Abstract
The trk1+ gene has been proposed as a component of the K+ influx system in the fission yeast Schizosaccharomyces pombe. Previous work from our laboratories revealed that trk1 mutants do not show significantly altered content or influx of K+, although they are more sensitive to Na+. Genome database searches revealed that S. pombe encodes a putative gene (designated here trk2+) that shows significant identity to trk1+. We have analyzed the characteristics of potassium influx in S. pombe by using trk1 trk2 mutants. Unlike budding yeast, fission yeast displays a biphasic transport kinetics. trk2 mutants do not show altered K+ transport and exhibit only a slightly reduced Na+ tolerance. However, trk1 trk2 double mutants fail to grow at low K+ concentrations and show a dramatic decrease in Rb+ influx, as a result of loss of the high-affinity transport component. Furthermore, trk1 trk2 cells are very sensitive to Na+, as would be expected for a strain showing defective potassium transport. When trk1 trk2 cells are maintained in K+-free medium, the potassium content remains higher than that of the wild type or trk single mutants. In addition, the trk1 trk2 strain displays increased sensitivity to hygromycin B. These results are consistent with a hyperpolarized state of the plasma membrane. An additional phenotype of cells lacking both Trk components is a failure to grow at acidic pH. In conclusion, the Trk1 and Trk2 proteins define the major K+ transport system in fission yeast, and in contrast to what is known for budding yeast, the presence of any of these two proteins is sufficient to allow growth at normal potassium levels.
In cell-walled eukaryotic cells, the intracellular concentration of K+ is quite constant (in the range of 10−1 M), whereas the concentration of Na+ varies from insignificant (less than 10−3 M) to values, in saline environments, very close to those of K+. In terrestrial environments, however, the levels of K+ and Na+ are highly variable; the norm is that potassium in the external media is several orders of magnitude less concentrated than inside the cell, whereas sodium is several times more concentrated. To maintain these asymmetric ionic distributions across the plasma membrane, different types of potassium transporters have evolved in cell-walled eukaryotic cells, all of them driven by the membrane potential created by the H+ pump ATPase (29).
Two different families of potassium transporters responsible for the uptake of the cation have been found in fungi. Transporters of the HAK type are present in mycelial fungi (10) and in the soil yeast Schwannyomyces occidentalis (4). The Hak1 gene product is a high-affinity transporter structurally related to the Escherichia coli Kup protein (4). The second family is defined by the TRK transporters. Genes encoding these transporters (TRK1 and TRK2) were initially isolated from Saccharomyces cerevisiae (7, 12). In S. cerevisiae, TRK1 appears as the major determinant for K+ uptake and probably is the only one participating in potassium uptake in physiological conditions (25). trk1 mutants display a dramatic decrease in potassium influx and a requirement for higher than normal levels of potassium (7, 23), whereas TRK2 contributes much less to the homeostasis of the cation, although this might be merely as a result of a lower expression level (6, 12, 25). trk1 and trk1 trk2 strains display an ectopic low-affinity potassium uptake, which is secondary to the hyperpolarization of the plasma membrane produced by the disruption of the TRK genes (15). The TRK system also allows uptake of Na+, and in the presence of this cation, the transporter increases its ability to discriminate potassium and sodium. Consequently, a proper function of the TRK cation uptake system is crucial for sodium tolerance, as it has been shown that S. cerevisiae trk1 mutants are hypersensitive to sodium ions (8, 9, 17).
Whereas the potassium uptake system in S. cerevisiae has been characterized in some detail, information on this process in the fission yeast Schizosaccharomyces pombe is almost nil. A gene designated SpTRK (referred here as trk1+) was found to encode a protein about 40% similar to S. cerevisiae Trk1 and Trk2 (13, 30). Evidence was also obtained that S. pombe trk1+ encoded a functional potassium transporter, on the basis that its overexpression was able to suppress the K+ uptake defect of a trk1 trk2 S. cerevisiae mutant (13). However, a functional analysis of trk1+ in fission yeast has been addressed only recently, through the construction of an S. pombe trk1 deletion mutant (3). Interestingly, we found that trk1 cells, although displaying a certain degree of sodium sensitivity, grew well even at relatively low K+ concentrations and did not show significantly altered content or influx of this cation. These data suggested that fission yeast ought to encode an efficient alternative K+ transport system. Search of the data bank provided by the systematic S. pombe genome sequencing project at the Sanger Center (Cambridge, England) revealed the existence of a possible homolog for trk1+. We have disrupted this second gene, here designated trk2+, and combined this mutation with that of trk1+ to characterize their contribution to cation homeostasis. Our data indicate that both genes contribute rather equally to potassium influx and that they define the major potassium uptake system in fission yeast.
MATERIALS AND METHODS
Media and growth of E. coli and S. pombe strains.
E. coli NM522 cells were grown at 37°C in Luria-Bertani medium containing 50 mg of ampicillin per ml for plasmid selection. S. pombe cells were grown at 28°C in YES medium (0.5% yeast extract, 3% glucose, 225 mg each of adenine, uracil, and leucine per ml) or essential minimal medium supplemented with the necessary requirements (18). The pH of the medium was buffered at 5.5 with 20 mM MES (2-(morpholino)ethanesulfonic acid). In some experiments mineral medium (containing 30 mM ammonium phosphate and 8 mM ammonium sulfate) with slight modifications (5× vitamins), supplemented with the auxotrophic requirements, was used (1). All S. pombe strains described in this report derive from the wild-type strain 117 (h− ade6-M210 ura4-D18 leu1-32).
Recombinant DNA techniques and gene disruptions.
E. coli cells were transformed by the standard calcium chloride method (28). S. pombe cells were transformed by a modification of the lithium acetate method (19). Standard recombinant DNA techniques were performed essentially as described elsewhere (28).
The construction of strain LB9 (trk1::LEU2) has been described previously (3). Gene disruptions were made by using the one-step gene disruption method (27). Disruption of the gene trk2+ was made as follows. A 4.76-kbp fragment containing the gene was amplified from genomic DNA obtained from strain 117 by PCR with oligonucleotides 5′-GCTGTTGGATGATTGAAGTTTCC-3′ and 5′-CATATAAGCATCATCCCAAATCG-3′ and cloned into plasmid pGEM-T (Promega) via overhanging A/T. The construct was digested with EcoRV and NheI, which remove residues 77 to 448 of the trk2+ coding region. To construct an ura4+ disruption cassette, the ura4+ gene was amplified by PCR from plasmid pUR18 (5) with oligonucleotides 5′-GCTAGCATTCTTTCTCTAAATAG-3′ and 5′-CCATGGTATTTTACATTCATC-3′ (added NheI and NcoI sites, respectively, are underlined) and cloned into pGEM-T (Promega). The 1.6-kbp marker was then released by digestion with NheI and NcoI and cloned into these sites of the above-mentioned trk2+ construct. The disruption cassette (4.4 kbp) was recovered with ClaI/PvuII (which yields the ura4+ marker flanked by 0.463 and 2.381 kbp of trk2+ sequences) and used to transform wild-type and LB9 cells to generate strains NG1 (trk2:: ura4+) and NG2 (trk1::LEU2 trk2:: ura4+), respectively.
To construct a LEU2 deletion cassette for trk2, a plasmid harboring the LEU2 marker (2) was cleaved with NcoI, blunt ended with the Klenow fragment, cleaved with NheI, and then ligated into the EcoRV/NheI sites of the above-mentioned trk2+ plasmid. The disruption cassette (3.7 kbp) was recovered with AccI (which yields the LEU2 marker flanked by 1.143 and 0.951 kbp of trk2+ sequences) and used to transform wild-type cells to yield strain NG3.
In all cases, positive clones were selected by plating in essential minimal medium plates lacking the appropriate supplement, and disruptions were verified at the molecular level by PCR analysis.
Determination of sodium sensitivity of yeast strains.
Sensitivity to sodium chloride was tested in liquid cultures by inoculating 96-well microtiter plates containing medium with different salt concentrations at an initial A620 of 0.015. Cells were grown for about 20 h with shaking, and growth rate determined by measuring the A620 of the cultures. Relative growth was calculated as the ratio between growth in the presence and absence of added salt and expressed as a percentage.
Uptake experiments and cation contents of the cells.
K+-starved cells were obtained by suspending cells with a normal K+ content in K+-free ammonium phosphate medium for 5 h (3). To study uptake of Rb+ (used as an analog of K+), K+-starved cells were resuspended in uptake buffer [10 mM MES brought to pH 5.5 with Ca(OH)2, containing 0.1 mM MgCl2 and 2% glucose]. RbCl (0.1 to 100 mM) was added at time zero; at different times, samples of cells were taken, filtered, and treated for determination of intracellular Rb+. Uptake was linear with time up to 15 min, and the initial uptake rate was obtained from the slope of the line. Kinetics parameters were deduced from Eadie-Hofstee plots of the experimental data.
Potassium loss was determined in cells grown in ammonium phosphate medium supplemented with 50 mM KCl up to an optical density at 550 nm (OD550) of 0.5. Cell were recovered by centrifugation and resuspended in the same medium lacking added KCl. Samples of cells were taken at different times, filtered, and treated for intracellular potassium content determination.
The intracellular cation (Rb+, K+, and Na+) content of the cells was determined as previously described (21, 25). Briefly, samples of cells were filtered, washed with 20 mM MgCl2, and treated with HCl, and the cations were analyzed by atomic absorption spectrophotometry.
RESULTS
S. pombe contains a gene encoding a protein structurally related to trk1+.
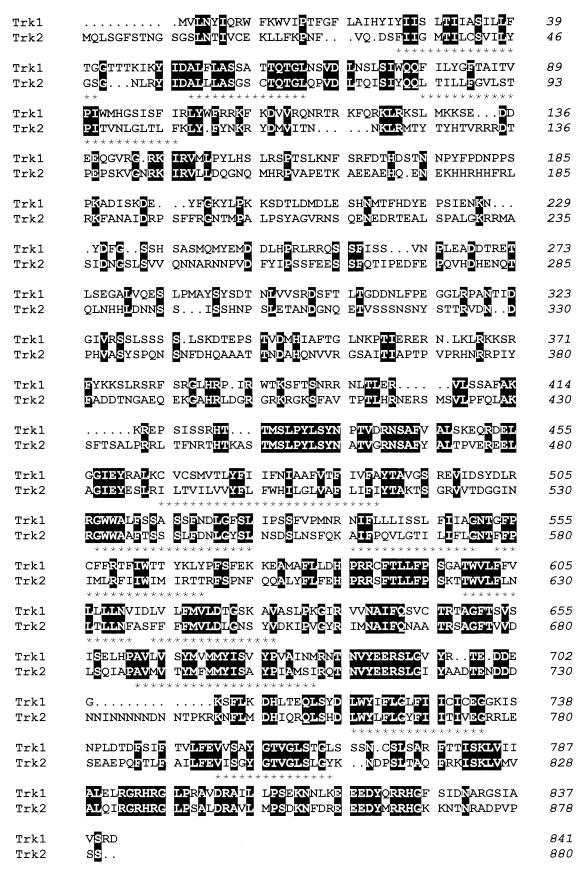
The finding that S. pombe cells lacking a functional trk1+ gene did not show an evident impairment in potassium uptake or altered potassium requirements prompted us to examine the S. pombe genomic database maintained at the Sanger Center. A BLAST search using the entire trk1+-encoded protein revealed the existence of a putative gene, located at chromosome I (accession no. Z68136), that codes for a 880-residue protein 34% identical (almost 50% similar) to the trk1+ gene product. A more detailed analysis (Fig. 1) revealed a number of features that supported the possibility that this putative gene, here called trk2+, might be a homolog of trk1+. For instance, both genes are roughly of the same size and display very similar hydrophobic profiles (not shown). Similarly to S. pombe Trk1 and S. cerevisiae Trk1 and Trk2, 12 transmembrane domains can be predicted for S. pombe Trk2. These elements are placed essentially at the same positions than those predicted for fission yeast Trk1 and define two regions within the Trk proteins: three transmembrane domains lie within the first 150 residues, whereas the remaining are grouped within the second half of the polypeptide. In fact, these two regions display the highest levels of identity between fission yeast Trk1 and Trk2 (45 and 51%, respectively).
FIG. 1.
Sequence comparison of the S. pombe Trk1 and Trk2 putative potassium transporters. Pairwise alignment of the Trk1 and Trk2 amino acid sequences (accession no. P47946 and Q10065, respectively) were performed by the Clustal W method (open gap penalty, 10; extended gap penalty, 0.2). Identical amino acids are boxed and highlighted. The residue number for each protein is indicated at the right. Asterisks denote putative transmembrane domains.
Lack of Trk1 and Trk2 results in increased potassium requirements.
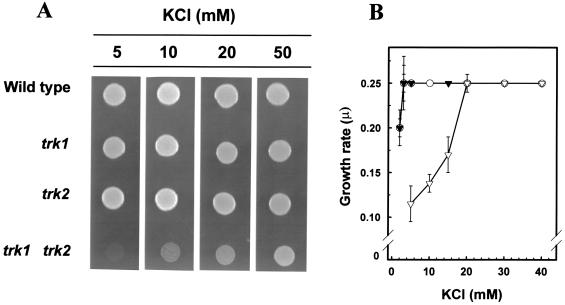
The striking similarities between fission yeast Trk1 and Trk2 led us to isolate trk2+ and to disrupt this gene in wild-type and trk1Δ cells. Here we present a detailed analysis of the growth characteristics of wild-type, trk1, trk2, and trk1 trk2 strains at different potassium concentrations in the medium. Figure 2A shows that under the conditions tested, the single mutants grew similarly to the wild-type strain, whereas growth of the double mutant was severely affected at low potassium. None of the strains were able to grow at external KCl concentrations as low as 1 mM (not shown). To confirm these observations, we performed experiments in liquid media containing different potassium concentrations and calculated the doubling times for the four strains (Fig. 2B). At concentrations as low as 2 mM KCl, wild-type, trk1, and trk2 strains, but not the double mutant, showed significant growth. In addition, the double mutant required about 6.5 times more K+ to reach the maximum growth rate (20 mM versus 3 mM).
FIG. 2.
Potassium dependence of trk mutants. (A) Wild type, LB9 (trk1::LEU2), NG1 (trk2::LEU2), and NG2 (trk1::URA3 trk2::LEU2) cells were grown on YES medium containing 50 mM KCl to an OD550 of 0.5, centrifuged, washed, and resuspended in sterile water to an OD550 of 0.05; 7 μl was deposited on ammonium phosphate plates containing the indicated concentrations of KCl. Growth was scored after 60 h. (B) Cells (wild type, ●; trk1, ○; trk2, ▾; trk1 trk2, ▿) were grown and processed as described above. Liquid ammonium phosphate medium containing different amounts of KCl was inoculated with the cells (OD550 of 0.05), and growth was monitored by measuring the OD550. The growth rate constant (μ) is defined as ln2 divided by the duplication time (h).
Fission yeast shows a biphasic kinetics for rubidium influx that is altered in trk1 trk2 mutants.
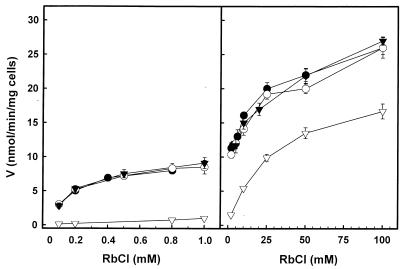
The increased K+ requirements of the trk1 trk2 strain raised the question of the characteristics of K+ transport in the different strains used in this study. Figure 3 shows the kinetics of Rb+ (used as an analog of potassium ions) influx in K+-starved cells. Interestingly, the kinetics of transport were essentially identical in the wild type and in the single mutants and showed a biphasic pattern: a transport process saturated in the micromolar range and a second phase observed at millimolar concentrations of Rb+. Both processes exhibit Michaelis-Menten kinetics (Vmax of 11 nmol/mg/min and K0.5 of 0.25 mM for the first phase; Vmax of 17 nmol/mg/min, and K0.5 of 6.8 mM for the second phase). However, in the case of the double mutant, the characteristics of Rb+ transport were completely different. Rb+ influx was hardly observed at micromolar concentrations, and the kinetics of transport was monophasic, with Vmax of 17 nmol/mg/min and K0.5 of 17 mM. Therefore, the lack of growth at low K+ concentrations observed for the trk1 trk2 strain can be explained on the basis of a failure to take up potassium when the external levels of this cation are too low.
FIG. 3.
Initial rates of Rb+ uptake as a function of Rb+ concentration. K+-starved cells (wild type, ●; trk1, ○; trk2, ▾; trk1 trk2, ▿) were resuspended in uptake buffer (see Materials and Methods). The required RbCl amounts were added, and samples of cells were taken at different times (0 to 15 min). The intracellular content of Rb+ was determined as described in the text.
trk1 trk2 mutants are highly sensitive to sodium ions.
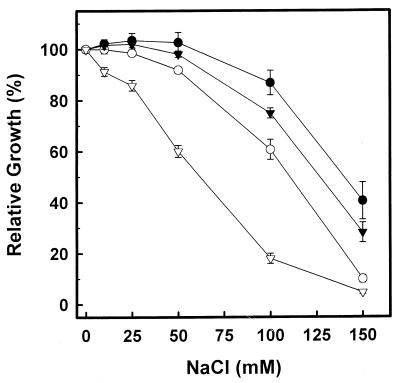
Previous work on characterization of the trk1 mutant (3) identified a phenotype of increased sensitivity to Na+ ions. We have extended this study to the whole set of trk mutants (Fig. 4). Under the conditions tested in this work, the wild-type strain shows a 50% inhibitory concentration (IC50) for NaCl of about 140 mM. This value is only slightly reduced (128 mM) in the trk2::LEU2 cells (strain NG1). The use of the marker ura4+ instead of LEU2 for disruption of trk2 (strain NG3) gave essentially the same results (not shown). The change observed is somewhat less prominent than that observed upon disruption of trk1 (IC50 of 110 mM). Interestingly, the double mutant was very sensitive to Na+ (IC50 of 62 mM), indicating that the lack of both Trk components results in a dramatic effect on growth under sodium stress. This growth defect is in keeping with the observation that cells lacking both trk1 and trk2 genes fail to maintain a proper Na+/K+ intracellular ratio. This has been evaluated by growing wild-type and double mutant NG2 cells in the presence of 10 mM KCl plus 100 mM NaCl. Under these conditions, the intracellular levels of sodium and potassium in wild-type cells were 35 ± 6 and 430 ± 30 nmol/mg of cells, respectively. Under the same conditions, NG2 cells accumulated 95 ± 8 nmol of sodium per mg of cells, with an intracellular concentration of potassium ions of only 160 ± 15 nmol/mg of cells.
FIG. 4.
Sensitivity to Na+ ions of trk mutants. Cells (wild type, ●; trk1, ○; trk2, ▾; trk1 trk2, ▿) were grown on YES medium supplemented with different concentrations of NaCl. Sodium sensitivity was determined as described in Materials and Methods. Data are means ± standard errors of the means from five to seven experiments.
Lack of Trk transporters might result in hyperpolarization of the plasma membrane.
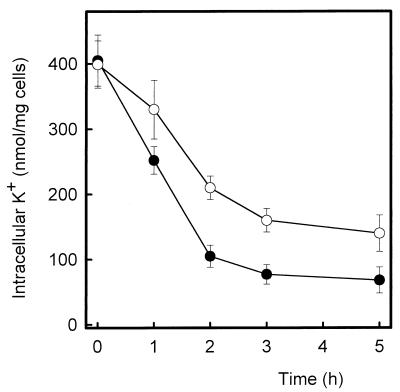
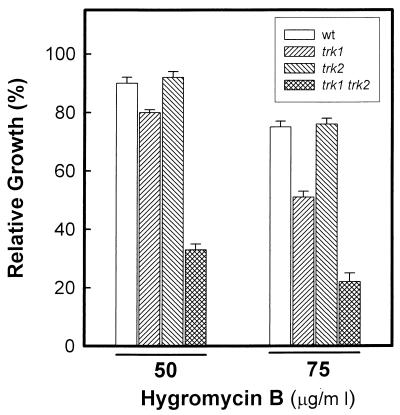
Wild-type, trk1, trk2, and trk1 trk2 cells contained similar amounts of internal potassium when grown at nonlimiting potassium concentrations (around 400 nmol of K+/mg of cells). When these cells are suspended in K+-free medium, potassium is immediately lost; after 3 to 5 h, the equilibrium is reached and internal potassium levels become stable (Fig. 5). In these conditions the cells were viable, and interestingly, wild-type and single mutants retained less potassium than the double mutant. This result could be explained on the basis of a hyperpolarized state of the plasma membrane of the trk1 trk2 strain. The resistance of yeast cells to the antibiotic hygromycin B has been also related to their membrane potential. If our hypothesis were correct, NG2 cells would display an altered tolerance to this compound. We tested (Fig. 6) hygromycin B tolerance in wild-type and trk mutants and found that the trk1 trk2 double mutant was clearly more sensitive to the antibiotic. Interestingly, whereas trk2 cells behaved like the wild-type strain, trk1 cells appeared to be somewhat more sensitive to the drug. It must be noted that these effects cannot be attributed to an altered function of the H+-ATPase, since measurement of proton efflux yielded essentially identical results (30 ± 2 and 28 ± 4 nmol of H+/mg of cells/min for wild-type and NG2 cells, respectively).
FIG. 5.
trk1 trk2 mutants retain more potassium than the wild-type strain. Cells (wild type, ●; trk1 trk2, ○) were grown in YES medium supplemented with 50 mM KCl and resuspended in ammonium phosphate K+-free medium. Samples of cells were taken at different times (up to 5 h). The intracellular K+ content was determined as described in Materials and Methods. Data are means ± standard errors of the means from three to six independent experiments.
FIG. 6.
trk1 trk2 mutants are sensitive to hygromycin B. Cells were inoculated (initial OD550 of 0.05) in ammonium phosphate medium supplemented with 50 mM KCl and grown in the absence or the presence of the indicated concentrations of hygromycin B. Sensitivity to the antibiotic was determined by measuring the OD550 of the cultures after 24 h of incubation. Relative growth was calculated as the ratio between growth in the presence and absence of the antibiotic and expressed as a percentage. Data are means ± standard errors of the means from three independent experiments. wt, wild type.
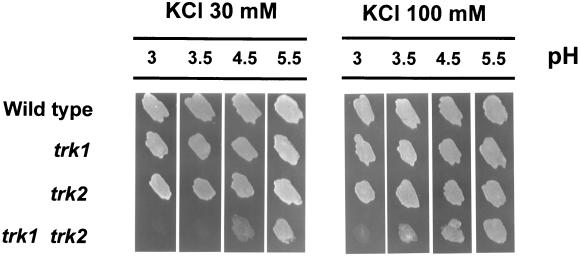
Low pH sensitivity requires the absence of both trk1 and trk2.
It has been reported that in budding yeast, a phenotype associated with lack of the TRK1 gene (but not of TRK2) is the hypersensitivity to low pH. This phenotype appears to be the consequence of the impaired potassium uptake, because high concentrations of K+ restore growth of these cells under low-pH conditions (11). We have tested the effect of acidic pH on growth of cells lacking the Trk components. The results (Fig. 7) indicate that in S. pombe, the lack of a single trk gene does not impair growth even at a pH as low as 3. However, the trk1 trk2 double mutant fails to grow even at pH 4.5 when the amount of potassium ions in the medium is relatively low (30 mM). Our data indicate that these cells cannot grow at pH 3 even in the presence of 100 mM KCl. Therefore, in contrast with the evidence described for budding yeast, lack of both trk genes is necessary to confer low pH sensitivity to fission yeast cells.
FIG. 7.
trk1 trk2 mutants are hypersensitive to acidic pHs. Cells were grown and processed as described for Fig. 2A; 7 μl of the cell suspension was deposited on ammonium phosphate plates adjusted at the desired pH with Tris citrate buffer (pH 3, 3.5, and 4.5) or MES (pH 5.5) and supplemented with 30 or 100 mM KCl, as indicated. Growth was scored after 60 h.
DISCUSSION
Our recent finding that disruption of the trk1+ gene in S. pombe did not result in significant changes in potassium requirements or in potassium influx (3) indicated that despite the ability of the Trk1 protein to behave like a potassium transporter in a heterologous system, alternative potassium uptake systems should exist in fission yeast. These observations drew our attention to a related, uncharacterized gene (trk2+) found during the S. pombe systematic sequencing project. We show here that deletion of trk2+ does not result in changes in potassium requirements or in potassium influx. Furthermore, sodium tolerance in trk2 mutants decreases only marginally. In fact, deletion of this gene fails to significantly alter sodium tolerance in the sodium-hypertolerant pzh1 (2) background (not shown). Interestingly, simultaneous deletion of both trk1 and trk2 results in a strong requirement for potassium in the medium, defective rubidium uptake, and dramatically increased sodium sensitivity. These results indicate that in S. pombe the Trk1 and Trk2 proteins have largely equivalent functions, since deletion of both genes is needed to observe a clear-cut mutant phenotype. This situation is different from what has been described for budding yeast, where, as mentioned above, the role of one of the Trk proteins (Trk1) is largely predominant. On the other hand, the strong Na+ sensitivity observed in trk1 trk2 mutants provides further support for the notion that in fission yeast, an efficient potassium transport is an important factor for sodium tolerance, a circumstance that has been previously documented for budding yeast (8).
We have analyzed the kinetics of rubidium transport in S. pombe and observed that this is a biphasic process, with a high-affinity and a low-affinity component. This is again different from the situation described for S. cerevisiae, which under the same conditions shows a high-affinity, monophasic potassium transport (24, 26), or even for other yeast types such as S. occidentalis (5) or Debaryomyces hansenii (21). Interestingly, a biphasic uptake process is well documented in fungi (22) and higher plants (14). Our data clearly show that trk1 and trk2 are similarly responsible for the high-affinity uptake in fission yeast, since this component is fully present in the single mutants and completely absent in cells lacking both genes. It should be stressed that our data do not rule out the possibility that the low-affinity process observed in trk1 trk2 mutants is different in nature from that observed in wild-type cells.
A remarkable observation is that after K+ starvation, trk1 trk2 cells retain more potassium than do wild-type cells. This could be explained if one assumes that the defect in potassium influx results in a hyperpolarization of the plasma membrane. This effect has been recently documented for S. cerevisiae (15) and is supported by our observation that S. pombe trk1 trk2 cells are more sensitive to hygromycin B, an aminoglycoside drug for which the resistance of the cells depends on their membrane potential (16, 20).
In conclusion, in this report we show that the kinetics of rubidium transport in fission yeast is more similar to that of higher plants than to that of other yeast cells such as S. cerevisiae, on the basis that it exhibits a biphasic process. The high-affinity component of this process is driven by the Trk1 and Trk2 transporters, which define the major uptake system in fission yeast. However, and in contrast to what has been described for budding yeast, the roles of the fission yeast Trk proteins are essentially equivalent.
ACKNOWLEDGMENTS
Nestor Gómez and Fernando Calero contributed equally to this work.
The contribution of L. Balcells at the earliest stage of this work as well as the skillful technical help of Anna Vilalta and Mireia Zaguirre are acknowledged. We thank Alonso Rodríguez-Navarro for fruitful discussion and the Sanger Center for maintaining and allowing free access to the S. pombe genome data bank.
This work was supported by grants PB98-0565-C4-02 and PB98-1036 (Dirección General de Investigación Científica y Técnica, Spain) to J.A. and J.R., respectively, by an Ajut de Suport als Grups de Recerca de Catalunya (SGR97-127) from the Generalitat de Catalunya to J.A., and by grant BIO4-CT97-2210, from the European Union to J.R.
REFERENCES
- 1.Asensio J, Ruiz-Argüeso T, Rodríguez-Navarro A. Sensitivity of yeasts to lithium. Antonie Leeuwenhoek. 1976;48:61–71. doi: 10.1007/BF00399443. [DOI] [PubMed] [Google Scholar]
- 2.Balcells L, Gómez N, Casamayor A, Clotet J, Ariño J. Regulation of salt tolerance in fission yeast by a PPZ-like Ser/Thr protein phosphatase. Eur J Biochem. 1997;250:476–483. doi: 10.1111/j.1432-1033.1997.0476a.x. [DOI] [PubMed] [Google Scholar]
- 3.Balcells L, Calero F, Gómez N, Ramos J, Ariño J. The Schizosaccharomyces pombe Pzh1 protein phosphatase regulates Na+ ion influx in a Trk1 independent fashion. Eur J Biochem. 1999;260:31–37. doi: 10.1046/j.1432-1327.1999.00129.x. [DOI] [PubMed] [Google Scholar]
- 4.Bañuelos M A, Klein R D, Alexander-Bowman S J, Rodríguez-Navarro A. A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 1995;14:3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbet N, Muriel W J, Carr A M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- 6.Bihler H, Gaber R F, Slayman C L, Bertl A. The presumed potassium carrier Trk2p in Saccharomyces cerevisiae determines an H+-dependent, K+-independent current. FEBS Lett. 1999;447:115–120. doi: 10.1016/s0014-5793(99)00281-1. [DOI] [PubMed] [Google Scholar]
- 7.Gaber R F, Styles C A, Fink G R. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2848–2859. doi: 10.1128/mcb.8.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez M J, Luyten K, Ramos J. The capacity to transport potassium influences sodium tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1996;135:157–160. doi: 10.1111/j.1574-6968.1996.tb07982.x. [DOI] [PubMed] [Google Scholar]
- 9.Haro R, Bañuelos M A, Quintero F J, Rubio F, Rodríguez-Navarro A. Genetic basis of sodium exclusion and sodium tolerance in yeast. A model for plants. Physiol Plant. 1993;89:868–874. [Google Scholar]
- 10.Haro R, Sainz L, Rubio F, Rodríguez-Navarro A. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:511–520. doi: 10.1046/j.1365-2958.1999.01192.x. [DOI] [PubMed] [Google Scholar]
- 11.Ko C H, Buckley A M, Gaber R F. TRK2 is required for low affinity K+ transport in Saccharomyces cerevisiae. Genetics. 1990;125:305–312. doi: 10.1093/genetics/125.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko C H, Gaber R F. TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichtenberg-Fraté H, Reid J D, Heyer M, Höfer M. The SpTRK gene encodes a potassium-specific transport protein THKp in Schizosaccharomyces pombe. J Membr Biol. 1996;152:169–181. doi: 10.1007/s002329900095. [DOI] [PubMed] [Google Scholar]
- 14.Maathuis F J M, Sanders D. Mechanisms of potassium absortion by higher plant roots. Physiol Plant. 1996;96:158–168. [Google Scholar]
- 15.Madrid R, Gómez M J, Ramos J, Rodríguez-Navarro A. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 16.McCusker J H, Perlin D S, Haber J E. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza I, Rubio F, Rodríguez-Navarro A, Pardo J M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–87966. [PubMed] [Google Scholar]
- 18.Moreno S, Amar K, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 19.Norbury C, Moreno S. Cloning cell cycle regulatory genes by trans-complementation in yeast. Methods Enzymol. 1997;283:44–59. doi: 10.1016/s0076-6879(97)83006-6. [DOI] [PubMed] [Google Scholar]
- 20.Perlin D S, Brown C L, Haber J E. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J Biol Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- 21.Prista C, Almagro A, Loureiro-Dias M C, Ramos J. Physiological basis for the high salt tolerance of Debaryomyces hansenii. Appl Environ Microbiol. 1997;63:4005–4009. doi: 10.1128/aem.63.10.4005-4009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos J, Rodríguez-Navarro A. Rubidum transport in Neurospora crassa. Biochim Biophys Acta. 1985;815:97–101. [Google Scholar]
- 23.Ramos J, Contreras P, Rodríguez-Navarro A. A potassium transport mutant of Saccharomyces cerevisiae. Arch Microbiol. 1985;143:88–93. [Google Scholar]
- 24.Ramos J, Haro R, Rodríguez-Navarro A. Regulation of potassium fluxes in S. cerevisiae. Biochim Biophys Acta. 1990;1029:211–217. doi: 10.1016/0005-2736(90)90156-i. [DOI] [PubMed] [Google Scholar]
- 25.Ramos J, Alijo R, Haro R, Rodríguez-Navarro A. TRK2 is not a low-affinity potassium transporter in Saccharomyces cerevisiae. J Bacteriol. 1994;176:249–252. doi: 10.1128/jb.176.1.249-252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Navarro A, Ramos J. Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988;947:1–28. doi: 10.1016/0304-4157(88)90017-2. [DOI] [PubMed] [Google Scholar]
- 30.Soldatenkov V A, Velasco J A, Avila M A, Dritschilo A, Notario V. Isolation and characterization of SpTRK, a gene from Schizosaccharomyces pombe predicted to encode a K+ transporter protein. Gene. 1995;161:97–101. doi: 10.1016/0378-1119(95)00274-a. [DOI] [PubMed] [Google Scholar]