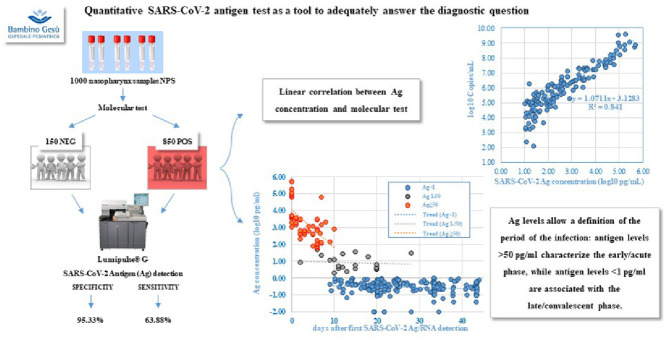
Graphical abstract
Dear Editor,
Fowler et al.1 proposed RNA RT-LAMP as a rapid and accurate tool to promptly identify highly contagious individuals during the pandemic era. In the current vaccination era, it would be useful to have a reliable tool to provide information on the infectivity/contagiousness of individuals.
FDA approved antigen (Ag) test as a fast and convenient alternative to PCR but, as known, this approach can be effective at symptoms onset,2 when viral antigen is abundant,3 otherwise false negative results can occur; moreover, positive antigenic results need to be confirmed by molecular test4.
These assays are mostly qualitative and, even when a numerical value is provided, no straightforward correlation with the virological and clinical parameters has ever been demonstrated.
We evaluated an Ag test based on chemiluminescence (CLEIA), Lumipulse®G SARS-CoV-2 Ag (Fujirebio INC), in an extensive population with different characteristics.
This comparative study included 1000 nasopharyngeal samples (NPS), analyzed during the period fall/winter 2020-2021 at the Virology laboratory of Bambino Gesù Pediatric Hospital.
NPS were collected in Universal Transport Medium (UTM, Copan) and immediately analyzed for molecular SARS-CoV-2 detection by AllplexTM SARS-CoV-2 Assay (Seegene), according to which, 850 samples resulted positive (Cycle threshold, Ct<40) and 150 negative. Referring to an external standard curve (y = −3.179x + 42.28; R2 = 0.939), defined on the basis of serial dilutions of a commercial standard (EDX SARS-CoV-2 Standard, Exact Diagnostics LLC), for each Ct, the corresponding RNA viral load was calculated and expressed in log10 copies/ml.
Antigen detection was performed by Lumipulse®G SARS-CoV-2 Ag, using the automated Lumipulse G1200 System (Fujirebio).
Samples were considered negative when SARS-CoV-2 Ag concentration was <1 pg/ml, in gray zone when ≥1.00 and <10 pg/ml and positive when ≥ 10 pg/ml, according to manufacturer's instruction.
Considering molecular test as the reference standard, Ag showed a specificity of 95.33% (143/150 samples resulted negative, 7/150 resulted positive, 6 of which, included in the gray zone) and a sensitivity of 64% (541/850 samples had a SARS-CoV-2 Ag concentration <1 pg/ml, 131/850 between 1.00 and < 10 pg/ml, and 178/850 ≥ 10 pg/ml).
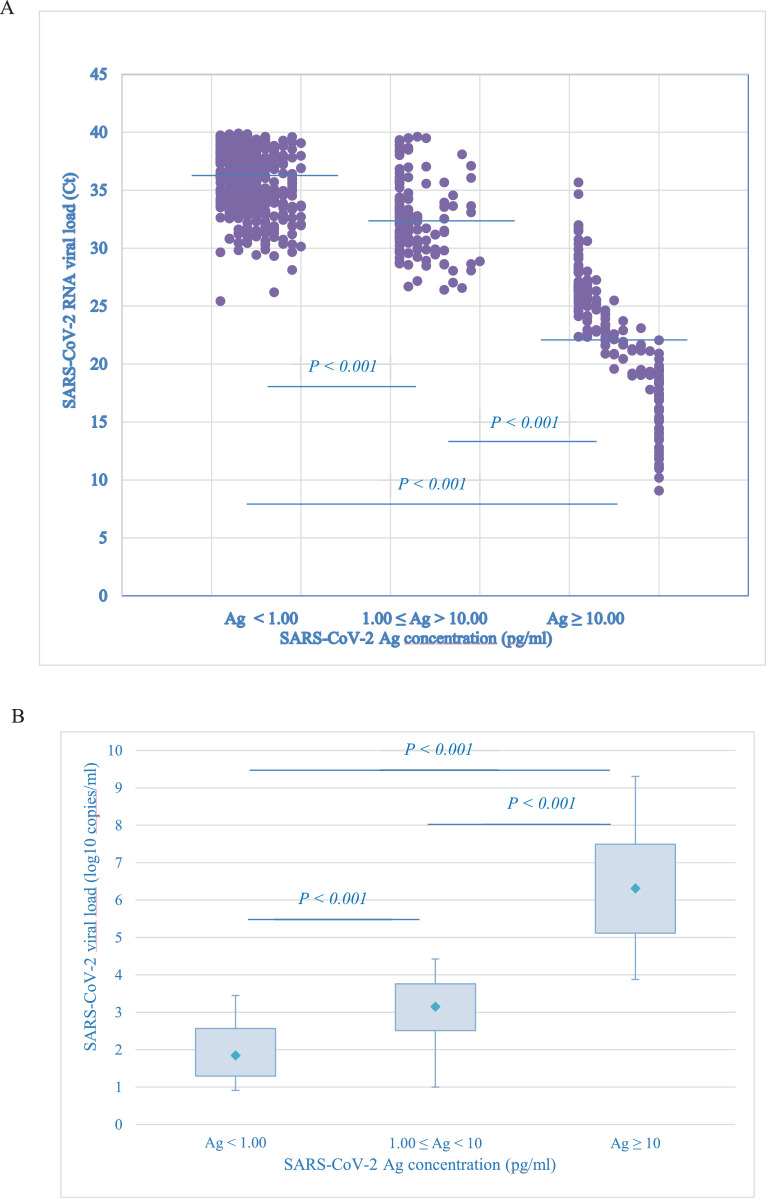
SARS-CoV-2 RNA viral load distribution versus antigen concentration, showed a clear difference in mean CTs and viral loads between the three groups (Fig. 1 ): negative (<1 pg/ml), gray-zone (≥ 1 and < 10 pg/ml) and positive (≥10 pg/ml) antigen, corresponded to 1.84, 3.15 and 6.31 log10 copies/ml, respectively (P value for trend < 0.001).
Fig. 1.
SARS-CoV-2 RNA viral load (Ct or copies/ml) versus Ag concentration (pg/ml)
Samples were grouped according to antigen concentration in: Ag <1.00 pg/ml, Ag ≥ 1.00 and <10 pg/ml and Ag ≥ 10 pg/ml.
(A) Lines represent median SARS-CoV-2 RNA viral load expressed in Ct in the three groups. It was 35.86 (IQR: Ct 34.12–38.19), 32.28 (IQR: Ct 30.37–34.30) and 22.23 (IQR: Ct 18.48–26.05), respectively. P value for trend <0.001
(B) Box plots represent medians and quartiles of SARS-CoV-2 RNA viral load expressed in copies/ml in the three groups. It was 1.84 log10 copies/ml (IQR: 1.29–2.57 log10 copies/ml), 3.15 log10 copies/ml (IQR: 2.51–3.76 log10 copies/ml) and 6.31 log10 copies/ml (IQR: 5.11–7.49 log10 copies/ml), respectively. The ends of the box are the upper and lower quartiles, the median is marked by a rhombus inside the box. The whiskers extend to 5–95%. P value for trend < 0.001.
IQR abbreviation for interquartile range.
Of interest, only 5/541 samples with a negative antigen value showed >4 log10 copies/ml RNA viral load (0.9%). Lumipulse Ag assay showed a remarkable high sensitivity (97.4%) when considering samples with medium-high viral load (>4 log10 copies/ml).
A strong positive correlation (R2 = 0.841) was evident between RNA viral load (log10 copies/ml) and antigen concentration (log10 pg/ml).
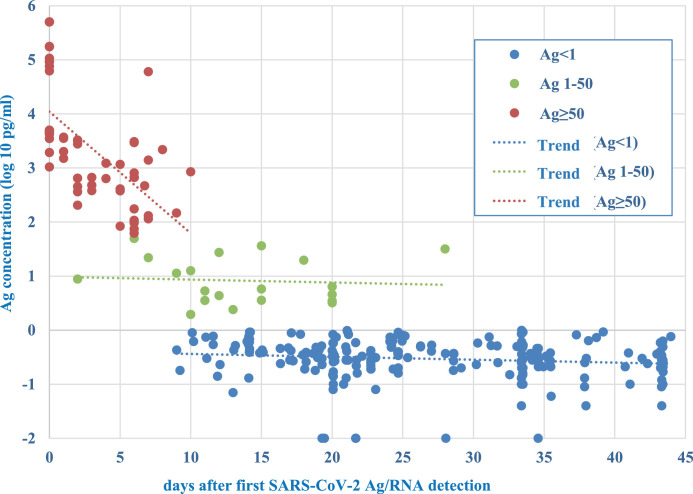
For 278 patients, it was possible to reconstruct the history of infection and to correlate antigen detection with days after first SARS-CoV-2 detection (Fig. 2 ): all 58 samples with antigen levels ≥ 50 pg/ml were collected from patients tested within ten days from first positivity. Of interest, only 3/58 samples referred to antigen detected from 8 to 10 days from first positivity.
Fig. 2.
Antigen concentration and timing of infection
Representation of the day of sample collection in relation to timing of infection and Ag concentration (log10 pg/ml) for 58 samples with Ag ≥ 50 pg/ml (orange dots), 20 samples with Ag ≥1 pg/ml and <50 pg/ml (gray dots) and 207 samples with Ag <1 pg/ml (blue dots). For each group, trend between Ag concentration and days after first SARS-CoV-2 Ag/RNA detection is indicated by dot line. P value for the two major groups < 0.001.
Conversely, 205/207 samples with Ag <1 pg/ml, referred to samples collected later than 10 days from the first SARS-CoV-2 detection, the remaining 2/207 samples were collected at the 9th day from diagnosis.
Finally, the 20 samples with antigen levels ≥1 pg/ml and <50 pg/ml, were distributed over a period ranging from the acute to the convalescence phase.
Both in symptomatic and asymptomatic subjects, SARS-CoV-2 RNA can be detectable up to 3,4 weeks or longer in nasopharynx.5 , 6 During convalescence, in presence of low amount of RNA (Ct > 35), only in 2–5% of cases virus isolation is possible and the risk to transmit infection is negligible.7 However, it is fundamental to find a tool able to give indication on timing of infection.
Our data indicate that Lumipulse antigen quantification allows a definition of the period of the infection: antigen levels > 50 pg/ml characterize the early/acute phase, while antigen levels <1 pg/ml the late/convalescent phase.
The 99% of samples presenting an antigen concentration <1 pg/ml (538/543) had a viral load <4 log10 copies/ml, reported as associated to a post-acute phase8 and were collected in a late/convalescent period of the infection. Analyzing the clinical course of the infection in the patients with a viral load >4 log10 copies/ml (5/543), negative antigen NPS were collected at least two weeks after the first SARS-CoV-2 detection, thus, in the post-acute phase.
Moreover, samples with Ag concentration ≥10 pg/ml showed a strong linear correlation with the corresponding RNA viral load (R2 = 0.841). Since high viral load is related to the early stages of infection,9 we could assume the same for antigen detection. In support to this hypothesis, all samples with antigen levels >50 pg/ml were taken within 10 days from the first positivity (infection onset).
Overall, Lumipulse® Ag results well correlate to the timing of infection, showing a net demarcation (P value < 0.001) between samples with Ag concentration >50 pg/ml, associated with early stages, and those with Ag concentration <1 pg/ml, related to late/convalescent phases.
Our results go beyond the classical utilization of the qualitative antigen test, as reported by Young et al. in their letter,10 and offer a new and clinically relevant role for the quantitative antigen, as a parameter able to define the timing of the infection. This might be particularly useful in those patients with unknown status of infection, and/or for those without a molecular test at symptoms onset, and/or for those asymptomatic with a positive molecular test and/or for vaccinated subjects with low viral shedding.
In conclusion, while real time RT-PCR remains the cornerstone for diagnosis of SARS-CoV-2 infection, Lumipulse quantitative Ag can be useful to define the stage of the disease. In particular, a positive molecular test with a negative Ag test can reasonably indicate a convalescent phase, identifying those subjects with low chances of being contagious.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.12.013.
Appendix. Supplementary materials
References
- 1.Fowler V.L., Armson B., Gonzales J.L., et al. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J Infect. 2021;82(1):117–125. doi: 10.1016/j.jinf.2020.10.039. Jandoi:Epub 2020 Nov 30. PMID: 33271166; PMCID: PMC7703389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.https://abbott.mediaroom.com/2020-08-26
- 3.Spearman P. Diagnostic testing for SARS-CoV-2/COVID19. Curr Opin Pediatr. 2021;33(1):122–128. doi: 10.1097/MOP.0000000000000972. Feb 1. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa T., Fukumori T., Nishihara Y., Sekine T., Okuda N., Nishimura T., et al. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104612. Octdoi:Epub 2020 Aug 25. PMID: 32871543; PMCID: PMC7445490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz A., Marklund E., Andersson M., Nilsson S., Andersson L.M., Lindh M., et al. Upper respiratory tract levels of severe acute respiratory syndrome coronavirus 2 RNA and duration of viral RNA shedding do not differ between patients with mild and severe/critical coronavirus disease 2019. J Infect Dis. 2021;223(1):15–18. doi: 10.1093/infdis/jiaa632. Jan 4doi:PMID: 33020822; PMCID: PMC7665561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han M.S., Choi E.H., Chang S.H., Jin B.L., Lee E.J., Kim B.N., et al. Clinical characteristics and viral RNA detection in children with coronavirus disease 2019 in the Republic of Korea. JAMA Pediatr. 2021;175(1):73–80. doi: 10.1001/jamapediatrics.2020.3988. Jan 1doi:PMID: 32857112; PMCID: PMC7455883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piralla A., Ricchi M., Cusi M.G., Prati P., Vicari N., Scarsi G., et al. Residual SARS-CoV-2 RNA in nasal swabs of convalescent COVID-19 patients: is prolonged quarantine always justified? Int J Infect Dis. 2021;102:299–302. doi: 10.1016/j.ijid.2020.10.072. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. Augdoi:Erratum in: Euro Surveill. 2021 Feb;26(7): PMID: 32794447; PMCID: PMC7427302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71(15):793–798. doi: 10.1093/cid/ciaa345. Juldoi:PMID: 32221523; PMCID: PMC7184442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young B.C., Eyre D.W., Jeffery K. Use of lateral flow devices allows rapid triage of patients with SARS-CoV-2 on admission to hospital. J Infect. 2021;82(6):276–316. doi: 10.1016/j.jinf.2021.02.025. Jundoi:Epub 2021 Mar 1. PMID: 33662408; PMCID: PMC7917467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.