Abstract
Bacteria in biofilm formations are up to 1000 times less susceptible to antibiotics than their planktonic counterparts. Recognition of the role of biofilms in ∼80% of chronic infections, their contribution to bacterial tolerance and development of antimicrobial resistance, and thus the search for compounds with antibiofilm properties, has increased greatly in recent years. The need for robust experimental methods is therefore critical but currently undermined by inappropriate controls when dimethyl-sulfoxide (DMSO) is used to enhance test compound solubility. DMSO is known to have a limited effect on planktonic growth, but emerging data indicates that the solvent can affect biofilm formation even at low concentrations. Here, we present both a literature review on the application of DMSO in in vitro antibiofilm studies, as well as a series of experiments and Bayesian hormetic dose-response modelling to define the effects of DMSO alone and in combination with standard antibiotics using two clinically important biofilm-forming bacteria. DMSO has been used in 76 published studies to solubilise a wide variety of synthesised and natural products, including plant extracts, isolated secondary metabolites, modified lead molecules and proteins, in in vitro antibiofilm assays. DMSO solvent concentrations to which biofilms were exposed ranged between <1 and 100% but unfortunately, 35% of articles did not specify the DMSO concentrations used, 50% of articles did not include solvent controls and, of those that did, 26% did not specify control concentrations, 47% did not report or discuss control data, and 53% omitted media controls. In a further 12 studies, DMSO is used as a biofilm treatment, demonstrating the antibiofilm properties of this solvent at higher concentrations. We provide evidence that DMSO (between 0.03 and 25%) significantly inhibits biofilm formation in Pseudomonas aeruginosa, but not Streptococcus pneumoniae, and acts synergistically with standard antibiotics at very low concentrations (<1%). Interestingly, intermediate concentrations of DMSO (∼6%) strongly promote the growth of P. aeruginosa biofilms. As the research community strives to identify bioactive antimicrobial compounds, there is a need for increased scientific rigour when using DMSO as a solvent in antibiofilm assays.
Keywords: Biofilm methods, Lipophilic compounds, Antibacterial, Drug discovery, In vivo screening, Bioassay, Solvent, DMSO, Hormesis
1. Introduction
Most antibiotic agents have been developed based on demonstrated effectiveness against free-living (planktonic) cells in vitro, but rarely do bacteria exist in this state [[1], [2], [3]]. Rather, the majority of bacterial cells are present as biofilms, that is, communities of microorganisms (bacteria and/or fungi) within a viscous, self-secreted matrix of polysaccharides and proteins, termed extracellular polymeric substance (EPS) [[4], [5], [6]]. This EPS matrix facilitates surface adhesion, gene transfer, cell-cell communication (quorum sensing), sorption of nutrients and water, and thus affords bacterial resistance to mechanical stressors, host immune defences and antimicrobial interventions [4,7]. Biofilms play a significant role in infections of the respiratory system (e.g., pneumonia, otitis media, sinusitis, and recurrent infections in cystic fibrosis and chronic obstructive pulmonary disease patients), as well as those associated with wounds and medical implants or devices (e.g., urinary catheters, prosthesis, pacemakers, intrauterine devices, and respiratory apparatus) [1,2,8]. Nonetheless, clinical biofilm-specific treatment options are limited; despite considerable research efforts in this field, high-dose antimicrobial combination therapy remains the recommended approach [9]. To address the issues of antimicrobial tolerance [10] and development of resistance [11,12] there is not only a need for new antimicrobial agents in the traditional bactericidal sense, but for compounds with novel mechanisms of action to attenuate biofilm formation and persistence (e.g., EPS disruption and dispersal, quorum sensing inhibition, antibiotic potentiation).
Solubility is one of the most important parameters in bioactivity screening, determining cellular (and ultimately, systemic) bioavailability and pharmacological response to a compound [13,14]. However, around half of all newly-discovered natural and synthetic products are hydrophobic, requiring either structural or chemical modifications to introduce them into a cellular system [13]. The addition of an organic solvent, most often dimethyl-sulfoxide (DMSO; Fig. 1) (and less often ethanol, methanol, acetone or N,N-Dimethylformamide) is therefore necessary.
Fig. 1.
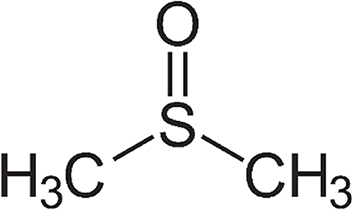
Dimethyl-sulfoxide (DMSO; [CH3]2S): the solvent-of-choice for the dissolution of small hydrophobic drug molecules.
DMSO is an aprotic organosulfur molecule with an amphipathic nature making it ideal for dissolving poorly soluble polar and non-polar drug molecules [15] (Fig. 1). During the past century, DMSO has been widely employed in toxicology and experimental pharmacology and is recommended, among in vitro and in vivo studies and standard protocols for antimicrobial screening of lipophilic synthetic and natural products [[16], [17], [18], [19], [20], [21], [22]]. DMSO is generally accepted as nontoxic below 10% (v/v) and in practice, the use of DMSO is regarded so ubiquitous and safe that applied concentrations (which are usually below 2%) are often unreported and biological effects are assumed negligible [15,23,24]. However, research is emerging to suggest that DMSO may be a potent biofilm inhibitor (and in some cases in fact promote biofilm formation) at very low concentrations (<2%), potentially interfering with interpretation of antibiofilm assay results [21,[25], [26], [27]].
Research interest into screening compounds for antibiofilm activity has increased markedly in recent years. This is anticipated to continue as biofilms are further understood. But, routine in vitro antimicrobial susceptibility testing (i.e., planktonic growth inhibition) has limited value in anticipating the effectiveness of a given agent against biofilm infection [28]. A timely review of methodological approaches and limitations in antibiofilm compound screening is therefore important, moving toward the development of standard antibiofilm screening protocols and the identification of bioactive compounds. Here, we provide a literature review on the application of DMSO in peer-reviewed in vitro antibiofilm studies. We also experimentally define the effects of DMSO alone and the effect of DMSO on the activity of standard antibiotics using two clinically important biofilm-forming bacteria. By using these discrete but complementary lines of evidence, we first identify then demonstrate the problem. We aim to encourage discussion and development of standardised approaches to solvent use and controls in antibiofilm assays which will improve the reliability and interpretation of data among the biomedical research community.
2. Methods
2.1. Literature review
The Scopus database was searched by article title, abstract and keywords without date limits (as of September 2021) using the string “biofilm AND (dimethyl sulfoxide OR dmso)” returning 191 hits. These were screened by relevance and finally 88 articles were reviewed in which DMSO was used as a solvent (n = 76 articles) or itself as a treatment (n = 12 articles) (Supplementary spreadsheet). Important variables extracted included compound-DMSO treatment preparation, experimental controls and control reporting, data calculation and results. Descriptive statistics were calculated in Excel.
2.2. Experimental work
2.2.1. Chemicals and media
Standard antibiotics– ampicillin trihydrate and gentamicin sulfate (CAS 7177-48-2 and 1405-41-0, Sigma-Aldrich) were reconstituted in phosphate buffered saline (PBS) to 64 μg/mL stocks for use against S. pneumoniae and P. aeruginosa, respectively [22]. DMSO, crystal violet, acetic acid, PBS, and other reagents used in this study were HPLC grade (Sigma-Aldrich). Cation-adjusted Mueller Hinton II Broth (CAMHB) (BD BBL™, Thermo Fisher) was prepared from dehydrated powder in MilliQ water and sterilised as per manufacturer instructions. CAMHB was used as growth media for P. aeruginosa. Defibrinated horse blood (Edwards Group, Australia) was lysed over five freeze-thaw cycles before addition to CAMHB at 5% v/v for use with Streptococcus pneumoniae.
2.2.2. Bacterial culture conditions
We used S. pneumoniae laboratory strain ATCC 51916. P. aeruginosa 385 was a mucoid strain serotype 2, phagetype 21/44/109/119X/1214 typed by the Central Public Health Laboratory, London, United Kingdom) originally isolated from sputum of a chronically infected patient with cystic fibrosis. Cryopreserved bacteria were maintained at −80° until being revived on horse blood agar (HBA) and incubated for 20–22 h at 37 °C with 5% CO2. Isolated colonies were suspended in 1 mL media and grown to the exponential phase in a shaking incubator at 37 °C with 5% CO2 for 3–4 h until blank-corrected absorbance was 0.06–0.17 for P. aeruginosa and 0.1–0.2 for S. pneumoniae; these absorbances were equivalent to ∼108 CFU/mL, predetermined experimentally and checked for suspensions prepared separately for each assay. Stock was diluted in media to achieve a working suspension of 106 CFU/mL, finally reduced to 5 × 105 CFU/mL in assays.
2.2.3. Antimicrobial-antibiofilm coupled assay
The liquid growth microdilution method using 96-well plates was applied as per Clinical and Laboratory Standards Institute (CLSI) procedures [22] for antimicriobial assays with some modifications for determination of biofilm inhibition. Three experiments were run for each bacteria species; these were designed to test: 1) exposure to DMSO alone across a range of DMSO concentrations (0.05–25% DMSO in log10 serial dilutions), 2) antibiotics (0.03–16 ug/ml) in combination with fixed 1% and 2% DMSO, and, 3) antibiotics (0.03–16 ug/ml) with varying proportional changes in DMSO concentration (0.03–12.5%), respectively.
The plates were prepared as follows: Experiment 1) 50 μL media was added to all wells, then 50 μL of pure DMSO was added to column A and ten-fold serial dilutions were made to consecutive rows before adding 50 μL of bacterial suspension; Experiment 2) 49 μL media and 48 μL media were added to rows D-E and G-H, respectively before adding 50 μL of antibiotic stocks to wells in column A and serially diluting 10-fold. 50 μL of bacterial suspension were added to all treatments and finally1 μL DMSO (1%) was added to rows D-E and 2 μL DMSO (2%) to rows G-H; Experiment 3) antibiotic stocks were reconstituted in 1:1 PBS:DMSO then plates were prepared with media and bacteria as per experiment 1. All plates included two positive-growth media controls, two blank media controls, and duplicate ten-fold dilutions of antibiotics (0% DMSO) as negative controls– control data were pooled across all experiments for comparisons.
Plates were incubated for 18–22 h at 37 °C with 5% CO2 then read spectrophotometrically at OD 600 to measure antimicrobial activity. The same plates were evaluated for inhibition of biofilm formation, similar to [[29], [30], [31]]. Planktonic bacteria and media were carefully aspirated from wells before 200 μL PBS was added to each well then discarded and repeated to rinse. Plates were sprayed liberally with 80% v/v ethanol and allowed to dry for 20 min to fix adhered biofilms, which were then stained with 200 μL 0.1% crystal violet over 20 min. Excess stain was discarded and plates were again twice rinsed with 200 μL PBS and tapped dry onto absorbent pads. Stained biofilms were solubilised with 200 μL 5% v/v glacial acetic acid and OD was measured at 570 nm.
Treatments were duplicated in each assay. Assays were repeated seven times in Experiment 1, five times in Experiment 2, and three times in Experiment 3 (total 15 assays per species). DMSO controls were not used to correct data because their purpose was to test the hypothesis that the addition of DMSO to standard antibiotic treatments results in significantly different dose-responses compared to antibiotics without DMSO, thereby demonstrating the importance of appropriate controls and data corrections. Blanks were used to correct all raw measurements and means of duplicate data from each assay were used in the statistical analysis. MIC values were recorded as the minimum treatment concentrations inhibiting growth relative to untreated (media-only) blanks, as per [22]. Biofilm inhibition was calculated as a percentage relative to the positive-growth control (100-[treatment/positive control] × 100).
2.2.4. Statistical analysis
We modelled the absorbance and inhibition data following inverse U-shaped and U-shaped Bayesian hormetic dose-response curves, respectively [32]. For the inverse U-shaped absorbance curves, the response (y) of observation i, for i = 1,2, …,n observations, was assumed normally distributed with expected value following:
And for the U-shaped inhibition curves, the response (y) of observation i, for 1,2, …,n, following [33]:
where xi is the concentration of the treatment, c and d are the expected response at infinite and 0 (baseline) concentration, respectively, f is the hormesis parameter (hormesis present for f > 0), is a rate parameter, e is the median effective concentration (EC50) in the absence of hormesis (providing a lower bound on the EC50 in the presence of hormesis), and b is the slope at e [32]. For Experiment 1 (DMSO-alone), we fit species-specific curves with x representing DMSO concentration for both absorbance and inhibition. Experiment 2 (antibiotic with fixed 1% and 2% DMSO) and Experiment 3 (antibiotics with variable DMSO concentrations) were analysed together and presented along with antibiotic controls (0% DMSO) to facilitate comparison of the effects of DMSO. We fit species- and treatment-specific curves with x representing respective antibiotic concentrations, for both absorbance and inhibition. For each model, we included covarying random trial effects. We used NIMBLE 0.12.2 [34] through R 4.1.0 [35] for Bayesian inference using Markov chain Monte Carlo (MCMC) methods to estimate posterior distributions of all model parameters, except for , which was fixed at 0.5 [32]; the R script is available at https://github.com/mhollanders/dmso. We used weakly informative priors: for absorbance models, c was fixed at 0, d ∼ N(0, 12), and f ∼ Half-N(0, 12); for inhibition modes, c was fixed at 100, d ∼ N(0, 1002), and f ∼ Exponential(1); for inhibition models, c was fixed at 100, d ∼ N(0, 1002), and f ∼ Exponential(0.01). For both models, e ∼ Uniform(0, xmax), where xmax is the highest concentration of x used, and b ∼ Exponential(0.1) For the residual error and random trial effects, we specified Exponential(1) and Exponential(0.1) priors for absorbance and inhibition models, respectively, and we used LKJ priors on the Cholesky factors of correlation matrices of random trial effects. To test for the presence of hormesis, we included reversible jump MCMC (RJMCMC; [36]) on the hormesis parameter. Briefly, RJMCMC samples models with different dimensions and can be used to test for the inclusion of model parameters. In the presence of hormetic effects, f will be included in the model and can take non-zero positive values; in the absence of hormesis, f will be excluded from the model and the corresponding value of f will be 0, replacing the models above with a standard log-logistic dose-response curve. We ran two chains for 60,000 iterations, discarded 10,000 as burn-in, and thinned remaining samples by 10, yielding 10,000 posterior samples per model. Convergence was confirmed by visual inspection of traceplots and Rhat values. We reported parameters as medians and 95% (equal-tailed) credible intervals (CIs), with RJMCMC inclusion probabilities where applicable. To identify differences in EC50 baseline inhibition between different treatments, we compared the posterior distributions of e and d, respectively, between groups in respective models. Differences were considered significant when 95% CIs of the differences did not overlap 0.
3. Results and discussion
3.1. Use of DMSO in the in vitro antibiofilm literature
3.1.1. Overall trends
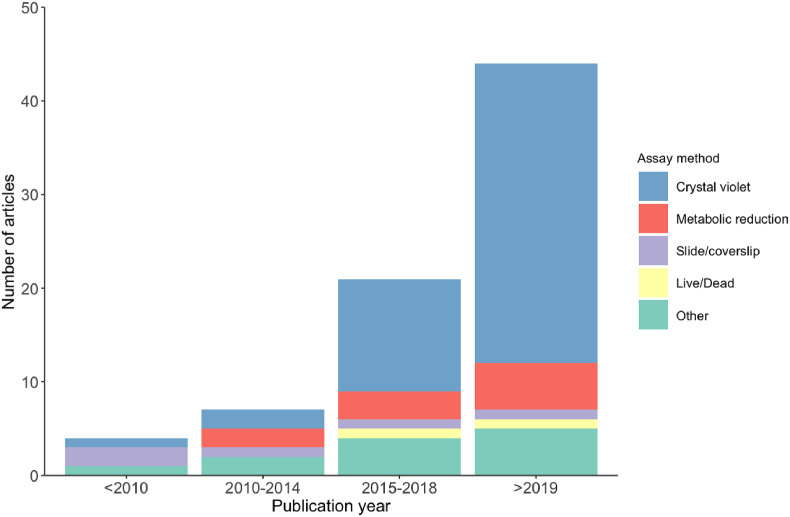
A total of 88 articles are included in this review, three quarters of which (n = 65) were published within the past five years (Fig. 2). The most common method (61%, n = 69 articles) used to assess antibiofilm activity was by colorimetric quantification of the formation/degradation of biofilms established in 96-well plates using crystal violet to stain adhered cells (the crystal violet assay) (Fig. 2). Alternatively, 14% of articles (n = 12) determined biofilm cell viability by colorimetric metabolic reduction assays using XTT ([2,3-bis-[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide]), MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) or resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide) (Fig. 2; Supplementary spreadsheet). Other methods included visualising biofilms grown on glass slides or coverslips by confocal or scanning electron microscopy [37,38], quorum sensing assays [[39], [40], [41]], and novel experimental approaches such as scraping or sonicating biofilms off growth surfaces and culturing on agar to determine viable colonies [37,[42], [43], [44], [45]].
Fig. 2.
In vitro antibiofilm studies, and the dominant assays used, incorporating DMSO published over the past decade (n = 88) reflecting an overall increase in biofilm research in recent years.
3.1.2. Articles using DMSO as a solvent
Since 2001, there have been at least 76 published articles using treatments incorporating DMSO to enhance the solubility of over 1500 different natural or synthetic products for in vitro antibiofilm screening (Table 1; Supplementary spreadsheet). Types of tested products range from crude or semi-purified extracts, mainly of plant material, to isolated phenols, terpenes, nanoparticles, proteins, aldehydes, azoles and fatty acids; targets have included a range of human and veterinary bacterial pathogens, as well as fungal Candida sp. (Supplementary spreadsheet). The concentrations of DMSO used to enhance extract/compound solubility to which biofilms were exposed ranged from <1 to 100% but were typically less than 10% (n = 43 articles) and most commonly less than 1% (n = 28 articles) (Table 1). While pure DMSO was often used to prepare stock solutions of test substances, only final exposure concentrations were of interest and were almost always lower than stocks following dilution with bacterial suspensions in media [[46], [47], [48]] (Supplementary spreadsheet). However, in assays measuring degradation of preformed biofilms whereby test solutions were added to effectively dry, adhered bacterial cells after media was discarded, high (50–100%) final DMSO exposures occurred if stocks were applied undiluted [49,50] (Table 1; Supplementary spreadsheet). Final DMSO exposure concentrations were directly specified or otherwise calculable based on information provided in methods in 66% of articles (n = 50) (Table 1). Final DMSO exposure concentrations were neither directly stated nor calculable and therefore considered altogether unspecified in 34% (n = 26 articles) (Table 1). In recognition of the potential confounding effects of the solvent, methods should always directly state the final DMSO concentrations to which treated biofilms are exposed and/or provide solution volumes and dilution factors to enable their calculation(the former is preferrable to avoid potential error when interpreting methods).
Table 1.
DMSO solvent concentrations and experimental controls reported in articles testing natural or synthetic extracts/compounds for antibiofilm activity in vitro.
| % DMSO used to solubilise treatments | Total number of articles | % of total articles (n = 76) |
|---|---|---|
| Stated or calculable: | ||
| <1% | 21 | 28 |
| 1.0–2.0% | 6 | 8 |
| 2.1–5.0% | 12 | 16 |
| 5.1–10.0% | 4 | 5 |
| 10.1–50.0% | 2 | 3 |
| 50.1–100% | 5 | 7 |
| Total stated or calculable | 50 | 66 |
| Unspecified: | 26 | 34 |
| Total: | 76 | |
| Was at least one DMSO control present? | % of total articles (n = 76) | |
| Yes | 38 | 50 |
| % of articles by QI | ||
| No | 38 | 50 |
| % of articles by QI | ||
| If present, was DMSO control concentration provided? | % of articles with DMSO control present (n = 38) | |
| Yes | 28 | 74 |
| No | 10 | 26 |
| If present, was DMSO control data reported? | % of articles with DMSO control present (n = 38) | |
| Yes | 20 | 53 |
| No | 18 | 47 |
| If DMSO control present, was there an additional media control? | % of articles with DMSO control present (n = 38) | |
| Yes | 18 | 47 |
| No | 20 | 53 |
Concerningly, only half of the studies using DMSO as a solvent included at least one DMSO control in the experimental design (Table 1). Those that did not include relevant DMSO controls (n = 38 articles) often used low concentrations (≤1% DMSO) to deliver treatments, which likely contributed to a reduced perceived need for a solvent control [47,[51], [52], [53], [54], [55]]. Studies that used higher DMSO concentrations without solvent controls were more concerning [49,50,[56], [57], [58], [59], [60], [61], [62], [63], [64]] as were studies that did not provide concentrations of DMSO used in treatments and did not include solvent controls [43,[65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77]]. Media-only control-treated biofilms were frequently used in the calculations [50,56,57,78,79] although it is most appropriate to compare treatments delivered with DMSO to DMSO-only control-treated biofilms normalised to 100% growth as in [41,[80], [81], [82]]. Alternatively, the range of treatments may be delivered with and without DMSO, as per Campbell et al. [83] who applied a range of ethyl 4-ethoxybenzoate (EEB) concentrations in media as well as the same EEB concentrations in media with 1% DMSO, to Staphylococcus aureus, Bacillus subtilis and Streptococcus mutans biofilms. EEB with 1% DMSO resulted in significant biofilm inhibition relative to the positive media control whereas EEB without DMSO did not, and the authors noted this potentiation effect (pp. 6).
Of the studies that did employ at least one DMSO control, 26% (n = 10 articles) did not specify the control concentration. However, when the concentration of DMSO used to solubilise treatments was provided, it can be reasonably assumed that the same DMSO concentration was used for the control [[84], [85], [86]] and vice versa [82,87]. More problematic were studies wherein both solvent (test solution) and control DMSO concentrations were not clearly specified [45,73,81,88]. While statements were occasionally included to imply that the experiments were controlled to a degree, greater transparency regarding the control conditions and any findings relative to the treatments would be best. It was also problematic when DMSO concentrations in treatments varied and a single control concentration was used [41,89], or when the DMSO exposure and control concentrations did not match [46]. Moving forward, DMSO control concentrations should reflect all DMSO concentrations that biofilms are exposed to by means of delivering the extract/compound of interest. Sometimes, DMSO solvent and/or control details were provided for certain assays/components of the study, but information was lacking as to whether these conditions were identical for the biofilm inhibition assays [48,65,69,80]; it can be assumed that conditions were the same, yet this is not always obvious or reliable. Despite these studies being evidently comprehensive, there is value in clearly explaining antibiofilm assay method to reduce any ambiguity in how to replicate it and interpret the results.
Of the studies that did include DMSO controls, 47% (n = 18) did not report or discuss the control data (even as supplementary) and additional media controls were frequently excluded (53% of studies; [[90], [91], [92]]), such that it was firstly difficult to interpret exactly how antibiofilm activity (i.e., % inhibition data) was calculated and secondly if any effects of DMSO were observed (Table 1; Supplementary spreadsheet). Whilst statements regarding the inclusion of a DMSO control and lack of effect thereof can provide reassurance that the potential for confounding solvent effects was considered, we did not consider ambiguous statements without supporting data as sufficient control reporting for the purpose of this review [43,54,84,93]. In future, the percent inhibition of treatments delivered in DMSO should always be benchmarked against the relevant solvent controls (not the media control [94]) to avoid potentially incorrect estimations of effective concentrations and all data should be reported.
Often, antibiotic controls were used in the antimicrobial assay but no equivalent control was used in the antibiofilm component [61,66,68,84]. This indicates an additional need to routinely include standard antibiofilm agents or antibiotics specifically known to inhibit biofilms, and ideally develop species-specific reference data similar to MIC breakpoints [16], for data quality assurance and to facilitate inter-laboratory comparison of data.
The issues aforementioned were detected across a wide variety and prestige of journals (Q1-Q4, Supplementary spreadsheet). This implies a fairly widespread lack of appreciation for the potential confounding effects of DMSO in in vitro antibiofilm experiments and a need to revisit the importance of adequate controls in any biological assay. It is necessary to collate further evidence for the direct effects of DMSO on biofilms and rigorously investigate potential synergistic effects when DMSO is used to solubilise potential or established antimicrobial agents.
3.1.3. Articles using DMSO as a treatment
There have been 12 studies (one published as early as 1989 [95], another in 1999 [96], but others only more recently – from 2014 onward [Supplementary spreadsheet]), that have used DMSO alone as a biofilm treatment thereby acknowledging it's potential antibiofilm properties (Table 2). DMSO applied between 0.01 and 100% has shown significant effects on biofilm formation in clinically important species including Gram-negative P. aeruginosa (4 studies), Escherichia coli (4 studies) and Salmonella typhimurium (4 studies), and Gram-positive Staphylococcus sp. (2 studies) (Table 2). Yahya et al. [26] forthrightly state that “DMSO…is regarded as an effective antibiofilm agent” (pp. 29) albeit at a relatively high concentration of 32%. In early antimicrobial/antibiofilm experiments Sampaio et al. [42] used DMSO to solubilise a methanolic plant extract but in a subsequent biofilm model used water as opposed to DMSO “to avoid interference [of the solvent] in cell adhesion” (pp. 293). Several authors appear to have discovered an effect of DMSO somewhat accidently, including Guo and colleagues [25] who unexpectedly found that DMSO (2%, v/v) exhibited significant antagonistic activities on the quorum sensing system of P. aeruginosa and warned that “[the impact of DMSO] on the virulence factors of bacterial pathogens complicates its usage as a solvent in biological and medicinal studies. At the least, the use of caution over such an effect is warranted when DMSO is used as a vehicle in antibiotic research” (pp. 7168). To our knowledge, Guo et al. [25] are the only other authors to identify and report the issue emphasised by this article.
Table 2.
Studies using dimethyl-sulfoxide (DMSO) as a treatment in in vitro antibiofilm assays: general results and proposed mechanisms of action. ATCC: American Type Culture Collection; EPS: exopolysaccharide; QS: quorum sensing.
| Target species | DMSO conc. | DMSO effect on biofilms | Proposed mechanism of action | Ref |
|---|---|---|---|---|
| Burkholderia cepacia; B. pyrrocinia (clinical isolate); Pseudomonas aeruginosa | 10–100% | DMSO dissociated double-stranded segments of cepacian (EPS) molecules leading to dispersion of polymeric chains and formation of a porous biofilm | DMSO induces disruption of polymer chain aggregation in polysaccharides | [102] |
| Staphylococcus aureus strains 72, 80, 510, ATCC 29213 | 1/1, 1/3, 1/9 v/v | When directly adding DMSO to a biofilm, a complete disruption of this biofilm was macroscopically observed | Not provided | [96] |
| P aeruginosa (PAO1), Escherichia coli | 2% v/v (10% v/v for model) | DMSO significantly attenuated a range of QS-controlled virulence factors and biofilm formation at a non-inhibitory growth concentration; DMSO did not affect antibiotic MICs up to 2%; DMSO treatment reduced mortality in a murine model of P. aeruginosa wound infection | Reduction of C4-HSL (N-butanoyl-l-homoserine lactone) involved in las and rhl QS systems was the main influence on virulence factors; “[the impact of DMSO] on virulence factors of bacterial pathogens complicates its usage as a solvent in biological and medicinal studies.” | [25] |
| E. coli UTI89, UTI89csgA, MC4100, MC4100csgA | 0.05–4% | At low concentrations (<1%) DMSO had no effect, at high concentrations (2–4%) DMSO (and ethanol, but to a lesser extent) increased cellular agglutination in broth and increased curli expression (adhesion molecule) to enhance biofilm formation | Effects currently not understood at the molecular and atomic level; “DMSO was not being metabolized or transformed by E. coli." | [108] |
| E. coli (n = 10), Klebsiella pneumoniae (n = 10), and P. aeruginosa (n = 8) isolates | 30% | DMSO significantly reduced preformed biofilm biomass and viable colony forming units; more effective than other tested agents (hypochlorous agents, ozone, antimicrobial peptide mimic); different efficacy depending on bacteria species | Not provided | [113] |
| Pseudomonas fluorescens (H2S) | 2% and 5% | “Treatment with DMSO produced different results in separate experiments, causing a slight decrease in biofilm thickness at 2% and at times an increase at 5% (data not shown)" | Not provided | [95] |
| Shewanella sp. (20 strains from various environmental and clinical sources) | 0.55–70 mM | DMSO (35 mM) increased biofilm production up to 3-fold in some isolates, but not in others, under different conditions- addition of nitrates (electron acceptors) resulted in a 3-fold reduction in biofilm formation at the same DMSO concentration | DMSO reduction is variable among certain isolates; respiration-driven biofilm formation may constitute a mechanism of niche colonization by specialized strains; a terminal DMSO reductase is involved in extracellular respiration and uses sulfoxides and N-oxides as substrates | [110] |
| Staphylococcus epidermidis (ATCC 35984) | 0.0039–1% | Biofilm formation stimulated by 12–42% (p < 0.05) with DMSO | Likely strain dependent; recommend use of <1% methanol as solvent as opposed to DMSO | [109] |
| Corynebacterium pseudotuberculosis (clinical isolate); Salmonella typhimurium ATCC 14028 | 50, 25, 12.5, 6.25, 3.13, 1.56% | DMSO significantly inhibited C. pseudotuberculosis biofilm formation at all concentrations relative to the control but the effect was similar between concentrations. | DMSO may inhibit functional linkages between glycolytic enzymes (hub proteins) | [103] |
| C. pseudotuberculosis (clinical isolate); S. typhimurium (ATCC 14028) | 50, 25, 12.5, 6.25, 3.13, 1.56% | All DMSO concentrations significantly inhibited C. pseudotuberculosis biofilm but not S. typhimurium and was the more effective than EDTA and EtOH | “Inhibition of bacterial growth by DMSO is known to involve membrane perturbation." | [114] |
| S. typhimurium (ATCC 14028) | 1–32% | DMSO (32%) inhibited pellicle formation, biofilm viability, biofilm biomass and several important components of the EPS matrix; planktonic bacteria were affected differentially by different DMSO concentrations | “Protein interaction network analysis identified several biological pathways to be affected, including glycolysis, PhoP–PhoQ phosphorelay signalling and flagellar biosynthesis; DMSO may inhibit multiple biological pathways to control biofilm formation." | [100] |
| E. coli ATCC 1299, P. aeruginosa (ATCC 10145), and S. typhimurium (ATCC 14029) | 1–32% | Significantly lower EPS protein conc with DMSO alone (32%) and afatanib + DMSO treatments; “planktonic fractions were affected differentially by DMSO” - killing effect at 10% DMSO | “DMSO, but not afatinib, is regarded as an effective antibiofilm agent [at 32%]. Chemical modification of EPS matrix may account for, at least, a part of the mode of action of DMSO.” | [26] |
DMSO has long been used in biomedical research and practice to treat a range of diseases; it is approved for cell/organ cryopreservation and treatment of interstitial cystitis, and to assist with topological and anti-inflammatory treatments in veterinary medicine [15,[97], [98], [99]]. But, the clinical utility of DMSO is controversial since the specific pharmacological mechanisms are largely unknown [99,100]. Nevertheless, the bioactivities of DMSO are considered functions of the polarity of the molecule and its ability to scavenge reactive oxygen species [101]. With reference to mechanisms of antibiofilm activity, it has been suggested that DMSO may disrupt polymer chain aggregation in polysaccharides (i.e., the major component of bacterial EPS matrices, and yeast cell walls comprised of glucan and mannan) [26,102], chemically reduce critical components of quorum sensing (i.e., regulating biofilm proliferation) pathways [25], inhibit functional protein linkages [100,103], and alter the electrostatic charge, solubility and interactions between polysaccharides [104] and proteins [105] to weaken the overall adhesion forces between the biofilm and the surface. This is in general agreement with certain clinical antibiofilm agents which effectively disperse/disassemble EPS components [106]. However, more research is warranted to fully ascertain the species-specific mechanisms by which DMSO affects biofilm formation as this information is mostly speculative at present (Table 2).
The relationship between DMSO concentration and biofilm inhibition/degradation (and planktonic growth above threshold concentrations) is, in most cases, inverse and high concentrations (generally >10%) show strong antimicrobial and antibiofilm activity [107] (Fig. 3; Fig. 4). However, microorganisms may respond differently to DMSO, particularly at low and intermediate concentrations, depending on the specific strain, their metabolic state, and growth conditions [108]. Four studies reported that relatively low DMSO concentrations actually stimulated biofilm formation: 2% DMSO caused a decrease in Pseudomonas fluorescens (Gram-negative) biofilm thickness, but 5% DMSO caused an increase in biofilm thickness [95]; Staphylococcus epidermidis (Gram-positive) biofilms were stimulated by 0.0039–1% DMSO [109]; 35 mM DMSO (equivalent to ∼0.25%) showed both stimulatory and inhibitory activity on Shewanella sp. (Gram-negative) biofilms, depending on the strain [110]; and 2–4% DMSO stimulated biofilm formation in uropathogenic Escherichia coli, but <0.1% DMSO showed no effect [108] (Table 2). This could reflect differential induction of biofilm formation in response to variable respiration pathways i.e. certain strains of bacteria may be able to use DMSO as an electron acceptor, particularly under anerobic conditions [108]. As well, there is limited evidence suggesting that low DMSO concentrations may provide a protective effect from reactive oxygen species, increasing bacterial tolerance to antimicrobial agents [111]. Our investigations confirmed strong stimulation of P. aeruginosa biofilm formation at 6.3% DMSO before causing significant inhibition (Fig. 4). This is a hormetic response whereby low doses of a xenobiotic agent induce a response opposite to that at high doses resulting in a biphasic dose-response relationship, which is a frequently encountered phenomenon in (eco)toxicological literature [112]. A hormetic or otherwise variable dose-response would only be detected/reported in the literature if a wide range of DMSO concentrations were tested, so it may be the case that the stimulatory effects of DMSO noted in selected articles were a function of limited DMSO test concentration ranges. Further studies are required to investigate why DMSO exerts a hormetic effect on P. aeruginosa biofilm formation. Regardless of the rationale or underlying mechanisms, the effect of DMSO is not always predictable nor linear and accordingly each DMSO concentration used in antibiofilm assays requires a discrete control.
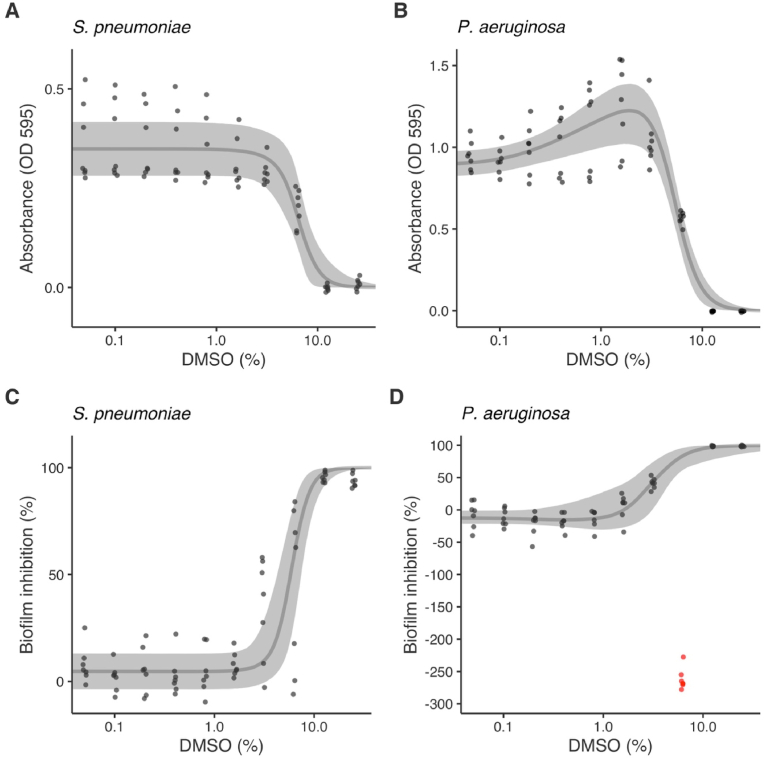
Fig. 3.
Predicted dose-response curves (median and 95% credible intervals) for antimicrobial activity (as absorbance) (A–B) and biofilm inhibition (as percentage reduction relative to 100% positive-growth control) (C–D) of Streptococcus pneumoniae and Pseudomonas aeruginosa as functions of dimethyl-sulfoxide (DMSO) concentration in media. Points are means of blank corrected measured data from repeated assays (n = 7 per species). The curve of D was fitted excluding points marked in red. (For interpretation of the references to color in this figure legend, the reader is referred to the online version of this article.)
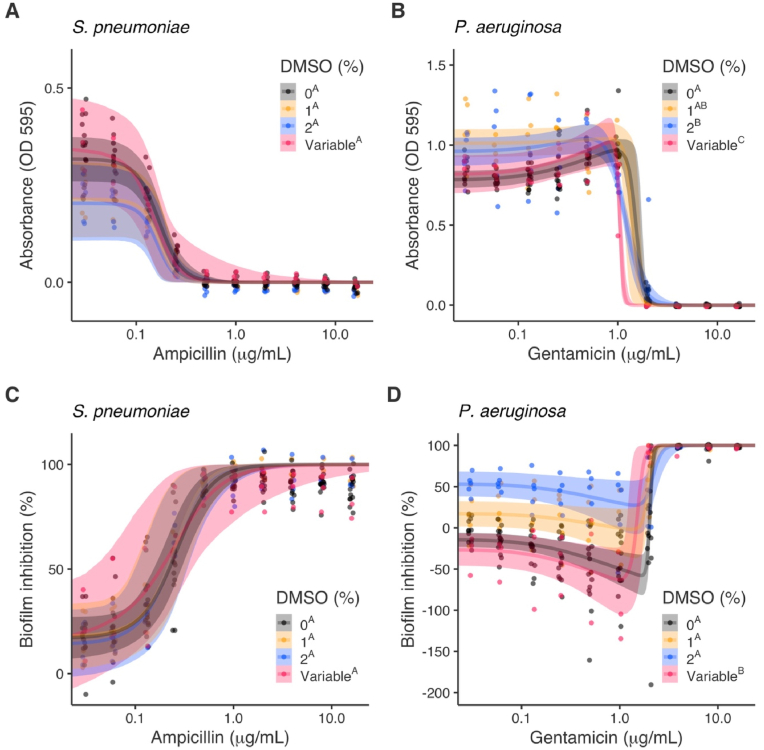
Fig. 4.
Predicted dose-response curves (median and 95% credible intervals) for antimicrobial activity (as reduction in absorbance) (A–B) and biofilm inhibition (as percentage reduction relative to 100% positive-growth control) (C–D) of Streptococcus pneumoniae and Pseudomonas aeruginosa as functions of antibiotic (ampicillin and gentamicin) concentration with different dimethyl-sulfoxide (DMSO) additions (i.e., 0% DMSO; antibiotic with 1% and 2% DMSO added to all treatments, antibiotic with variable proportionate changes in DMSO). Points are means of blank corrected measured data from repeated assays (n = 5 for 1% and 2% DMSO, n = 3 for variable DMSO, n = 15 for 0% DMSO). Superscripted letters in the legends show significant differences in 50% effective concentration (EC50) values between DMSO treatments.
3.2. Experimental work
3.2.1. QA/QC
In all assays, MICs were 0.25 μg/mL ampicillin for S. pneumoniae and 2 μg/mL gentamicin for P. aeruginosa providing data quality assurance (CLSI MIC breakpoints ≤0.25 μg/mL ampicillin for Streptococcus sp., ≤4 μg/mL gentamicin for sensitive P. aeruginosa) [16].
3.2.2. Experiment 1: The effect of DMSO alone
3.2.2.1. Antimicrobial activity (planktonic growth inhibition)
The MIC for DMSO alone was 12.5% (±0.00) against both S. pneumoniae and P. aeruginosa. Using Bayesian hormetic dose-response curves [32] we estimated antimicrobial EC50 values for DMSO alone of 6.44% (95% credible intervals [CI]: 4.037, 7.374) for S. pneumoniae and 5.34% (95% CI: 4.704, 8.052) for P. aeruginosa, and the susceptibility of the species was similar (difference in EC50 of 1.11% [95% CI: -0.77, 2.93]) (Fig. 3A–B, Table 3). A hormetic response was detected (Reversible Jump Markov chain Monte Carlo [RJMCMC] inclusion: 100%, f = 0.72 [95% CI: 0.29, 1.18]) for P. aeruginosa with DMSO at intermediate concentrations causing an increase in planktonic growth before it declined at ≥6.3% DMSO (Fig. 3A–B). There were no hormetic responses detected for S. pneumoniae (RJMCMC inclusion: 0%).
Table 3.
Posterior distributions of 50% effective concentrations (EC50) values (median, standard deviation [SD], and 95% credible intervals [CI]) for DMSO and antibiotics (Amp: ampicillin, Gent: gentamicin) estimated using Bayesian hormetic dose-response curves based on in vitro antimicrobial-antibiofilm assays using Streptococcus pneumoniae and Pseudomonas aeruginosa (per species: n = 7 for DMSO alone, n = 5 for DMSO 1% and 2%, n = 3 for variable DMSO, n = 15 for ampicillin/gentamicin alone).
| Bacteria sp. | Experiment | Treatment | EC50 values for antimicrobial activity |
EC50 values for biofilm inhibition (%) |
||||
|---|---|---|---|---|---|---|---|---|
| Median | SD | 95% CI | Median | SD | 95% CI | |||
| S. pneumoniae | 1 | DMSO alone | 6.474% DMSO | 0.797 | [4.915, 8.088] | 5.904% DMSO | 0.815 | [4.308, 7.561] |
| 2 | Amp alone (0% DMSO) | 0.168 μg/mL Amp | 0.015 | [0.139, 0.200] | 0.258 μg/mL Amp | 0.039 | [0.183, 0.336] | |
| Amp + 1% DMSO | 0.168 μg/mL Amp | 0.03 | [0.114, 0.229] | 0.246 μg/mL Amp | 0.068 | [0.112, 0.382] | ||
| Amp + 2% DMSO | 0.16 μg/mL Amp | 0.03 | [0.106, 0.223] | 0.235 μg/mL Amp | 0.064 | [0.110, 0.366] | ||
| Amp + variable DMSO | 0.143 μg/mL Amp | 0.031 | [0.081, 0.206] | 0.196 μg/mL Amp | 0.087 | [0.038, 0.377] | ||
| P. aeruginosa | 1 | DMSO alone | 5.226% DMSO | 0.442 | [4.377, 6.141] | 2.907% DMSO | 0.785 | [1.374, 4.431] |
| 2 | Gent alone (0% DMSO) | 1.505 μg/mL Gent | 0.121 | [1.329, 1.759] | 2.032 μg/mL Gent | 0.041 | [1.953, 2.114] | |
| Gent + 1% DMSO | 1.378 μg/mL Gent | 0.134 | [1.116, 1.672] | 1.943 μg/mL Gent | 0.084 | [1.752, 2.087] | ||
| Gent + 2% DMSO | 1.172 μg/mL Gent | 0.047 | [1.088, 1.272] | 2.179 μg/mL Gent | 0.149 | [1.960, 2.535] | ||
| Gent + variable DMSO | 1.048 μg/mL Gent | 0.029 | [1.004, 1.118] | 1.504 μg/mL Gent | 0.154 | [1.203, 1.793] | ||
3.2.2.2. Biofilm inhibition
DMSO inhibited the formation of S. pneumoniae and P. aeruginosa biofilms with EC50 values of 5.904% (95% CI: 4.308, 7.561) and 2.907% (95% CI: 1.374, 2.907), respectively, which differed significantly between the species (2.994% [95% CI: 0.835, 5.337]) (Fig. 3C–D; Table 3). P. aeruginosa biofilms were therefore almost twice as susceptible to DMSO than S. pneumoniae. There was some evidence for a hormetic response in P. aeruginosa (RJMCMC inclusion: 60.3%, f = 17.904 [95% CI: 0, 78.727]) but not in S. pneumoniae (RJMCMC inclusion: 4.9%).
P. aeruginosa biofilm formation was inhibited at low DMSO concentrations (mean 5% and 40% biofilm inhibition was observed at 1.6% DMSO and 3.1% DMSO, respectively) and high DMSO concentrations (>98% inhibition observed at ≥12.5% DMSO) but interestingly, the intermediate concentration of 6.3% DMSO had a strong promotion effect with a mean 262% increase in biofilm formation (i.e., 262% decreased inhibition, Fig. 3D). These data deviated considerably from the observed trend and were excluded from the dose-response model to enable calculation of EC50 values though were nonetheless remarkable and highly repeatable (Fig. 3D).
3.2.3. Experiments 2 & 3: The effect of DMSO on the activity of standard antibiotics
3.2.3.1. Antimicrobial activity (planktonic growth inhibition)
DMSO had limited impact on the antimicrobial activity of ampicillin against S. pneumoniae with no significant differences in EC50 between treatments (Table 3, Fig. 4A). Although 95% CIs marginally included 0, there appeared to be differences in baseline antimicrobial activity (parameter d in dose-response curves, Table 3) between DMSO treatments: 1% and 2% DMSO treatments had borderline significantly lower antimicrobial activity at baseline compared to 0% and variable DMSO treatments (Fig. 4A). No hormetic effects were detected for S. pneumoniae with low RJMCMC inclusions probabilities and 95% CIs of hormetic effects (f) encompassing 0.
DMSO had a more considerable effect on the antimicrobial activity of gentamicin against P. aeruginosa. Antimicrobial EC50 values were reduced from 1.505 μg/mL (95% CI: 1.329, 1.759) for gentamicin alone (0% DMSO) (Exp. 1, 2, 3) to 1.378 μg/mL (95% CI: 1.116, 1.672) with 1% DMSO, 1.172 μg/mL (95% CI: 1.088, 1.272) with 2% DMSO (Exp. 2), and 1.048 μg/mL (95% CI: 1.004, 1.118) with variable DMSO (Exp. 3). EC50 values for 2% and variable DMSO treatments were significantly lower than gentamicin alone (0% DMSO) (Fig. 4B, Table 3). These data suggest an increase in the activity of gentamicin with each treatment and thus the importance of DMSO controls in antimicrobial activity assays using P. aeruginosa even when DMSO concentrations are reasonably low. Hormetic effects were detected for all DMSO treatments with high RJMCMC inclusion probabilities and all 95% CIs not encompassing 0.
3.2.3.2. Biofilm inhibition
Antibiofilm EC50 values for ampicillin against S. pneumoniae decreased stepwise with 1% (0.246 μg/mL [CI: 0.112, 0.382]), 2% (0.235 μg/mL [CI: 0.110, 0.366]) and variable (0.196 μg/mL [CI: 0.038, 0.377]) DMSO implying stronger activity than ampicillin alone (0% DMSO; 0.258 μg/mL [CI: 0.183, 0.336]) (Fig. 4C; Table 3). However, in alignment with results for antimicrobial activity, none of these differences in EC50 or baseline inhibition (d) were significant (Fig. 4C, Table 2). No hormetic effects were detected with low RJMCMC inclusions probabilities and hormetic effect posterior distributions encompassing 0.
Results for P. aeruginosa were more notable since low DMSO concentrations definitely interfered with the measured activity of gentamicin. Variable DMSO significantly reduced the EC50 value for gentamicin (EC50 1.504 μg/mL [95% CI: 1.203, 1.793]) compared to gentamicin alone (0% DMSO; EC50 2.032 μg/mL [95% CI: 1.953, 2.114]) and gentamicin with 1% and 2% DMSO (1.943 μg/mL [95% CI: 1.752, 2.087] and 2.179 μg/mL [95% CI: 1.96, 2.535], respectively) (Fig. 4D, Table 3). Variable DMSO also acted synergistically to promote biofilm formation at low gentamicin concentrations (Fig. 4D). Baseline inhibition (d) increased with 1% (17.7% [95% CI: 4.79, 30.9]) and 2% (53% [95% CI: 39.8, 66]) relative to 0% (−14.4% [95% CI: -22.4, −6.36]) DMSO indicating stronger activity at lower gentamicin concentrations with these treatments (Fig. 4D).
P. aeruginosa is the most common biofilm model organism (used in 30% [n = 26] of articles reviewed) owing to the clinical significance and virulence of biofilms formed by this species and the relative ease of establishing them in vitro. P. aeruginosa biofilms are also most susceptible to the effects of DMSO, based on the literature [[25], [26], [113]] and our data (Fig. 3, Fig. 4). As some explanation, Yahya et al. [26] demonstrated a significant reduction in total EPS protein with DMSO treatment [26] and Guo et al. [25] demonstrated that DMSO attenuated quorum sensing controlled biofilm formation. Acknowledging that DMSO significantly affects P. aeruginosa biofilm formation (alone and in combination with standard antibiotics) and that this effect is non-linear and variable, it is of paramount importance that DMSO solvent concentrations are as low as possible and DMSO controls are precise when using this species. A caveat to this point is that, considering our non-significant findings for S. pneumoniae versus significant findings for P. aeruginosa (Figs. 3 and 4) and the differences between responses reported in the literature (Table 2), the effect of DMSO is highly species dependent (and probably also influenced by the different extract/compound it is delivering). This may be due to differences in EPS polysaccharide/protein composition, concentration and chemical nature [115] and quorum sensing molecules between species and Gram-type. So, assumptions ought not be made as to whether the use of DMSO is concerning and/or whether controls should or should not be used based on the species of bacteria: when DMSO is used as a solvent, controls must be unequivocally included as standard practice in all antibiofilm assays.
4. Conclusion and recommendations
Screening for antibiofilm activity is an important area of pre-clinical research and product solubility challenges are commonly overcome using carrier solvents. By convention, controls should always be performed in any biological assay to exclude the solvent as a confounding factor. Unfortunately, insufficient DMSO controls undermine the validity of some recently published studies investigating the antibiofilm activity of many products with potential pharmacological utility. Our new experimental data confirms that DMSO can significantly affect biofilm formation in the model organism P. aeruginosa. Even at low concentrations, DMSO potentially alters the activity (somewhat unpredictably) of the antibiofilm agent of interest. Moving forward, the following criteria are essential when using DMSO as a solvent in in vitro antibiofilm assays:
-
1.
Specify the DMSO concentrations used to solubilise treatments and whether this is consistent or variable across dilutions; provide volumes and dilution factors to enable clear calculation of DMSO concentrations if not directly stated
-
2.
Use positive-growth DMSO controls at specified concentrations corresponding to each of those used as the solvent for treatments
-
3.
Include a positive-growth media control and compare to DMSO controls; provide control data
-
4.
Include media-only blanks for data correction
-
5.
Include species-specific standard antimicrobial controls for data QA/QC
-
6.
Calculate biofilm inhibition relative to DMSO and media controls (normalised to 100% growth) to accurately determine the activity of the compound of interest. As a percentage, applicable to most colorimetric (crystal violet and metabolic reduction) assays, the formula for calculating standardised percent biofilm inhibition (and % degradation) is:
where:
| Treatment = compound of interest including DMSO at concentration x% |
| Pos growth DMSO control = DMSO x% corresponding to treatment DMSO x% with biofilm growth (normalised to 100% biofilm growth) |
| Blank = media only (no biofilm growth) |
In which case, the positive-growth DMSO controls may be separately compared to the positive-growth media control to elucidate the effect of the solvent alone, if of interest. Alternatively, the following may be used to incorporate all variables:
where:
| Treatment = compound of interest including DMSO at concentration x% |
| Pos growth DMSO control = DMSO x% corresponding to treatment DMSO x% with biofilm growth |
| Pos growth media control = media with biofilm growth (normalised to 100% biofilm growth) |
| Blank = media only (no biofilm growth) |
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials and at https://github.com/mhollanders/dmso.
CRediT authorship contribution statement
Kate Summer: Conceptualization, Methodology, Investigation, Literature review, Formal analysis, Writing – original draft, Writing – review & editing. Jessica Browne: Supervision, Conceptualization, Methodology. Matthijs Hollanders: Formal analysis, Writing – review & editing. Kirsten Benkendorff: Supervision, Conceptualization, Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
K.S. is the recipient of an Australian Government Research Training Program (RTP) stipend. Project funding and research facilities were provided by the SCU Faculty of Science and Engineering and SCU Faculty of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2022.100081.
Contributor Information
Kate Summer, Email: kate.summer@scu.edu.au.
Jessica Browne, Email: jesssica.browne@scu.edu.au.
Matthijs Hollanders, Email: matthijs.hollanders@gmail.com.
Kirsten Benkendorff, Email: kirsten.benkendorff@scu.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.van Tilburg Bernardes E., Lewenza S., Reckseidler-Zenteno S. Current research approaches to target biofilm infections. Postdoc J : J Postdoc Res Postdoc Aff. 2015;3(6):36–49. doi: 10.14304/surya.jpr.v3n6.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill E.E., Franco O.L., Hancock R.E.J.C.b., design d. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des. 2015;85(1):56–78. doi: 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackman L.D., Qu Y., Cass P., Locock K.E.S. Approaches for the inhibition and elimination of microbial biofilms using macromolecular agents. Chem Soc Rev. 2021;50(3):1587–1616. doi: 10.1039/d0cs00986e. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Liang E., Cheng Y., Mahmood T., Ge F., Zhou K., Bao M., Lv L., Li L., Yi J., Lu C., Tan Y. Is combined medication with natural medicine a promising therapy for bacterial biofilm infection? Biomed Pharmacother. 2020;128 doi: 10.1016/j.biopha.2020.110184. [DOI] [PubMed] [Google Scholar]
- 6.Penesyan A., Paulsen I.T., Gillings M.R., Kjelleberg S., Manefield M.J. Secondary effects of antibiotics on microbial biofilms. Front Microbiol. 2020;11(2109) doi: 10.3389/fmicb.2020.02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart P.S., Costerton J. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 8.Wu H., Moser C., Wang H.-Z., Høiby N., Song Z.-J. Strategies for combating bacterial biofilm infections. Int J Oral Sci. 2015;7(1):1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi Y., Xia G., Shi C., Wan J., Liu L., Chen Y., Wu Y., Zhang W., Zhou M., He H., Liu R. Therapeutic strategies against bacterial biofilms. Fundam Res. 2021;1(2):193–212. [Google Scholar]
- 10.Windels E.M., Michiels J.E., Bergh B.V.d., Fauvart M., Michiels J., Epstein S., Rubin E.J. Antibiotics: combatting tolerance to stop resistance. mBio. 2019;10(5) doi: 10.1128/mBio.02095-19. e02095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organisation Antibiotic resistance. 2020. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance Available from.
- 12.Tillotson G. Antimicrobial resistance: what's needed. Lancet Infect Dis. 2015;15(7):758–760. doi: 10.1016/S1473-3099(15)00081-X. [DOI] [PubMed] [Google Scholar]
- 13.Savjani K.T., Gajjar A.K., Savjani J.K. ISRN Pharmaceutics; 2012. Drug solubility: importance and enhancement techniques. 2012. 195727-195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 15.Verheijen M., Lienhard M., Schrooders Y., Clayton O., Nudischer R., Boerno S., Timmermann B., Selevsek N., Schlapbach R., Gmuender H., Gotta S., Geraedts J., Herwig R., Kleinjans J., Caiment F. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci Rep. 2019;9(1):4641. doi: 10.1038/s41598-019-40660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI; Wayne, PA, U.S.A: 2020. [Google Scholar]
- 17.Su P.-W., Yang C.-H., Yang J.-F., Su P.-Y., Chuang L.-Y.J.M. Antibacterial activities and antibacterial mechanism of Polygonum cuspidatum extracts against nosocomial drug-resistant pathogens. Molecules. 2015;20(6):11119–11130. doi: 10.3390/molecules200611119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyrda G., Boniewska-Bernacka E., Man D., Barchiewicz K., Słota R. The effect of organic solvents on selected microorganisms and model liposome membrane. Mol Biol Rep. 2019;46(3):3225–3232. doi: 10.1007/s11033-019-04782-y. [DOI] [PubMed] [Google Scholar]
- 19.Miller B.W., Torres J.P., Tun J.O., Flores M.S., Forteza I., Rosenberg G., Haygood M.G., Schmidt E.W., Concepcion G.P. Synergistic anti-methicillin-resistant Staphylococcus aureus (mrsa) activity and absolute stereochemistry of 7,8-dideoxygriseorhodin C. J Antibiot. 2020;73:290–298. doi: 10.1038/s41429-019-0275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadhwani T., Desai K., Patel D., Lawani D., Bahaley P., Joshi P., Kothari V. Effect of various solvents on bacterial growth in context of determining MIC of various antimicrobials. Internet J Microbiol. 2009;7(1):1–8. [Google Scholar]
- 21.Modrzyński J.J., Christensen J.H., Brandt K.K. Evaluation of dimethyl sulfoxide (DMSO) as a co-solvent for toxicity testing of hydrophobic organic compounds. Ecotoxicology. 2019;28(9):1136–1141. doi: 10.1007/s10646-019-02107-0. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. eleventh ed. CLSI; Wayne, PA, U.S.A: 2018. p. 112. [Google Scholar]
- 23.Food and Drug Authority (FDA) FDA Centre for Drug Evaluation and Research, and Centre for Biologics Evaluation and Research; 2011. Q3C impurities: residual solvents.https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q3c-impurities-residual-solvents_2011 Available from. [Google Scholar]
- 24.Galvao J., Davis B., Tilley M., Normando E., Duchen M.R., Cordeiro M.F. Unexpected low-dose toxicity of the universal solvent DMSO. Faseb J. 2014;28(3):1317–1330. doi: 10.1096/fj.13-235440. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q., Wu Q., Bai D., Liu Y., Chen L., Jin S., Wu Y., Duan K. Potential use of dimethyl sulfoxide in treatment of infections caused by Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(12):7159–7169. doi: 10.1128/AAC.01357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yahya M., Alias Z., Karsani S.A. Antibiofilm activity and mode of action of DMSO alone and its combination with afatinib against gram-negative pathogens. Folia Microbiol. 2018;63(1):23–30. doi: 10.1007/s12223-017-0532-9. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee S.K., Farber J.M. Trend and pattern of antimicrobial resistance in molluscan Vibrio species sourced to Canadian estuaries. Antimicrob Agents Chemother. 2018;62(10) doi: 10.1128/AAC.00799-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A., Kumar D., Dahiya K., Hawthorne S., Jha S.K., Jha N.K., Nand P., Girgis S., Raj S., Srivastava R., Goswami V.K., Gregoriou Y., El-Zahaby S.A., Ojha S., Dureja H., Gupta G., Singh S., Chellappan D.K., Dua K. Advances in pulmonary drug delivery targeting microbial biofilms in respiratory diseases. Nanomedicine. 2021;16(21):1905–1923. doi: 10.2217/nnm-2021-0057. [DOI] [PubMed] [Google Scholar]
- 29.Azeredo J., Azevedo N.F., Briandet R., Cerca N., Coenye T., Costa A.R., Desvaux M., Di Bonaventura G., Hébraud M., Jaglic Z., Kačániová M., Knøchel S., Lourenço A., Mergulhão F., Meyer R.L., Nychas G., Simões M., Tresse O., Sternberg C. Critical review on biofilm methods. Crit Rev Microbiol. 2017;43(3):313–351. doi: 10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- 30.Ishwarya R., Iswarya A., Thangaviji V., Sivakamavalli J., Esteban M.A., Thangaraj M.P., Vaseeharan B. Immunological and antibiofilm property of haemocyanin purified from grooved tiger shrimp (Penaeus semisulcatus): an in vitro and in silico approach. Microb Pathog. 2020;147 doi: 10.1016/j.micpath.2020.104253. [DOI] [PubMed] [Google Scholar]
- 31.Maselli V., Galdiero E., Salzano A.M., Scaloni A., Maione A., Falanga A., Naviglio D., Guida M., Di Cosmo A., Galdiero S. Octopartenopin: identification and preliminary characterization of a novel antimicrobial peptide from the suckers of Octopus vulgaris. Mar Drugs. 2020;18(8) doi: 10.3390/md18080380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cedergreen N., Ritz C., Streibig J. Improved empirical models describing hormesis. Environ Toxicol Chem. 2005;24(12):3166–3172. doi: 10.1897/05-014r.1. [DOI] [PubMed] [Google Scholar]
- 33.Nweke C.O., Ogbonna C.J.J.E., Contamination E. Statistical models for biphasic dose-response relationships (hormesis) in toxicological studies. Ecotoxicol Environ Contam. 2017;12(1):39–55. [Google Scholar]
- 34.de Valpine P., Turek D., Paciorek C.J., Anderson-Bergman C., Lang D.T., Bodik R. Programming with models: Writing statistical algorithms for general model structures with nimble. J Comput Graph Stat. 2017;26(2):403–413. [Google Scholar]
- 35.Team R.C. R foundation for statistical computing; 2022. R: a language and environment for statistical computing.https://www.r-project.org/ 2022. Available from: [Google Scholar]
- 36.Green P.J. Reversible Jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82(4):711–732. [Google Scholar]
- 37.Jafri H., Banerjee G., Khan M.S.A., Ahmad I., Abulreesh H.H., Althubiani A.S. Synergistic interaction of eugenol and antimicrobial drugs in eradication of single and mixed biofilms of Candida albicans and Streptococcus mutans. AMB Express. 2020;10(1) doi: 10.1186/s13568-020-01123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijayakumar A., Sarveswari H.B., Vasudevan S., Shanmugam K., Solomon A.P., Neelakantan P. Baicalein inhibits Streptococcus mutans biofilms and dental caries-related virulence phenotypes. Antibiotics. 2021;10(2):1–13. doi: 10.3390/antibiotics10020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J.W., Jia A.Q., Jiang H., Li P.L., Chen H., Tan X.J., Liu E.Q. 1-(4-amino-2-hydroxyphenyl)ethanone from Phomopsis liquidambari showed quorum sensing inhibitory activity against Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2021;105(1):341–352. doi: 10.1007/s00253-020-11013-z. [DOI] [PubMed] [Google Scholar]
- 40.Juárez-Rodríguez M.M., Cortes-López H., García-Contreras R., González-Pedrajo B., Díaz-Guerrero M., Martínez-Vázquez M., Rivera-Chávez J.A., Soto-Hernández R.M., Castillo-Juárez I. Tetradecanoic acids with anti-virulence properties increase the pathogenicity of Pseudomonas aeruginosa in a murine cutaneous infection model. Front Cell Infect Microbiol. 2021;10 doi: 10.3389/fcimb.2020.597517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabir S., Suresh D., Subramoni S., Das T., Bhadbhade M., Black D.S., Rice S.A., Kumar N. Thioether-linked dihydropyrrol-2-one analogues as pqsr antagonists against antibiotic resistant Pseudomonas aeruginosa. Bioor Med Chem. 2021;31 doi: 10.1016/j.bmc.2020.115967. [DOI] [PubMed] [Google Scholar]
- 42.Sampaio F.C., Pereira M.d.S.V., Dias C.S., Costa V.C.O., Conde N.C.O., Buzalaf M.A.R. In vitro antimicrobial activity of caesalpinia ferrea martius fruits against oral pathogens. J Ethnopharmacol. 2009;124(2):289–294. doi: 10.1016/j.jep.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz H.K., Serrano D.R., Dea-Ayuela M.A., Bilbao-Ramos P.E., Bolás-Fernández F., Torrado J.J., Molero G. New amphotericin b-gamma cyclodextrin formulation for topical use with synergistic activity against diverse fungal species and Leishmania spp. Int J Pharm. 2014;473(1–2):148–157. doi: 10.1016/j.ijpharm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Park Y.N., Srikantha T., Daniels K.J., Jacob M.R., Agarwal A.K., Li X.C., Solla D.R. Protocol for identifying natural agents that selectively affect adhesion, thickness, architecture, cellular phenotypes, extracellular matrix, and human white blood cell impenetrability of Candida albicans biofilms. Antimicrob Agents Chemother. 2017;61(11) doi: 10.1128/AAC.01319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teo S.P., Bhakta S., Stapleton P., Gibbons S. Bioactive compounds from the bornean endemic plant Goniothalamus longistipetes. Antibiotics. 2020;9(12):1–11. doi: 10.3390/antibiotics9120913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halawany H.S., Abraham N.B., Siddiqui Y.M., Balto H.A., Jacob V. Antimicrobial efficacy of Salvadora persica extracts on a monospecies biofilm on orthodontic brackets in vitro. Oral Health Prev Dent. 2016;14(2):149–155. doi: 10.3290/j.ohpd.a33926. [DOI] [PubMed] [Google Scholar]
- 47.Hympanova M., Terlep S., Markova A., Prchal L., Dogsa I., Pulkrabkova L., Benkova M., Marek J., Stopar D. The antibacterial effects of new n-alkylpyridinium salts on planktonic and biofilm bacteria. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.573951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L.P., An M.M., Shen H., Huang X., Yao X., Liu J., Zhu F., Zhang S.Q., Chen S.M., He L.J., Zhang J., Zou Z., Jiang Y.Y. The non-geldanamycin hsp90 inhibitors enhanced the antifungal activity of fluconazole. Am J Tourism Res. 2015;7(12):2589–2602. [PMC free article] [PubMed] [Google Scholar]
- 49.Amalia R., Utami Dewi S., Margono A., Usman M. Antibacterial effects of Cuminum cyminum extract against Enterococcus faecalis biofilms from clinical isolates. Pesqui Bras em Odontopediatria Clínica Integr. 2019;19(1) [Google Scholar]
- 50.Fitri M., Nazar K., Meidyawati R., Azmi R. Antibacterial effect of xanthorrhizol (Curcuma xanthorrhiza Roxb.) against the biofilm of Fusobacterium nucleatum. Int J Appl Pharm. 2020;12(Special Issue 2):57–61. [Google Scholar]
- 51.Cusicanqui Méndez D.A., Gutierres E., José Dionisio E., Afonso Rabelo Buzalaf M., Cardoso Oliveira R., Andrade Moreira Machado M.A., Cruvinel T. Curcumin-mediated antimicrobial photodynamic therapy reduces the viability and vitality of infected dentin caries microcosms. Photodiagnosis Photodyn Ther. 2018;24:102–108. doi: 10.1016/j.pdpdt.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Durães F., Resende D.I.S.P., Palmeira A., Szemerédi N., Pinto M.M.M., Spengler G., Sousa E. Xanthones active against multidrug resistance and virulence mechanisms of bacteria. Antibiotics. 2021;10(5) doi: 10.3390/antibiotics10050600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kincses A., Szabó S., Rácz B., Szemerédi N., Watanabe G., Saijo R., Sekiya H., Tamai E., Molnár J., Kawase M., Spengler G. Benzoxazole-based metal complexes to reverse multidrug resistance in bacteria. Antibiotics. 2020;9(10):1–12. doi: 10.3390/antibiotics9100649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siles S.A., Srinivasan A., Pierce C.G., Lopez-Ribot J.L., Ramasubramanian A.K. High-throughput screening of a collection of known pharmacologically active small compounds for identification of Candida albicans biofilm inhibitors. Antimicrob Agents Chemother. 2013;57(8):3681–3687. doi: 10.1128/AAC.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohd-Said S., Kweh W.W., Than C.Y., Zainal-Abidin Z., Adnan S.N.A., Baharin S.A., Soo E. vol. 7. 2018. (Vitro inhibitory and biofilm disruptive activities of ginger oil against Enterococcus faecalis F1000Research). [Google Scholar]
- 56.Bhandari S., Khadayat K., Poudel S., Shrestha S., Shrestha R., Devkota P., Khanal S., Marasini B.P. Phytochemical analysis of medicinal plants of Nepal and their antibacterial and antibiofilm activities against uropathogenic Escherichia coli. BMC Compl Med Therap. 2021;21(1) doi: 10.1186/s12906-021-03293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khalate S. Biofilm inhibition of uti pathogens using Terminalia arjuna and Ipomea carnea plant extract. Indian J Sci Technol. 2020;13:2452–2462. [Google Scholar]
- 58.Kim H.R., Eom Y.B. Synergistic activity of equol and meropenem against carbapenem-resistant Escherichia coli. Antibiotics. 2021;10(2):1–13. doi: 10.3390/antibiotics10020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers S., Honma K., Mang T.S. Confocal fluorescence imaging to evaluate the effect of antimicrobial photodynamic therapy depth on P. gingivalis and T. denticola biofilms. Photodiagnosis Photodyn Ther. 2018;23:18–24. doi: 10.1016/j.pdpdt.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 60.Santezi C., Reina B.D., de Annunzio S.R., Calixto G., Chorilli M., Dovigo L.N. Photodynamic potential of curcumin in bioadhesive formulations: optical characteristics and antimicrobial effect against biofilms. Photodiagnosis Photodyn Ther. 2021;35 doi: 10.1016/j.pdpdt.2021.102416. [DOI] [PubMed] [Google Scholar]
- 61.Shariati A., Asadian E., Fallah F., Azimi T., Hashemi A., Sharahi J.Y., Moghadam M.T. Evaluation of nano-curcumin effects on expression levels of virulence genes and biofilm production of multidrug-resistant pseudomonas aeruginosa isolated from burn wound infection in Tehran, Iran. Infect Drug Resist. 2019;12:2223–2235. doi: 10.2147/IDR.S213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Šilha D., Hurdálková B., Papajová M., Šilhová L. In vitro activity of spice extracts against Arcobacter spp. and influence on their biofilm formation. J Microbiol Biotechnol Food Sci. 2019;9(3):552–556. [Google Scholar]
- 63.Stecoza C.E., Drăghici C., Căproiu M.T., Pîrcălăbioru G.G., Măruțescu L. Synthesis and evaluation of the antimicrobial and antibiofilm activity of novel dibenzothiepines. FARMACIA. 2020;68(6):1099–1105. [Google Scholar]
- 64.Trigo Gutierrez J.K., Zanatta G.C., Ortega A.L.M., Balastegui M.I.C., Sanitá P.V., Pavarina A.C., Barbugli P.A., De Oliveira Mima E.G. Encapsulation of curcumin in polymeric nanoparticles for antimicrobial photodynamic therapy. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Araniciu C., Oniga S., Oniga O., Palage M., Chifiriuc M.C., Maruţescu L. Antimicrobial and anti-pathogenic activity evaluation of some 2-(trimethoxyphenyl)-4-ar1-5-r2-thiazoles. FARMACIA. 2015;63(1):40–45. [Google Scholar]
- 66.Atalan E., Bulbul A.S., Ceylan Y. Cephalaria syriaca (l.): Investigation of antimicrobial, antibiofilm, antioxidant potential and seed morphology. Fresenius Environ Bull. 2020;29(5):3641–3649. [Google Scholar]
- 67.Bădiceanu C.D., Nuță D.C., Missir A.V., Hrubaru M., Delcaru C., Dițu L.M., Chifiriuc M.C., Limban C. Synthesis, structural, phisico-chemical characterization and antimicrobial activity screening of new thiourea derivatives. FARMACIA. 2018;66(1):149–156. [Google Scholar]
- 68.Chen L., Yu K., Chen L., Zheng X., Huang N., Lin Y., Jia H., Liao W., Cao J., Zhou T. Synergistic activity and biofilm formation effect of colistin combined with pfk-158 against colistin-resistant gram-negative bacteria. Infect Drug Resist. 2021;14:2143–2154. doi: 10.2147/IDR.S309912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gawron G., Krzyczkowski W., Lemke K., Ołdak A., Kadziński L., Banecki B. Nigella sativa seed extract applicability in preparations against methicillin-resistant Staphylococcus aureus and effects on human dermal fibroblasts viability. J Ethnopharmacol. 2019;244 doi: 10.1016/j.jep.2019.112135. [DOI] [PubMed] [Google Scholar]
- 70.Gonçalves J., Luís Â., Gradillas A., García A., Restolho J., Fernández N., Domingues F., Gallardo E., Duarte A.P. Ayahuasca beverages: phytochemical analysis and biological properties. Antibiotics. 2020;9(11):1–21. doi: 10.3390/antibiotics9110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazemian H., Ghafourian S., Heidari H., Amiri P., Yamchi J.K., Shavalipour A., Houri H., Maleki A., Sadeghifard N. Antibacterial, anti-swarming and anti-biofilm formation activities of chamaemelum nobile against Pseudomonas aeruginosa. Rev Soc Bras Med Trop. 2015;48(4):432–436. doi: 10.1590/0037-8682-0065-2015. [DOI] [PubMed] [Google Scholar]
- 72.Koenig H.N., Durling G.M., Walsh D.J., Livinghouse T., Stewart P.S. Novel nitro-heteroaromatic antimicrobial agents for the control and eradication of biofilm-forming bacteria. Antibiotics. 2021;10(7) doi: 10.3390/antibiotics10070855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schroeder T.H., Zooid T., Pier G.B. Lack of adherence of clinical isolates of Pseudomonas aeruginosa to asialo-gm1 on epithelial cells. Infect Immun. 2001;69(2):719–729. doi: 10.1128/IAI.69.2.719-729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shahid S.A., Anwar F., Shahid M., Majeed N., Azam A., Bashir M., et al. Laser-assisted synthesis of mn0.50zn0.50fe2o4 nanomaterial: characterization and in vitro inhibition activity towards Bacillus subtilis biofilm. J Nanomater. 2015:1–6. 896185. [Google Scholar]
- 75.Song H.S., Choi T.R., Bhatia S.K., Lee S.M., Park S.L., Lee H.S., Kim Y.G., Kim J.S., Kim W., Yang Y.H. Combination therapy using low-concentration oxacillin with palmitic acid and span85 to control clinical methicillin-resistant Staphylococcus aureus. Antibiotics. 2020;9(10):1–11. doi: 10.3390/antibiotics9100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tekintaş Y., Temel A., Ateş A., Eraç B., Metin D.Y., Hilmioğlu Polat S., Hoşgör Limoncu M. Antifungal and antibiofilm activities of selective serotonin reuptake inhibitors alone and in combination with fluconazole. Turk J Pharmaceut Sci. 2020;17(6):667–672. doi: 10.4274/tjps.galenos.2019.65481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wuren T., Toyotome T., Yamaguchi M., Takahashi-Nakaguchi A., Muraosa Y., Yahiro M., Wang D.N., Watanabe A., Taguchi H., Kamei K. Effect of serum components on biofilm formation by Aspergillus fumigatus and other Aspergillus species. Jpn J Infect Dis. 2014;67(3):172–179. doi: 10.7883/yoken.67.172. [DOI] [PubMed] [Google Scholar]
- 78.Talukdar P.K., Turner K.L., Crockett T.M., Lu X., Morris C.F., Konkel M.E. Inhibitory effect of puroindoline peptides on Campylobacter jejuni growth and biofilm formation. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.702762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang S., Wang J., Xu W., Liu Y., Wang W., Wu K., Wang Z., Zhang X. Antibacterial effects of traditional Chinese medicine monomers against Streptococcus pneumoniae via inhibiting pneumococcal histidine kinase (vick) Front Microbiol. 2015;6(MAY) doi: 10.3389/fmicb.2015.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Y., Liu S., Xi X., Ma C., Wang L., Chen X., Shi Z., Chen T., Shaw C., Zhou M. Study on the structure-activity relationship of an antimicrobial peptide, brevinin-2gub, from the skin secretion of hylarana guentheri. Antibiotics. 2021;10(8) doi: 10.3390/antibiotics10080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maura D., Rahme L.G. Pharmacological inhibition of the Pseudomonas aeruginosa mvfr quorum-sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob Agents Chemother. 2017;61(12) doi: 10.1128/AAC.01362-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banerjee M., Moulick S., Bhattacharya K.K., Parai D., Chattopadhyay S., Mukherjee S.K. Attenuation of Pseudomonas aeruginosa quorum sensing, virulence and biofilm formation by extracts of andrographis paniculata. Microb Pathog. 2017;113:85–93. doi: 10.1016/j.micpath.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 83.Campbell M., Zhao W., Fathi R., Mihreteab M., Gilbert E.S. Rhamnus prinoides (gesho): a source of diverse anti-biofilm activity. J Ethnopharmacol. 2019;241 doi: 10.1016/j.jep.2019.111955. [DOI] [PubMed] [Google Scholar]
- 84.Chaverra Daza K.E., Gómez E.S., Moreno Murillo B.D., Wandurraga H.M. Natural and enantiopure alkylglycerols as antibiofilms against clinical bacterial isolates and quorum sensing inhibitors of Chromobacterium violaceum ATCC12472. Antibiotics. 2021;10(4) doi: 10.3390/antibiotics10040430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hakimi Alni R., Ghorban K., Dadmanesh M. Combined effects of Allium sativum and Cuminum cyminum essential oils on planktonic and biofilm forms of Salmonella typhimurium isolates. 3 Biotech. 2020;10(7) doi: 10.1007/s13205-020-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sainudeen S., Nair V.S., Zarbah M., Abdulla A.M., Najeeb C.M., Ganapathy S. Can herbal extracts serve as antibacterial root canal irrigating solutions? Antimicrobial efficacy of Tylophora indica, Curcumin longa, Phyllanthus amarus, and sodium hypochlorite on Enterococcus faecalis biofilms formed on tooth substrate: In vitro study. J Pharm BioAllied Sci. 2020;12(5):S423–S429. doi: 10.4103/jpbs.JPBS_127_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bindiya E.S., Tina K.J., Sasidharan R.S., Bhat S.G. Bacf3: highly thermostable bacteriocin from Bacillus amyloliquefaciens btss3 antagonistic on food-borne pathogens. 3 Biotech. 2019;9(4) doi: 10.1007/s13205-019-1639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Shiekh R.A., Hassan M., Hashem R.A., Abdel-Sattar E. Bioguided isolation of antibiofilm and antibacterial pregnane glycosides from Caralluma quadrangula: disarming multidrug-resistant pathogens. Antibiotics. 2021;10(7) doi: 10.3390/antibiotics10070811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.She P., Wang Y., Li Y., Zhou L., Li S., Zeng X., Liu Y., Xu L., Wu Y. Drug repurposing: In vitro and in vivo antimicrobial and antibiofilm effects of bithionol against Enterococcus faecalis and Enterococcus faecium. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.579806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gandee L., Hsieh J.T., Sperandio V., Moreira C.G., Lai C.H., Zimmern P.E. The efficacy of immediate versus delayed antibiotic administration on bacterial growth and biofilm production of selected strains of uropathogenic Escherichia coli and Pseudomonas aeruginosa. Int Braz J Urol. 2015;41(1):67–77. doi: 10.1590/S1677-5538.IBJU.2015.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.D'Almeida R.E., Molina R.R.D.I., Viola C.M., Luciardi M.C., Nieto Peñalver C., Bardón A., Arena M.E. Comparison of seven structurally related coumarins on the inhibition of quorum sensing of Pseudomonas aeruginosa and Chromobacterium violaceum. Bioorg Chem. 2017;73:37–42. doi: 10.1016/j.bioorg.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 92.de Almeida J., Pimenta A.L., Pereira U.A., Barbosa L.C.A., Hoogenkamp M.A., van der Waal S.V., Crielaard W., Felippe W.T. Effects of three γ-alkylidene-γ-lactams on the formation of multispecies biofilms. Eur J Oral Sci. 2018;126(3):214–221. doi: 10.1111/eos.12411. [DOI] [PubMed] [Google Scholar]
- 93.Mochtar C.F., Sholikhah E.N., Nugrahaningsih D.A.A., Nuryastuti T., Nitbani F.O., Jumina Inhibitory and eradication activities of 1-monolaurin as anti-biofilm on monospecies and polymicrobial of Staphylococcus epidermidis and Candida tropicalis. Int J Pharmaceut Res. 2020;13(1):550–560. [Google Scholar]
- 94.Ali I.A.A., Cheung B.P.K., Matinlinna J., Lévesque C.M., Neelakantan P. Trans-cinnamaldehyde potently kills Enterococcus faecalis biofilm cells and prevents biofilm recovery. Microb Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104482. [DOI] [PubMed] [Google Scholar]
- 95.Marshall P.A., Loeb G.I., Cowan M.M., Fletcher M. Response of microbial adhesives and biofilm matrix polymers to chemical treatments as determined by interference reflection microscopy and light section microscopy. Appl Environ Microbiol. 1989;55(11):2827–2831. doi: 10.1128/aem.55.11.2827-2831.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gracia E., Fernández A., Conchello P., Alabart J.L., Pérez M., Amorena B. In vitro development of Staphylococcus aureus biofilms using slime-producing variants and atp-bioluminescence for automated bacterial quantification. Luminescence. 1999;14(1):23–31. doi: 10.1002/(SICI)1522-7243(199901/02)14:1<23::AID-BIO513>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 97.Lim Y.N., Dwyer P., Murray C., Karmakar D., Rosamilia A., Thomas E. Long-term outcomes of intravesical dimethyl sulfoxide/heparin/hydrocortisone therapy for interstitial cystitis/bladder pain syndrome. Int Urogynecol J. 2017;28(7):1085–1089. doi: 10.1007/s00192-016-3232-0. [DOI] [PubMed] [Google Scholar]
- 98.Awan M., Buriak I., Fleck R., Fuller B., Goltsev A., Kerby J., Lowdell M., Mericka P., Petrenko A., Petrenko Y., Rogulska O., Stolzing A., Stacey G.N. Dimethyl sulfoxide: a central player since the dawn of cryobiology, is efficacy balanced by toxicity? Regen Med. 2020;15(3):1463–1491. doi: 10.2217/rme-2019-0145. [DOI] [PubMed] [Google Scholar]
- 99.Jacob S.W., Wood D.C. Dimethyl sulfoxide (DMSO) toxicology, pharmacology, and clinical experience. Am J Surg. 1967;114(3):414–426. doi: 10.1016/0002-9610(67)90166-3. [DOI] [PubMed] [Google Scholar]
- 100.Yahya M.F.Z.R., Alias Z., Karsani S.A. Subtractive protein profiling of Salmonella typhimurium biofilm treated with DMSO. Protein J. 2017;36(4):286–298. doi: 10.1007/s10930-017-9719-9. [DOI] [PubMed] [Google Scholar]
- 101.Ali B.H. Dimethyl sulfoxide: recent pharmacological and toxicological research. Vet Hum Toxicol. 2001;43(4):228–231. [PubMed] [Google Scholar]
- 102.Herasimenka Y., Cescutti P., Sampaio Noguera C.E., Ruggiero J.R., Urbani R., Impallomeni G., Zanetti F., Campidelli S., Prato M., Rizzo R. Macromolecular properties of cepacian in water and in dimethylsulfoxide. Carbohydr Res. 2008;343(1):81–89. doi: 10.1016/j.carres.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 103.Yaacob M.F., Abdullah F.F.J., Jamil N.M., Yunus N.M., Aazmi S., Yahya M.F.Z.R. The effect of dimethyl sulfoxide on corynebacterium pseudotuberculosis biofilm: an in silico prediction and experimental validation. J Phys Conf. 2021 [Google Scholar]
- 104.Antoniou E., Buitrago C.F., Tsianou M., Alexandridis P. Solvent effects on polysaccharide conformation. Carbohydr Polym. 2010;79(2):380–390. [Google Scholar]
- 105.Hansen J., Platten F., Wagner D., Egelhaaf S.U. Tuning protein–protein interactions using cosolvents: specific effects of ionic and non-ionic additives on protein phase behavior. Phys Chem Chem Phys. 2016;18(15):10270–10280. doi: 10.1039/c5cp07285a. [DOI] [PubMed] [Google Scholar]
- 106.Roy R., Tiwari M., Donelli G., Tiwari V. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9(1):522–554. doi: 10.1080/21505594.2017.1313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ansel H.C., Norred W.P., Roth I.L. Antimicrobial activity of dimethyl sulfoxide against Escherichia coli, Pseudomonas aeruginosa, and Bacillus megaterium. J Pharmaceut Sci. 1969;58(7):836–839. doi: 10.1002/jps.2600580708. [DOI] [PubMed] [Google Scholar]
- 108.Lim J.Y., May J.M., Cegelski L. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl Environ Microbiol. 2012;78(9):3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu X., Santos R.R., Fink-Gremmels J. Staphylococcus epidermidis biofilm quantification: effect of different solvents and dyes. J Microbiol Methods. 2014;101(1):63–66. doi: 10.1016/j.mimet.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 110.Martín-Rodríguez A.J., Reyes-Darias J.A., Martín-Mora D., González J.M., Krell T., Römling U. Reduction of alternative electron acceptors drives biofilm formation in shewanella algae. npj Biofilms Microbiom. 2021;7(1) doi: 10.1038/s41522-020-00177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mi H., Wang D., Xue Y., Zhang Z., Niu J., Hong Y., Drlica K., Zhao X. Dimethyl sulfoxide protects Escherichia coli from rapid antimicrobial-mediated killing. Antimicrob Agents Chemother. 2016;60(8):5054–5058. doi: 10.1128/AAC.03003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Agathokleous E., Calabrese E.J. Hormesis: the dose response for the 21st century: the future has arrived. Toxicology. 2019;425 doi: 10.1016/j.tox.2019.152249. [DOI] [PubMed] [Google Scholar]
- 113.Loncar K.D., Ferris R.A., McCue P.M., Borlee G.I., Hennet M.L., Borlee B.R. In vitro biofilm disruption and bacterial killing using nonantibiotic compounds against gram-negative equine uterine pathogens. J Equine Vet Sci. 2017;53:94–99. [Google Scholar]
- 114.Yaacob M.F., Murata A., Nor N.H.M., Jesse F.F.A., Raja Yahya M.F.Z. Biochemical composition, morphology and antimicrobial susceptibility pattern of Corynebacterium pseudotuberculosis biofilm. J King Saud Univ Sci. 2021;33(1) [Google Scholar]
- 115.Vu B., Chen M., Crawford R.J., Ivanova E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14(7):2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials and at https://github.com/mhollanders/dmso.