2011). Quercetin induces apoptosis in the methotrexate-resistant osteosarcoma cell line U2-OS/MTX300 via mitochondrial dysfunction and dephosphorylation of Akt. Oncol. Rep. 26, 687–693. 10.3892/or.2011.1328 [DOI] [PubMed] [Google Scholar]
- Xie Y., Shi X., Sheng K., Han G., Li W., Zhao Q., et al. (2019). PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 19, 783–791. 10.3892/mmr.2018.9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. G., Xie Y., Dong Q. R. (2013). Inhibition of the mammalian target of rapamycin leads to autophagy activation and cell death of MG63 osteosarcoma cells. Oncol. Lett. 6, 1465–1469. 10.3892/ol.2013.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Herschman H. R. (2019). A tumor agnostic therapeutic strategy for hexokinase 1-null/hexokinase 2-positive cancers. Cancer Res. 79, 5907–5914. 10.1158/0008-5472.CAN-19-1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. N., Yang R. Z., Zheng H. L., Jiang L. S., Jiang S. D. (2020). NDUFA4L2 regulated by HIF-1α promotes metastasis and epithelial-mesenchymal transition of osteosarcoma cells through inhibiting ROS production. Front. Cell. Dev. Biol. 8, 515051. 10.3389/fcell.2020.515051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li Y., Chen X., Xiang F., Deng Y., Li Z., et al. (2021). TGF-β protects osteosarcoma cells from chemotherapeutic cytotoxicity in a SDH/HIF1α dependent manner. BMC Cancer 21, 1200. 10.1186/s12885-021-08954-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Shu B., Tian Y., Wang G., Wang Y., Wang J., et al. (2018). Oleanolic acid induces osteosarcoma cell apoptosis by inhibition of Notch signaling. Mol. Carcinog. 57, 896–902. 10.1002/mc.22810 [DOI] [PubMed] [Google Scholar]
- Yalcin A., Telang S., Clem B., Chesney J. (2009). Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatases in cancer. Exp. Mol. Pathol. 86, 174–179. 10.1016/j.yexmp.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Yan M., Ni J., Song D., Ding M., Huang J. (2015). Activation of unfolded protein response protects osteosarcoma cells from cisplatin-induced apoptosis through NF-κB pathway. Int. J. Clin. Exp. Pathol. 8, 10204–10215. [PMC free article] [PubMed] [Google Scholar]
- Yan X., Yang C., Hu W., Chen T., Wang Q., Pan F., et al. (2020). Knockdown of KRT17 decreases osteosarcoma cell proliferation and the Warburg effect via the AKT/mTOR/HIF1α pathway. Oncol. Rep. 44, 103–114. 10.3892/or.2020.7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Li L., Tong G., Zeng Z., Tan J., Su Z., et al. (2021). Circular RNA circ_001422 promotes the progression and metastasis of osteosarcoma via the miR-195-5p/FGF2/PI3K/Akt axis. J. Exp. Clin. Cancer Res. 40, 235. 10.1186/s13046-021-02027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. S., Lin C. W., Hsieh Y. S., Cheng H. L., Lue K. H., Yang S. F., et al. (2013). Selaginella tamariscina (Beauv.) possesses antimetastatic effects on human osteosarcoma cells by decreasing MMP-2 and MMP-9 secretions via p38 and Akt signaling pathways. Food Chem. Toxicol. 59, 801–807. 10.1016/j.fct.2013.06.028 [DOI] [PubMed] [Google Scholar]
- Yang Q. C., Zeng B. F., Dong Y., Shi Z. M., Jiang Z. M., Huang J. (2007). Overexpression of hypoxia-inducible factor-1alpha in human osteosarcoma: Correlation with clinicopathological parameters and survival outcome. Jpn. J. Clin. Oncol. 37, 127–134. 10.1093/jjco/hyl137 [DOI] [PubMed] [Google Scholar]
- Yang W., Xia Y., Cao Y., Zheng Y., Bu W., Zhang L., et al. (2012). EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell. 48, 771–784. 10.1016/j.molcel.2012.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chin A., Zhang L., Lu J., Wong R. W. (2014). The role of traditional Chinese medicines in osteogenesis and angiogenesis. Phytother. Res. 28, 1–8. 10.1002/ptr.4959 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yujiao W., Fang W., Linhui Y., Ziqi G., Zhichen W., et al. (2020). The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol. Res. 53, 40. 10.1186/s40659-020-00309-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Wei J. J., Wang Z. Y., Ding H. M., Li D., Yan S. C., et al. (2013). Perifosine induces cell apoptosis in human osteosarcoma cells: New implication for osteosarcoma therapy? Cell. biochem. Biophys. 65, 217–227. 10.1007/s12013-012-9423-5 [DOI] [PubMed] [Google Scholar]
- Ye B., Qiao K., Zhao Q., Jiang Z., Hu N., Wang F. (2022). Tanshinone I restrains osteosarcoma progression by regulating circ_0000376/miR-432-5p/BCL2 axis. Mol. Cell. Biochem. 477, 1–13. 10.1007/s11010-021-04257-4 [DOI] [PubMed] [Google Scholar]
- Yi M., Ban Y., Tan Y., Xiong W., Li G., Xiang B. (2019). 6-Phosphofructo-2-kinase/fructose-2, 6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 20, 1–13. 10.1016/j.molmet.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W., Tu M. J., Liu Z., Zhang C., Batra N., Yu A. X., et al. (2020). Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharm. Sin. B 10, 159–170. 10.1016/j.apsb.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Fan J., Zhou W., Ding Q., Zhang J., Wu X., et al. (2017). ERK inhibition sensitizes CZ415-induced anti-osteosarcoma activity in vitro and in vivo . Oncotarget 8, 82027–82036. 10.18632/oncotarget.18303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Lagos D., Enver T., Boshoff C. (2014). Kaposi's sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog. 10, e1004400. 10.1371/journal.ppat.1004400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G. J. (2018). Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 37, 173. 10.1186/s13046-018-0835-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Zhang F., Liu L., Liang Y., Tang X., Peng Y., et al. (2021). The novel prognostic risk factor STC2 can regulate the occurrence and progression of osteosarcoma via the glycolytic pathway. Biochem. Biophys. Res. Commun. 554, 25–32. 10.1016/j.bbrc.2021.03.067 [DOI] [PubMed] [Google Scholar]
- Yuan G., Zhao Y., Wu D., Gao C. (2017). Mir-150 up-regulates Glut1 and increases glycolysis in osteosarcoma cells. Asian pac. J. Cancer Prev. 18, 1127–1131. 10.22034/APJCP.2017.18.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Yu H., Chen J., Song X., Sun L. (2020). Antitumor effect of miR-1294/pyruvate kinase M2 signaling cascade in osteosarcoma cells. Onco. Targets. Ther. 13, 1637–1647. 10.2147/OTT.S232718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Yu H., Chen J., Song X., Sun L. (2018). Knockdown of pyruvate kinase type M2 suppresses tumor survival and invasion in osteosarcoma cells both in vitro and in vivo . Exp. Cell. Res. 362, 209–216. 10.1016/j.yexcr.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Zeng M., Zhou J., Wen L., Zhu Y., Luo Y., Wang W. (2021). The relationship between the expression of Ki-67 and the prognosis of osteosarcoma. BMC Cancer 21, 210. 10.1186/s12885-021-07880-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N., Boutros M. (2017). Wnt signaling in cancer. Oncogene 36, 1461–1473. 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Yan T., Wang K., Huang Z., Liu J. (2017). PI3Kα isoform-dependent activation of RhoA regulates Wnt5a-induced osteosarcoma cell migration. Cancer Cell. Int. 17, 27. 10.1186/s12935-017-0396-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Gou X., He W., Yang H., Yin H. (2020). A glycolysis-based 4-mRNA signature correlates with the prognosis and cell cycle process in patients with bladder cancer. Cancer Cell. Int. 20, 177. 10.1186/s12935-020-01255-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yang L., Geng Y. D., An F. L., Xia Y. Z., Guo C., et al. (2016). Icariside II, a natural mTOR inhibitor, disrupts aberrant energy homeostasis via suppressing mTORC1-4E-BP1 axis in sarcoma cells. Oncotarget 7, 27819–27837. 10.18632/oncotarget.8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Guo X., Feng X., Wang T., Hu Z., Que X., et al. (2017a). MiRNA-543 promotes osteosarcoma cell proliferation and glycolysis by partially suppressing PRMT9 and stabilizing HIF-1α protein. Oncotarget 8, 2342–2355. 10.18632/oncotarget.13672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jiang C., Li M., Wang X., Tian F., Fang X., et al. (2017b). CXCR4 enhances invasion and proliferation of bone marrow stem cells via PI3K/AKT/NF-κB signaling pathway. Int. J. Clin. Exp. Pathol. 10, 9829–9836. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yu X. H., Yan Y. G., Wang C., Wang W. J. (2015). PI3K/Akt signaling in osteosarcoma. Clin. Chim. Acta. 444, 182–192. 10.1016/j.cca.2014.12.041 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Cheng S., Mu Y., Liu Y., Yi X., et al. (2020). LAIR-1 overexpression inhibits epithelial-mesenchymal transition in osteosarcoma via GLUT1-related energy metabolism. World J. Surg. Oncol. 18, 136. 10.1186/s12957-020-01896-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang L., Cao L., Wei T. (2021a). Novel circular RNA circATRNL1 accelerates the osteosarcoma aerobic glycolysis through targeting miR-409-3p/LDHA. Bioengineered 12, 9965–9975. 10.1080/21655979.2021.1985343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wu J., Zhang X., Cao L., Wu Y., Miao X. (2021b). Transcription factor ELK1 accelerates aerobic glycolysis to enhance osteosarcoma chemoresistance through miR-134/PTBP1 signaling cascade. Aging (Albany NY) 13, 6804–6819. 10.18632/aging.202538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun S., Chen J., Ren P., Hu Y., Cao Z., et al. (2014). Oxymatrine induces mitochondria dependent apoptosis in human osteosarcoma MNNG/HOS cells through inhibition of PI3K/Akt pathway. Tumour Biol. 35, 1619–1625. 10.1007/s13277-013-1223-z [DOI] [PubMed] [Google Scholar]
- Zhao S. J., Shen Y. F., Li Q., He Y. J., Zhang Y. K., Hu L. P., et al. (2018). SLIT2/ROBO1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell. Death Dis. 9, 390. 10.1038/s41419-018-0419-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Jiang F., Ge D., Tang J., Chen H., Yang J., et al. (2019). LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed. Pharmacother. 112, 108695. 10.1016/j.biopha.2019.108695 [DOI] [PubMed] [Google Scholar]
- Zheng W. D., Zhou F. L., Lin N. (2015). MicroRNA-26b inhibits osteosarcoma cell migration and invasion by down-regulating PFKFB3 expression. Genet. Mol. Res. 14, 16872–16879. 10.4238/2015.December.14.14 [DOI] [PubMed] [Google Scholar]
- Zheng X. M., Xu C. W., Wang F. (2018). MiR-33b inhibits osteosarcoma cell proliferation through suppression of glycolysis by targeting Lactate Dehydrogenase A (LDHA). Cell. Mol. Biol. 64, 31–35. 10.14715/cmb/2018.64.11.6 [DOI] [PubMed] [Google Scholar]
- Zhong H., de Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., et al. (1999). Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59, 5830–5835. [PubMed] [Google Scholar]
- Zhou R., Zhang Z., Zhao L., Jia C., Xu S., Mai Q., et al. (2011). Inhibition of mTOR signaling by oleanolic acid contributes to its anti-tumor activity in osteosarcoma cells. J. Orthop. Res. 29, 846–852. 10.1002/jor.21311 [DOI] [PubMed] [Google Scholar]
- Zhu R., Li X., Ma Y. (2019). miR-23b-3p suppressing PGC1α promotes proliferation through reprogramming metabolism in osteosarcoma. Cell. Death Dis. 10, 381. 10.1038/s41419-019-1614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. R., Zhou X. Z., Zhu L. Q., Yao C., Fang J. F., Zhou F., et al. (2016). The anti-cancer activity of the mTORC1/2 dual inhibitor XL388 in preclinical osteosarcoma models. Oncotarget 7, 49527–49538. 10.18632/oncotarget.10389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo B., Li Y., Li Z., Qin H., Sun Q., Zhang F., et al. (2015). PI3K/Akt signaling mediated Hexokinase-2 expression inhibits cell apoptosis and promotes tumor growth in pediatric osteosarcoma. Biochem. Biophys. Res. Commun. 464, 401–406. 10.1016/j.bbrc.2015.06.092 [DOI] [PubMed] [Google Scholar]
- Zonta F., Borgo C., Quezada Meza C. P., Masgras I., Rasola A., Salvi M., et al. (2021). Contribution of the CK2 catalytic isoforms alpha and alpha' to the glycolytic phenotype of tumor cells. Cells 10, 181. 10.3390/cells10010181 [DOI] [PMC free article] [PubMed] [Google Scholar]