Abstract
The regulation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl degradation in Pseudomonas azelaica is mediated by the regulatory gene, hbpR. The hbpR gene encodes a 63-kDa protein belonging to the NtrC family of prokaryotic transcriptional activators and having the highest homology to members of the XylR/DmpR subclass. Disruption of the hbpR gene in P. azelaica and complementation in trans showed that the HbpR protein was the key regulator for 2-hydroxybiphenyl metabolism. Induction experiments with P. azelaica and Escherichia coli containing luxAB-based transcriptional fusions revealed that HbpR activates transcription from a promoter (PhbpC) in front of the first gene for 2-hydroxybiphenyl degradation, hbpC, and that 2-hydroxybiphenyl itself is the direct effector for HbpR-mediated activation. Of several compounds tested, only the pathway substrates 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl and structural analogs like 2-aminobiphenyl and 2-hydroxybiphenylmethane were effectors for HbpR activation. HbpR is therefore, to our knowledge, the first regulator of the XylR/DmpR class that recognizes biaromatic but not monoaromatic structures. Analysis of a spontaneously occurring mutant, P. azelaica HBP1 Prp, which can grow with the non-wild-type effector 2-propylphenol, revealed a single mutation in the hbpR gene (T613C) leading to a Trp→Arg substitution at amino acid residue 205. P. azelaica HBP1 derivative strains without a functional hbpR gene constitutively expressed the genes for 2-hydroxybiphenyl degradation when complemented in trans with the hbpR-T613C gene. This suggests the importance of this residue, which is conserved among all members of the XylR/DmpR subclass, for interdomain repression.
2-Hydroxybiphenyl has been widely used in disinfectant and preservative formulations, as an intermediate in the synthesis of dyes, resins, and rubbers (71), and as a fungicide to control postharvest diseases of various fruits (15). From 1915 to 1978, 2-hydroxybiphenyl and 4-hydroxybiphenyl appeared as major by-products of the industrial synthesis of phenol. In the United States, dumping of the by-products on the production sites led to groundwater and surface water contamination with hydroxybiphenyls at nearby locations (71). 2-Hydroxybiphenyl is also formed during microbial desulfurization of dibenzothiophene in fossil fuels (26, 29).
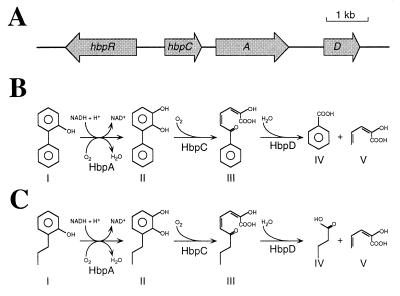
Different bacterial strains are able to use hydroxybiphenyls as sole carbon and energy sources (23, 30). One of these strains, Pseudomonas azelaica HBP1, degrades 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl through a meta-cleavage pathway (30, 31). The initial metabolism of these compounds is catalyzed by enzymes encoded by the hbpCAD genes (57) (Fig. 1). HbpA, a flavin adenine dinucleotide-containing 2-hydroxybiphenyl-3-monooxygenase, catalyzes the NADH-dependent ortho hydroxylation of 2-hydroxy- and 2,2′-dihydroxybiphenyl to 2,3-dihydroxy- and 2,2′,3-trihydroxybiphenyl, respectively (30, 31, 68). Next, HbpC, a 2,3-dihydroxybiphenyl-1,2-dioxygenase, catalyzes the meta cleavage, resulting in 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid and 2-hydroxy-6-oxo-6-(2-hydroxyphenyl)-2,4-hexadienoic acid, respectively (31, 57). The last two compounds are hydrolyzed by the meta-cleavage product hydrolase HbpD to 2-hydroxy-2,4-pentadienoic acid and either benzoic acid or salicylic acid (31). Benzoic acid and salicylic acid are further converted by benzoate 1,2-dioxygenase and salicylate monooxygenase, respectively, to catechol, which is the substrate for the lower meta-cleavage pathway (30, 31).
FIG. 1.
(A) Genetic organization of the hbp genes in P. azelaica HBP1. The orientation and sizes of the genes are indicated by arrows; the solid line represents noncoding DNA. (B) Pathway for the initial metabolism of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl in P. azelaica HBP1. The responsible enzymes are indicated below each conversion step. The names for the different compounds are as follows (in the case where the initial substrate is 2,2′-dihydroxybiphenyl, names are given in brackets): I, 2-hydroxybiphenyl (2,2′-dihydroxybiphenyl); II, 2,3-dihydroxybiphenyl (2,2′,3-trihydroxybiphenyl); III, 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid [2-hydroxy-6-oxo-6-(2-hydroxyphenyl)-2,4-hexadienoic acid]; IV, benzoic acid (salicylic acid); V, 2-hydroxy-2,4-pentadienoic acid. (C) Pathway for the initial metabolism of 2-propylphenol in P. azelaica HBP1 Prp. The responsible enzymes are the same as used for the initial metabolism of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl. The names for the different compounds are as follows: I, 2-propylphenol; II, 3-propylcatechol; III, 2-hydroxy-6-oxo-2,4-nonadienoic acid; IV, butyric acid; V, 2-hydroxy-2,4-pentadienoic acid.
Although the breakdown of 2-hydroxybiphenyl in P. azelaica HBP1 had been well characterized on the biochemical level, little is known about the regulation of this pathway. Activity measurements in cell extracts revealed that the hbpCAD genes are specifically expressed in the presence of 2-hydroxybiphenyl or 2,2′-dihydroxybiphenyl (30, 31). From DNA sequence information of regions near the hbpCAD genes, hints about the possible location of a regulatory gene upstream of hbpC were obtained (57). The amino acid sequence encoded by this open reading frame (ORF) showed homology to members of the NtrC family of prokaryotic transcriptional activators. Members of this family activate gene expression in concert with the RNA polymerase holoenzyme containing the ς54 subunit (RNAP-ς54) (reviewed in references 34 and 40). RNAP-ς54 is unable to catalyze the isomerization to open transcriptional complexes after having formed stable closed complexes with distinct promoters containing a −24(GG)/−12(GC) motif. The process can proceed only when it is coupled to the ATPase activity of an NtrC-type regulator which contacts RNAP-ς54 (34); triggering the ATPase activity in such regulators depends on biochemical or physiological stimuli (40).
The intricate mechanism by which members of the NtrC family activate gene transcription is reflected in their modular design. The amino-terminal A domain is the signal receptor module, the central C domain has an ATPase activity and is responsible for contacting promoter-bound RNAP-ς54, and the carboxy-terminal D domain contains a DNA binding motif (40). On the basis of sequence similarities within the A domain, different subclasses within the NtrC family can be distinguished, reflecting the different mechanisms underlying activity modulation of these regulators (58). In the subclass containing the response regulators of two-component systems, signal transduction is performed by a separate sensor histidine protein kinase mediating the phosphorylation of a conserved Asp residue in the A domain (reviewed in reference 64). Another subclass is reserved for NifA and analogous proteins. Members of this subclass are specifically inhibited by a second protein factor, such as NifL for NifA (5). The third group we will refer to as the XylR/DmpR subclass, after the two first-described and best-understood members forming this group (25, 59). Regulators of this subclass are activated by direct interaction with an effector molecule, without involvement of a sensor kinase, which is normally the substrate molecule for the catabolic pathways they control (58). In XylR and DmpR the A domain acts as a specific interdomain inhibitor which occludes the otherwise constitutive ATPase activity of the central C domain (18, 42, 47). In the current model of activation, the binding of an effector molecule leads to a conformational change in the A domain which is transmitted through a short flexible interdomain linker hinge region, the Q linker (72), in such a way that the ATPase activity of the C domain is being derepressed (58).
In this paper, we characterize the hbpR gene, demonstrate that HbpR is necessary for transcriptional activation of the 2-hydroxybiphenyl pathway genes, and analyze its phylogenetic relation to members of the XylR/DmpR subclass within the NtrC family. The capability of different monoaromatic and biaromatic compounds to act as effectors for HbpR-mediated transcription activation is investigated. Finally, the effect of a spontaneous point mutation in a conserved residue within the C-terminal region of the A domain of HbpR on HbpR-mediated transcription activation is illustrated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are presented in Table 1. P. azelaica HBP1 is able to use 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl as the sole source of carbon and energy (30). P. azelaica HBP1 Prp is a spontaneous mutant of strain HBP1 that is capable of growing with 2-propylphenol (32). The plasmids used in this study are listed in Table 2, and the structures of the most relevant constructed plasmids are shown in Fig. 2. Plasmid pJAMA8 (Fig. 2) was constructed to make transcriptional fusions with the luxAB genes of Vibrio harveyi (11, 27).
TABLE 1.
Bacterial strains used in this work
| Strain | Relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| BL21(DE3) | lon ompT hsdS, lysogenized with DE3, a λ phage carrying the T7 RNA polymerase gene under the control of the lacUV5 promoter; host strain for overexpression of genes cloned in vector pET3d | 65 |
| CC118λpir | Rifr, araD139 Δ(ara-leu) araD ΔlacX74 galE galK phoAΔ20 thi-1 rpsE rpoB argE recA1, lysogenized with λpir; host strain to propagate plasmids with a R6K origin of replication | 22 |
| DH5α | F−supE44 (φ80dlacZΔM15) Δlac(lacZYA-argF)U169 hsdR17 recA1 endA1 gyrA96 (Nalr) thi-1 relA deoR; host strain in routine cloning experiments | Gibco BRL, Life Technologies |
| HB101 | Smr, supE44 hsdS20 recA13 leuB6 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 7 |
| S17-1λpir | Smr Tpr, supE44 endA1 recA thi pro hsdR RP4-2-Tc::Mu-Km::Tn7, RP4 derivative integrated in the chromosome, lysogenized with λpir, donor strain in biparental matings to mobilize RP4 oriT containing plasmids lacking res sites | 14 |
| P. azelaica | ||
| HBP1 | 2-HBP+, 2-PP−, wild-type | 30 |
| HBP1 Prp | 2-HBP+, 2-PP+, spontaneous mutant obtained from strain HBP1 | 32 |
| HBP104 | Kmr, 2-HBP+, 2-PP−, contains mini-Tn5 (PhbpC− luxAB res-npt-res) | This work |
| HBP104 Prp | Kmr, 2-HBP+, 2-PP+, contains mini-Tn5 (PhbpC− luxAB res-npt-res) | This work |
| HBP121 | Smr Tcr, 2-HBP−, 2-PP−, plasmid pHYBP121 integrated in hbpR | This work |
| HBP127 | Kmr Smr Tcr, 2-HBP+, 2-PP−, plasmid pHYBP121 integrated in hbpR; contains mini-Tn5 (hbpR res-npt-res) | This work |
| HBP129 | Kmr Smr Tcr, 2-HBP+, 2-PP+, plasmid pHYBP121 integrated in hbpR; contains mini-Tn5 (hbpR-T613C res-npt-res) | This work |
| HBP104121 | Kmr Smr Tcr, 2-HBP−, 2-PP−, plasmid pHYBP121 integrated in hbpR; contains mini-Tn5 (PhbpC-luxAB res-npt-res) | This work |
2-HBP+/−, ability/inability to grow on 2-hydroxybiphenyl; 2-PP+/−, ability/inability to grow on 2-propylphenol; hbpR-T613C, hbpR gene isolated from strain HBP1 Prp; res-npt-res, element with the kanamycin resistance gene of Tn5 (npt) flanked by tandem multimer resolution sequences (res) of RP4 (33).
TABLE 2.
Plasmids used in this work
| Plasmid | Relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| pCK218 | Apr Kmr, R6K, RP4oriT (Mob), delivery vector for mini-Tn5 (luxAB res-npt-res) | 33 |
| pET3d | Apr, ColE1, overexpression vector containing the φ10 promoter | 66 |
| pGEM-7Zf(+) | Apr, ColE1, general cloning vector | Promega Corp. |
| pGEM-T Easy | Apr, ColE1, linearized pGEM-5Zf(+) with single 3′-thymidine overhangs to facilitate cloning of PCR products | Promega Corp. |
| pGreenTIR | Apr, ColE1, contains a mutant gfp (F64L/S65T) cloned into the EcoRI site of the symmetric polylinker of pUC1813 (28) | 39 |
| pHG171-luxAB | Apr, ColE1, contains the luxAB genes of V. harveyi | 2 |
| pHP45Ω | Apr Smr Spr, ColE1, contains the Ω fragment, a 2.0-kb DNA region with the aadA gene of the R100.1 plasmid conferring Smr and Spr, flanked by transcriptional termination signals of bacteriophage T4 gene 32 | 49 |
| pKK232-8 | Apr, ColE1, CAT based promoter-probe vector containing the rrnB rRNA T1 terminator | 9 |
| pLysS | Cmr, p15A, contains the T7 phage lysS gene encoding T7 lysozyme which inhibits T7 RNA polymerase | 66 |
| pRK2013 | Kmr, ColE1, RP4tra+, RP4oriT (Mob), helper plasmid in triparental matings | 19 |
| pSUP202 | Apr Cmr Tcr, ColE1, RP4oriT (Mob), mobilizable plasmid | 62 |
| pUC18Not | Apr, ColE1, general cloning vector based on pUC18 (74) but with polylinker flanked by NotI sites | 22 |
| pUC28 | Apr, ColE1, general cloning vector | 6 |
| pT7Blue(R) T-vector | Apr, ColE1, linearized pT7Blue(R) with single 3′-thymidine overhangs to facilitate cloning of PCR products | Novagen |
| pHBP130 | Apr, ColE1, contains a 7.8-kb MluI-SalI fragment from P. azelaica HBP1 with hbpR and hbpC | 57 |
| pHYBP100 | Apr, ColE1, pT7Blue(R) T vector carrying 0.7-kb PCR fragment containing the hbpR-hbpC intergenic region | This work |
| pHYBP103 | Apr, ColE1, pJAMA8 carrying the 0.7-kb SphI-XbaI fragment of pHYBP100; contains PhbpC-luxAB fusion | This work |
| pHYBP104 | Apr Kmr, R6K, RP4oriT (Mob), PCK218 carrying the 3.1-kb NotI fragment of pHYBP103; delivery vector for mini-Tn5 (PhbpC-luxAB res-npt-res) | This work |
| pHYBP109 | Apr, ColE1, pHYBP103 carrying the 2.3-kb PstI-NcoI fragment of pHBP130; contains hbpR-PhbpC-luxAB fusion | This work |
| pHYBP110 | Apr, ColE1, pHYBP109 with a destroyed SphI site in hbpR; contains hbpRΔ-PhbpC-luxAB fusion | This work |
| pHYBP111 | Apr, ColE1, pGEM-7Zf(+) carrying the 0.9-kb AatII-NsiI fragment of pHBP130 containing middle part of hbpR | This work |
| pHYBP119 | Apr Smr Spr, ColE1, pHYBP111 containing the 2.0-kb HindIII fragment of pHP45Ω; contains middle part of hbpR interrupted by the Smr Spr gene of the Ω fragment | This work |
| pHYBP120 | Apr Smr Spr, ColE1, pUC1813 containing the 2.9-kb ApaI-NsiI fragment of pHYBP119 flanked by HindIII, PstI, SalI, XbaI, and BamHI sites | This work |
| pHYBP121 | Cmr Smr Spr Tcr, ColE1, RP4oriT (Mob), pSUP202 containing the 2.9-kb PstI fragment of pHYBP120 | This work |
| pHYBP122 | Apr, ColE1, pUC28 containing the 2.8-kb NotI-XbaI fragment of pHYBP109; contains hbpR and the hbpR-hbpC intergenic region | This work |
| pHYBP126 | Apr, ColE1, pUC18Not containing the 2.8-kb EcoRI-PstI fragment of pHYBP122 | This work |
| pHYBP127 | Apr Kmr, R6K, RP4oriT (Mob), PCK218 carrying the 2.8-kb NotI fragment of pHYBP126; delivery vector for mini-Tn5 (hbpR res-npt-res) | This work |
| pHYBP128 | Apr, ColE1, pUC18Not containing the 2.7-kb PstI fragment of pHBP130; contains hbpR and the hbpR-hbpC intergenic region | This work |
| pHYBP128prp | Apr, ColE1, pUC18Not containing the 2.7-kb PstI fragment of pWSprp; contains hbpR-T613C and the hbpR-hbpC intergenic region | This work |
| pHYBP129 | Apr Kmr, R6K, RP4oriT (Mob), pCK218 carrying the 2.7-kb NotI fragment of pHYBP128prp; delivery vector for mini-Tn5 (hbpR-T613C res-npt-res) | This work |
| pHYBP130 | Apr, ColE1, pGEM-T Easy carrying 0.5-kb PCR fragment containing the N-terminal part of the coding region of hbpR | This work |
| pHYBP132 | Apr, ColE1, pET3d containing hbpR under the φ10 promoter | This work |
| pHYBP133 | Apr, ColE1, pET3d containing hbpRΔ under the φ10 promoter | This work |
| pJAMA3 | Apr, ColE1, pUC18Not carrying the 2.2-kb HindIII-BglII fragment of pHG171-luxAB | This work |
| pJAMA8 | Apr, ColE1, pJAMA3 carrying the 0.2-kb EcoRI fragment of pKK232-8; luxAB based promoter-probe vector | This work |
| pWSprp | Apr, ColE1, contains a 6.8-kb EcoRI fragment from P. azelaica HBP1 Prp with hbpR-T613C and hbpC | 67 |
CAT, chloramphenicol acetyltransferase; hbpR-T613C, hbpR gene isolated from strain HBP1 Prp; gfp, gene encoding green fluorescent protein from Aequorea victoria; res-npt-res, see Table 1 footnote.
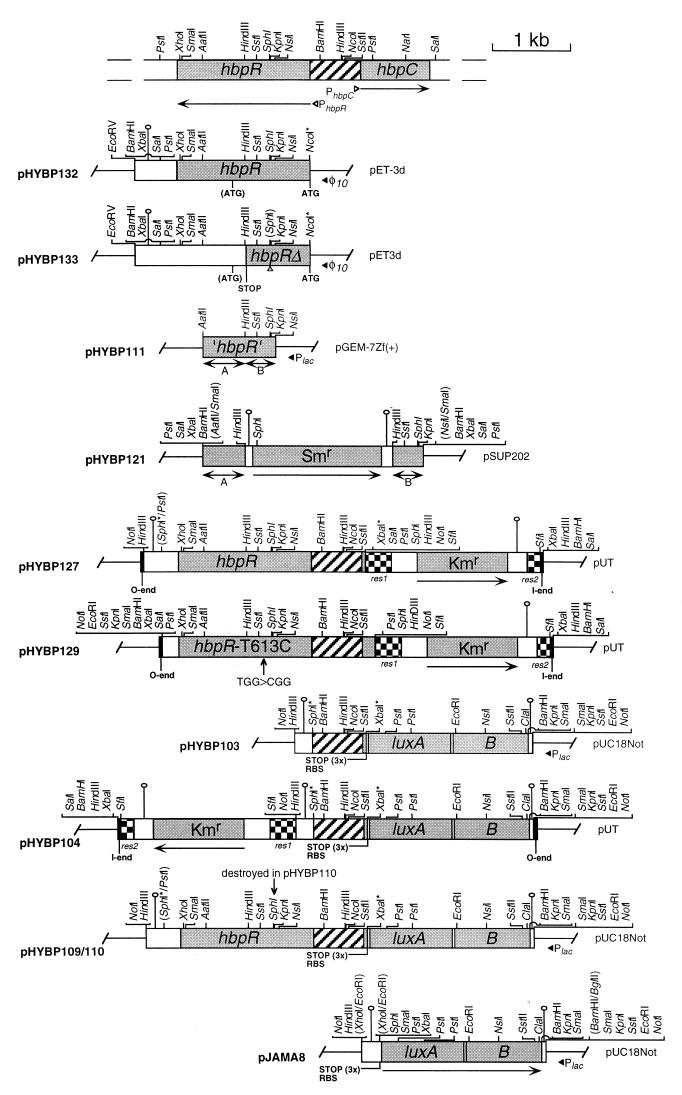
FIG. 2.
Restriction maps of the most relevant plasmids used in this study. All plasmids were constructed as described in Materials and Methods. At the top, the relative location of the hbpR and hbpC genes on the chromosome of P. azelaica HBP1 is depicted. All plasmid constructs are shown relative to this DNA fragment. For each plasmid the vector from which it was derived is indicated together with the abbreviation and the orientation of vector-situated promoters. All relevant restriction sites are shown except for the 2.1-kb SfiI fragment in pHYBP104, pHYBP127, and pHYBP129 containing the res-npt-res element (33). Restriction sites in parentheses were destroyed during cloning. PCR-introduced restriction sites are indicated by asterisks. Abbreviations: ‘hbpR’, internal region of the hbpR ORF; hbpRΔ, hbpR containing a frameshift mutation; hbpR-T613C, hbpR gene isolated from strain HBP1 Prp; A and B, indicate the 3′ and the 5′ region of ‘hbpR’, which are separated by the Smr marker in pHYBP121, respectively; ATG, start codon; (ATG), internal start codon in hbpR; RBS, ribosome binding site; STOP, stop codon(s); PhbpC, ς54-dependent promoter of the hbpC gene (M. C. M. Jaspers, unpublished data); PhbpR, putative ς70-dependent promoter of the hbpR gene. Arrows indicate the direction of transcription of the corresponding genes; shaded boxes indicate structural genes or fragments thereof; hatched boxes indicate the hbpR-hbpC intergenic region; checked boxes indicate resolution (res) sites of RP4; black boxes indicate the 19-bp I and O ends of Tn5 (not drawn to scale); white boxes indicate noncoding DNA; black solid lines indicate vector-derived DNA; black triangles indicate the orientation of vector-situated promoters; white triangles indicate the orientation of putative promoters in the hbpR-hbpC intergenic region; the grey triangle (in pHYBP133) indicates the introduced frameshift mutation in the hbpR ORF; hairpin-like structures represent transcriptional terminators.
Media and growth conditions.
Escherichia coli strains were grown at 37°C on Luria-Bertani medium (54). P. azelaica strains were grown at 30°C on Pseudomonas mineral medium (MM) (21), containing 10 mM succinate, 500 mg of 2-hydroxybiphenyl per liter (2.9 mM), or 250 mg of 2-propylphenol per liter (1.8 mM). When required, the media were supplemented with the following antibiotics at the indicated concentrations: ampicillin, 100 μg/ml (E. coli); chloramphenicol, 25 μg/ml (E. coli); kanamycin, 50 μg/ml (E. coli and P. azelaica); rifampin, 50 μg/ml (E. coli); streptomycin, 50 μg/ml (E. coli) or 200 μg/ml (P. azelaica); and tetracycline, 10 μg/ml (E. coli) or 30 μg/ml (P. azelaica). When necessary, the media were supplemented with 0.004% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) or 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Preparation of cell extract and enzyme activity measurements.
Cell extracts were prepared from 50-ml cultures of P. azelaica HBP1, HBP1-Prp, HBP121, HBP127, and HBP129 pregrown for 24 h in MM containing 10 mM succinate and then exposed for 10 h to 2-hydroxybiphenyl (1.2 mM), 2-propylphenol (1.2 mM), or succinate (1.5 mM), as described previously (30, 32). The activity of 2-hydroxybiphenyl 3-monooxygenase (HbpA) was measured in cell extracts by monitoring the disappearance of NADH at 340 nm as described elsewhere (30). The activities of 2,3-dihydroxybiphenyl dioxygenase (HbpC) and 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid hydrolase (HbpD) were measured in cell extracts by monitoring 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid (the meta-cleavage product) formation and disappearance at 434 nm, respectively, as described previously (31). Protein concentrations were determined by the method of Bradford (8) with bovine serum albumin as a standard.
Recombinant DNA techniques and DNA sequencing.
Plasmid DNA isolations, restriction endonuclease digestions, ligations, transformations, and other DNA manipulations were carried out by well-established procedures (54). DNA amplification by PCR was performed with Taq DNA polymerase (Gibco BRL, Life Technologies, Inc., Gaithersburg, Md.) as described elsewhere (37).
Chromosomal DNA was isolated from P. azelaica strains by the method of Marmur (38). Southern analysis was carried out with radioactively labeled DNA fragments as described elsewhere (36). Double-stranded template sequencing was performed as described previously (50).
Sequence analysis.
Comparisons of sequence data with published sequences in the nonredundant (NR) protein sequence database were performed with version 2.0 of the BLAST program via the Internet at http://www.ncbi.nlm.nih.gov/BLAST (3). Pairwise comparisons between protein sequences were made with the BLAST 2 sequences algorithm. A clustal alignment of the predicted amino acid sequences of the HbpR protein with other proteins was made with the program MegAlign from the Lasergene package (DNASTAR, Inc.). Phylogenetic inferences were derived on a set of aligned amino acid sequences with significant homology to HbpR, prepared with the PILEUP routine of the Wisconsin sequence analysis software package 8.0 (Genetics Computer Group, Madison, Wis.). Bootstrapping (100 replicas), protein distance calculations (PROTDIST), maximum-parsimony analysis (PROTPARS), and neighbor-joining analysis (NEIGHBOR) were carried out with the routines in the program package PHYLIP (version 3.5c) (17).
Overexpression of hbpR in E. coli.
For overexpression of the hbpR gene in E. coli, plasmid pHYBP132, in which the ATG triplet as present in the NcoI site of plasmid pET3d (66) was used as the start codon for hbpR, was constructed. To do this, a 481-bp fragment of hbpR was amplified by PCR on P. azelaica HBP1 total DNA to introduce an NcoI site at the start of hbpR, which was then cloned with the remaining part of hbpR in several steps into pET3d (pHYBP132 [Fig. 2]). Plasmid pHYBP133 is similar to pHYBP132 except for an interruption of the ORF of hbpR by a 4-bp deletion at the internal SphI site (giving the smaller ORF hbpRΔ) (Fig. 2). To test HbpR expression, cultures of E. coli BL21(DE3)(pLysS) harboring plasmid pHYBP132 (hbpR), pHYBP133 (hbpRΔ), or pET3d were induced with IPTG. Crude extracts were prepared and analyzed as described previously (50).
Disruption of the chromosomal hbpR gene copy and trans complementation.
To disrupt the ORF of the hbpR gene on the P. azelaica chromosome, we used single recombination with a nonreplicating plasmid (pHYBP121) containing an internal fragment of hbpR, which was interrupted by an Smr marker (Fig. 2). Plasmid pHYBP121 was transferred from E. coli S17-1λpir to P. azelaica in a biparental mating carried out on filter disks as described elsewhere (22). Selection for P. azelaica recombinants was done on MM plates supplemented with streptomycin and 10 mM succinate. A single recombinant, P. azelaica HBP121, was purified and verified by Southern hybridizations on total DNA.
To complement P. azelaica HBP121, a functional hbpR copy was introduced in trans on the chromosome by mini-Tn5 delivery with plasmid pHYBP127 (resulting in P. azelaica HBP127) (Table 2 and Fig. 2). Similarly, the hbpR gene from strain HBP1 Prp (hbpR-T613C) was cloned into plasmid pHYBP129 to complement strain HBP121 (resulting in P. azelaica HBP129). Triparental filter matings were performed to mobilize plasmids pHYBP127 or pHYBP129 from E. coli CC118λpir to the recipient P. azelaica strain with the help of E. coli HB101(pRK2013) (19) as described elsewhere (22). Selection for P. azelaica exconjugants containing a mini-Tn5 transposon derivative was done on MM plates plus kanamycin and 2.9 mM 2-hydroxybiphenyl. Proper insertion was verified by Southern hybridization on total DNA.
Construction of a transcriptional fusion of the hbpRC intergenic region with luxAB.
A 704-bp DNA fragment containing the intergenic region between the hbpR and hbpC genes, as well as the first 47 nucleotides of the hbpC coding region, was obtained by PCR on P. azelaica HBP1 total DNA with primers HBP-1 (5′-GCATGCGATGGTTCAGGTCCGG-3′; the SphI site is underlined) and HBP-2 (5′-TCTAGATCACTTACACCTAAAACG-3′; the XbaI site is underlined). This fragment was cloned into pT7Blue(R)-T, resulting in plasmid pHYBP100. The insert of pHYBP100 was sequenced and confirmed to be identical to the original sequence. The hbpR-hbpC intergenic region was recovered from pHYBP100 as a 0.7-kb SphI-XbaI fragment and ligated with pJAMA8 digested with SphI and XbaI (producing pHYBP103). The 3.1-kb NotI fragment of pHYBP103 containing the luxAB fusion was inserted into PCK218 (33) by replacing its 3.2-kb NotI fragment with the 3.1-kb NotI fragment of pHYBP103. This resulted in plasmid pHYBP104, which was used for chromosomal insertion of the luxAB-based transcriptional fusion in P. azelaica HBP1 or HBP1 Prp. Proper insertions were verified by Southern hybridization (strains HBP104 and HBP104Prp [Table 1]).
To test inducible expression from the hbpC promoter in E. coli, we constructed plasmids pHYBP109 and pHYBP110, containing hbpR or hbpRΔ, respectively, plus the hbpRC intergenic region transcriptionally fused to the luxAB genes (Fig. 2).
Luciferase assays.
Activity of the hbpC promoter in E. coli and P. azelaica strains was analyzed by measuring luciferase activity. For reasons of convenience and reproducibility, frozen stocks were prepared from each strain to be tested. These stocks were prepared by adding dimethyl sulfoxide (DMSO) (5% [vol/vol]) to washed cultures which had grown to mid-log phase (optical density at 600 nm of 0.60). Stocks were stored at −80°C until further use and were used within 2 months, a period during which no decrease of the inducibility of the hbpC promoter could be observed.
Cells were induced in 7-ml glass vials that were tightly closed with a screw-cap PTFE liner (Supelco, Bellefonte, Pa.) to avoid possible evaporation or adsorption of volatile hydrophobic compounds. For P. azelaica strains, each assay mixture contained 1.8 ml of antibiotic-free MM, 166 μl of cell suspension, and 20 μl of a stock solution of 2-hydroxybiphenyl or another potential inducer compound dissolved in DMSO (assay concentration, 0.2 mM). For E. coli DH5α strains, each assay mixture contained 1.9 ml antibiotic-free MM [supplemented with 0.01% tryptone, 0.005% yeast extract, and 10 mM d-(+)-glucose], 33 μl of cell suspension, and 20 μl of the inducer stock solution. The negative control contained 20 μl of DMSO. The frozen cell suspensions were thawed in a water bath at 25°C for 2 min and then placed back on ice until immediately prior to inoculation of the assay mixture. Induction experiments were started by addition of the cells to the prewarmed (30°C) assay mixture. During incubation, the glass vials with their contents were incubated at 30°C on a rotary shaker at 200 rpm. Bioluminescence was measured at 30°C at a final n-decanal concentration of 2 mM in a MicroLumat LB 96 P luminometer (Berthold AG, Regensdorf, Switzerland) as described previously (63).
Synthetic oligonucleotides and chemicals.
Primers labeled with the fluorescent dye IRD-800 at the 5′ end were purchased from MWG-BIOTECH GmbH (Ebersberg, Germany); all other primers were obtained from Microsynth GmbH (Balgach, Switzerland). 2,3-Dihydroxybiphenyl was obtained from Wako Chemicals GmbH (Neuss, Germany), 2-chlorobiphenyl was obtained from Johnson Matthey GmbH (Karlsruhe, Germany), and 3-hydroxybiphenyl was obtained from Eastman Kodak Co. (Rochester, New York). 2,2′-Dihydroxybiphenyl, 2,5-dihydroxybiphenyl, 4,4′-dihydroxybiphenyl, 2-ethylphenol, 2-hydroxydiphenylmethane, 3-methylcatechol, and 2-propylphenol were purchased from Aldrich-Chemie (Steinheim, Germany). All other organic chemicals were obtained from FLUKA Chemie AG (Buchs, Switzerland).
Nucleotide sequence accession number.
The nucleotide sequence of hbpR has been deposited in the GenBank database under accession no. U73900.
RESULTS
DNA sequence and overexpression of hbpR.
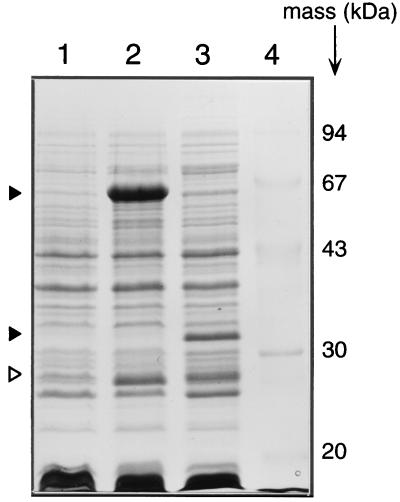
Recently, the hbpCAD genes, which encode the enzymes responsible for the initial metabolism of 2-hydroxybiphenyl in P. azelaica HBP1, were cloned and sequenced (57). Analysis of the DNA region located upstream of the hbpC gene revealed a large ORF which was oriented in the opposite direction to the hbpCAD genes (Fig. 1A). The nucleotide sequence of this ORF (tentatively named hbpR [see below]) was 1,710 bp, corresponding to a protein of 570 amino acids (aa) with a predicted molecular mass of 63,004 Da. To confirm the integrity of the ORF and the actual size of the hbpR-encoded protein, the hbpR coding region was fused with the ATG start codon present on plasmid pET3d (pHYBP132) and overexpressed in E. coli BL21(DE3)(pLysS). A polypeptide of approximately 63 kDa could be seen on sodium dodecyl sulfate-polyacrylamide gel electrophoresis in crude extracts prepared from induced E. coli BL21(DE3)(pLysS) harboring plasmid pHYBP132 (Fig. 3, lane 2). This size corresponded to that predicted from the sequence of hbpR. The 63-kDa protein was absent in extracts from induced BL21 cultures containing pHYBP133, which showed a 31-kDa protein instead (lane 3). This shorter protein was the result of the introduced frameshift mutation at the unique internal SphI restriction site at nucleotide 520 in hbpR. This mutated hbpR gene (hbpRΔ) would code for a shorter protein of 272 aa with a predicted molecular mass of 30,679 Da. Both the 63- and 31-kDa proteins were absent in crude extract prepared from E. coli BL21(DE3)(pLysS) carrying pET3d itself (Fig. 3, lane 1). A second specifically induced protein band of 26 kDa was observed in extracts of E. coli BL21(DE3)(pLysS) harboring pHYBP132 or pHYBP133 (lanes 2 and 3). This protein may have been produced from an internal ATG codon at nucleotide 1006 of hbpR (downstream of the introduced frameshift mutation) (Fig. 2).
FIG. 3.
Overexpression of the hbpR gene in E. coli. Shown is a Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel of crude extracts from E. coli BL21(DE3)(pLysS) cultures induced with IPTG and harboring pET3d (lane 1), pHYBP132 (intact hbpR; lane 2), or pHYBP133 (frameshift in hbpR, hbpRΔ; lane 3). Lane 4 shows molecular markers with their masses indicated. The black triangles in lanes 2 and 3 indicate the full-length HbpR (63 kDa) and the out-of-frame truncated HbpR (31 kDa), respectively. The white triangles in lanes 2 and 3 point to the shorter (26-kDa) in-frame product possibly originating from an internal hbpR ATG codon. The indicated protein bands were not visible under noninduced conditions (data not shown).
HbpR is a member of the NtrC family of bacterial transcriptional activators.
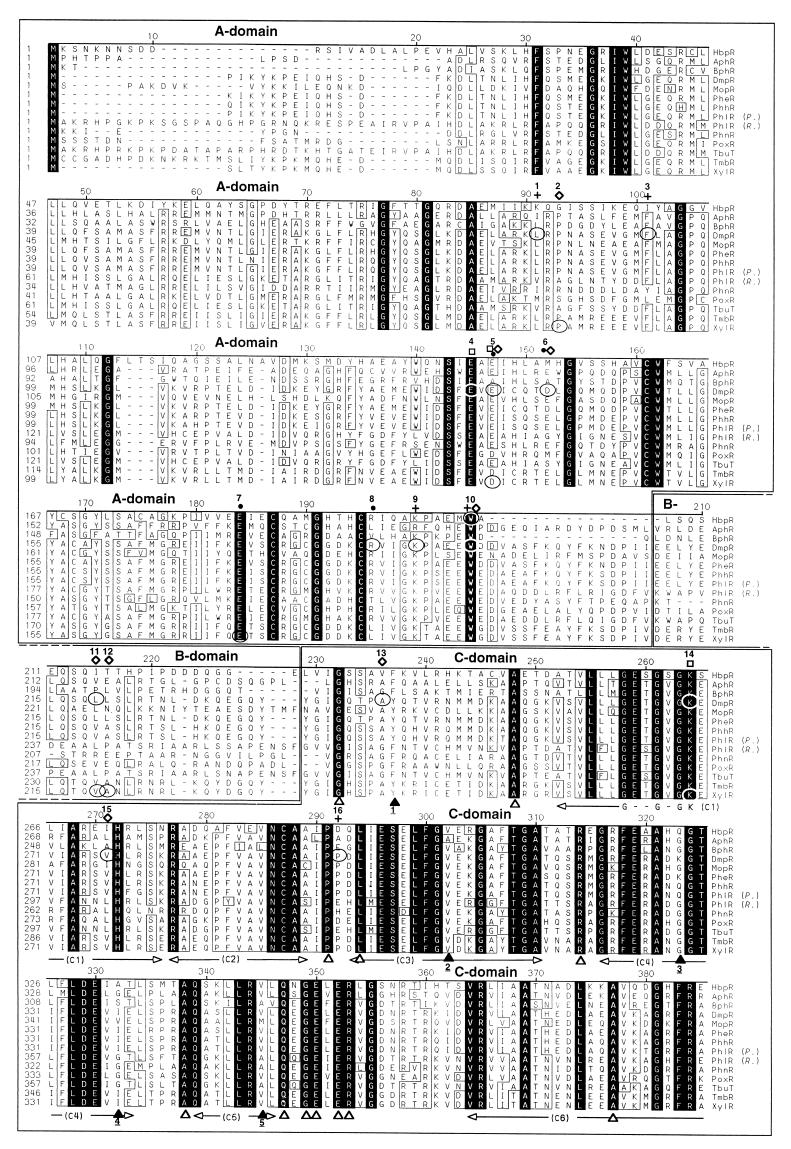
Comparison of the deduced amino acid sequence of the HbpR protein with other sequences in the nonredundant (NR) protein sequence database revealed that HbpR is homologous to members of the NtrC family of bacterial transcriptional activators. The homology of HbpR to most NtrC family members was restricted to the central C domain, with the exception of 13 proteins within the XylR/DmpR subclass that had an overall homology to HbpR (Fig. 4). Pairwise comparisons revealed that within this subclass, HbpR showed the highest overall homology to TbuT, the regulator of the toluene-3-monooxygenase operon in Burkholderia pickettii PKO1 (10) (now renamed Ralstonia pickettii PKO1 [73]), with 42% identity and 58% similarity in a 553-aa overlap. HbpR showed the lowest overall homology to XylR, the activator for both the upper-pathway genes and the regulatory gene xylS on plasmid pWW0 in Pseudomonas putida mt-2 (25), with 37% identity and 53% similarity in a 549-aa overlap. Considering the different domains, the extent of homology between HbpR and the other members of the XylR/DmpR subclass was higher for the C domain (48 to 56% identity, 65 to 71% similarity) and the D domain (35 to 57% identity, 67 to 76% similarity) than for the A domain (31 to 37% identity, 45 to 56% similarity).
FIG. 4.
Alignment of the predicted amino acid sequences of the HbpR protein and the 13 most similar members of the NtrC family of transcriptional activators in the nonredundant (NR) protein sequence database. The alignment was made with the program MegAlign from the Lasergene package (DNASTAR, Inc.) and further improved manually. The ruler shown above the sequences corresponds to the amino acid sequence of the HbpR protein. Conserved amino acid residues in all proteins are indicated against a black background, whereas residues conserved in at least 10 of 14 proteins appear boxed. Gaps introduced to maximize the alignment are dashed. The regions corresponding to the different domains are boxed, and their names are displayed above the sequences. The borders of the A, C, and D domains in the different proteins were set according to those proposed for the XylR protein (25). The sections C1 to C7, shown below the sequences, correspond to the conserved regions within the C domain as described by Morett and Segovia (40). The Walker A motif within regions C1 (55, 69), involved in ATP binding, and a putative helix-turn-helix in the D domain (40) are indicated below the sequences. U represents hydrophobic residues. White triangles below the sequences mark fully conserved residues within the C and D domains of the XylR/DmpR subclass members but not fully conserved in an alignment made by Morett and Segovia, which included 27 members of the NtrC family, including XylR (40). Black triangles below the sequence mark positions which were not conserved in at least 10 of 14 proteins but appeared to be conserved in the alignment made by Morett and Segovia (40). Underlined numbers below the black triangles indicate which residues replaced the conserved ones: 1, F or Y instead of L or M; 2, A or V instead of H; 3, G, K or Q instead of only G: 4, A, G, I, S, or V instead of only G; 5, A, M, or V instead of I or V; 6, A or P instead of only P; 7, C, L, or M instead of L or M; 8, L, M or V instead of L or M. Encircled residues mark the positions of described point mutations within XylR/DmpR subclass members. The phenotypes of described point mutations is indicated above the sequence: black dots, altered effector specificity (ALT); open diamonds, constitutive or semiconstitutive transcription-promoting activity (CON); empty squares, abolition of transcription-promoting activity (NON); crosses, suppressor mutation of a semiconstitutive mutation (SUP); or a combination of these symbols. Numbers above these symbols are used to describe all known mutations for the corresponding position and their resulting phenotype(s): 1, DmpR-L83P, SUP selected for DmpR-E135A (42); 2, XylR-P85S, CON (13); 3, DmpR-F93L, SUP selected for DmpR-E135D (42); 4, DmpR-E133K, NON (61); 5, DmpR-E135A, ALT CON (61); DmpR-E135D, ALT CON (61); DmpR-E135K, ALT (46); DmpR-E135R, ALT (61); XylR-D135E, NON (53); XylR-D135N, CON (13); XylR-D135Q, CON (53); 6, DmpR-D140K, ALT CON (61); 7, XylR-E172K, ALT (12); 8, DmpR-R184W, ALT (12); 9, DmpR-K188E, SUP selected for DmpR-E135D (42); 10, HbpR-W205R, CON (this study); DmpR-W193R, SUP for DmpR-V276A (42); 11, DmpR-L219P, CON (61); XylR-V219DA220P (double mutation), CON (18); 12, XylR-V219DA220P (double mutation), CON (18); 13, DmpR-A241T, CON (61); 14, DmpR-G268S, NON (42); XylR-G268N, NON (48); 15, DmpR-V276A, CON (61); DmpR-V276G, CON (61); 16, DmpR-P297R, SUP selected for DmpR-E135D (42); 17, DmpR-F424L, CON (61); 18, XylR-R453H, NON (48). The database entries for the different proteins are given in the legend of Fig. 6.
Disruption of the hbpR gene in P. azelaica HBP1.
Southern analysis of total DNA isolated from P. azelaica HBP1 with a probe against the hbpR gene indicated that hbpR was present in only one copy on the chromosome (data not shown). The hbpR gene was disrupted in strain HBP1 by single recombination with the introduced plasmid pHYBP121. Attempts to obtain double recombinants by plating out serial dilutions of cultures of single recombinants on MM containing streptomycin, followed by counterselection against the Tcr marker, were not successful in our hands. However, every type of single and double recombination of the truncated hbpR gene on pHYBP121 with the chromosomal hbpR copy would lead to disruption of the hbpR ORF. Southern analysis of the total DNA from one purified recombinant, designated strain HBP121, showed that in this strain pHYBP121 had integrated in region B (Fig. 2). Therefore, it contained two partial copies of hbpR, the first copy lacking the 5′ part and the second copy disrupted by the Smr marker (data not shown).
Whereas growth of strain HBP1 with 2-hydroxybiphenyl as the sole source of carbon and energy occurred at a maximum specific growth rate (μmax) of 0.51 h−1 (doubling time [td] = 1.4 h), strain HBP121 had completely lost the ability to grow on 2-hydroxybiphenyl. Measurements of HbpA, HbpC, and HbpD activities in cell extracts also indicated that whereas 2-hydroxybiphenyl induced the formation of HbpA, HbpC, and HbpD in strain HBP1, there was no inducible activity in strain HBP121 (with disrupted hbpR) (Table 3). We then complemented P. azelaica HBP121 in trans on the chromosome with a functional hbpR gene by mini-Tn5 delivery with pHYBP127. Southern analysis of the total DNA from one exconjugant, designated HBP127, revealed that it contained the two partial copies of the hbpR gene, as in P. azelaica HBP121, and in addition a complete hbpR gene copy (data not shown). Strain HBP127 could indeed again use 2-hydroxybiphenyl as the sole carbon and energy substrate, albeit at a slightly reduced μmax of 0.46 h−1 (td = 1.5 h) compared to strain HBP1. Enzyme activities for HbpA, HbpC, and HbpD in strain HBP127 were again inducible with 2-hydroxybiphenyl (Table 3). The fact that a functional hbpR gene provided in trans could overcome the growth constraints of strain HBP121 (lacking a complete hbpR gene) identified the HbpR protein as a key element involved in the activation of the hbpCAD genes.
TABLE 3.
Induction of HbpA, HbpC and HbpD activities with 2-hydroxybiphenyl and 2-propylphenol in P. azelaica HBP1, HBP121, HBP127, HBP129, and HBP1 Prp
| Strain | Inducing compound | Enzyme sp act (μmol of substrate min−1 mg of protein−1)
|
||
|---|---|---|---|---|
| HbpA monooxygenase | HbpC dioxygenase | HbpD hydrolase | ||
| HBP1 | Succinate | 0 | 0 | 0 |
| 2-Hydroxybiphenyl | 0.154 | 5.88 | 0.294 | |
| 2-Propylphenol | 0 | 0 | 0 | |
| HBP121 | Succinate | 0 | 0 | 0 |
| 2-Hydroxybiphenyl | 0 | 0 | 0 | |
| 2-Propylphenol | 0 | 0 | 0 | |
| HBP127 | Succinate | 0 | 0 | 0 |
| 2-Hydroxybiphenyl | 0.064 | 5.46 | 0.193 | |
| HBP129 | Succinate | 0.072 | 4.51 | 0.458 |
| 2-Propylphenol | 0.082 | 4.41 | 0.344 | |
| HBP1 Prp | Succinate | 0.045 | 4.73 | 0.429 |
| 2-Propylphenol | 0.161 | 11.4 | 0.524 | |
HbpR activates transcription from the hbpC promoter in the presence of 2-hydroxybiphenyl.
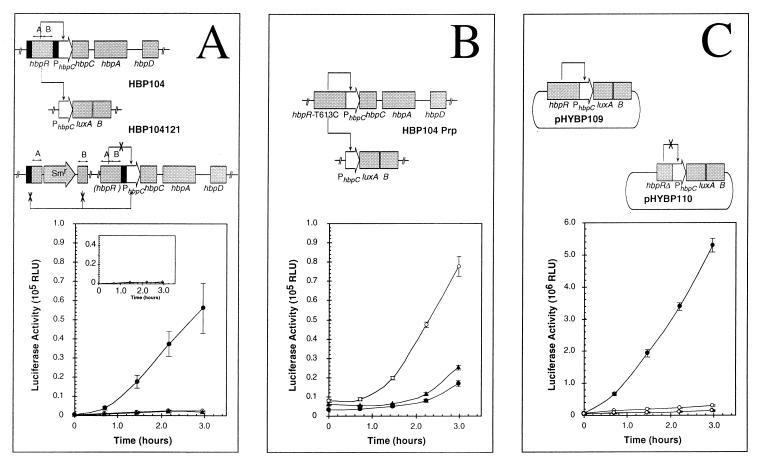
To establish whether HbpR was indeed able to activate gene transcription from a promoter upstream of the hbpC gene and whether 2-hydroxybiphenyl was the necessary effector for activation, a transcriptional fusion between the hbpRC intergenic region and the luxAB genes of V. harveyi was inserted into the chromosome of P. azelaica HBP1. The resulting strain, P. azelaica HBP104, showed a 23-fold-increased bioluminescence after a 3-h incubation in the presence of 0.2 mM 2-hydroxybiphenyl (Fig. 5A). This indicated that the hbpRC intergenic region contained a regulatable promoter (called the hbpC promoter, PhbpC) and again demonstrated the role of 2-hydroxybiphenyl as an effector. When the hbpR gene in strain HBP104 was disrupted by homologous recombination (P. azelaica HBP104121) with plasmid pHYBP121 at site A (Fig. 2 and data not shown), inducible expression from the hbpC promoter was abolished (Fig. 5A, inset). Adding 2-hydroxybiphenyl to strain HBP104121 even reduced the levels of observed bioluminescence by 37% after 3 h.
FIG. 5.
HbpR-mediated in vivo transcriptional activation from PhbpC in a homologous (P. azelaica HBP104, HBP104121, HBP104 Prp) or heterologous (E. coli) host system. (A) Activation from PhbpC in P. azelaica HBP104 containing a functional hbpR gene. Induction took place with 2-hydroxybiphenyl (black circles), 2-propylphenol (black triangles), or DMSO only (white circles). Inset: Activation from PhbpC in P. azelaica HBP104121 containing a disrupted hbpR gene. Induction took place with 2-hydroxybiphenyl (black diamonds) or DMSO only (white diamonds). (B) Activation from PhbpC in P. azelaica HBP104 Prp containing the mutated hbpR gene (hbpR-T613C). Induction took place with 2-hydroxybiphenyl (black circles), 2-propylphenol (black triangles), or DMSO only (white circles). (C) Activation from PhbpC in E. coli with functional hbpR (pHYBP109, circles) or dysfunctional hbpR (pHYBP110, diamonds). Induction took place with 2-hydroxybiphenyl (black symbols) or DMSO only (white symbols). The concentrations of 2-hydroxybiphenyl and 2-propylphenol used in the assays were 0.2 mM. The error bars indicate the standard deviation in two independent experiments each carried out in triplicate. Structures of the relevant hbp configurations are depicted above the respective panels. RLU, relative light units.
Taken together, these results provided strong evidence that HbpR directly mediated transcriptional activation from the hbpC promoter in P. azelaica HBP1. To exclude the possibility that activation from the hbpC promoter was an indirect effect of HbpR activation, the expression from PhbpC was evaluated in E. coli DH5α. Induction experiments were carried out with E. coli harboring plasmid pHYBP109 containing the hbpR-PhbpC-luxAB fusion and with E. coli harboring pHYBP110, which is similar to pHYBP109 except for a frameshift mutation in hbpR (Fig. 2). Upon addition of 2-hydroxybiphenyl, E. coli cells harboring pHYBP109 showed increased bioluminescence, which was not detected in cells containing pHYBP110 (Fig. 5C). This suggested that HbpR alone is capable of activating transcription from PhbpC. Moreover, since E. coli cannot metabolize 2-hydroxybiphenyl, these results identified 2-hydroxybiphenyl as the actual effector for HbpR.
HbpR possesses a limited effector range.
Several compounds other than 2-hydroxybiphenyl, including different monoaromatics, hydroxybiphenyls, alkylphenols, and polycyclic aromatic compounds, were tested (at 0.2 mM) for their ability to activate luciferase expression from PhbpC in P. azelaica HBP104 (Table 4). Significant induction was found only for a small group of structurally similar compounds, including 2-hydroxybiphenyl, 2,2′-dihydroxybiphenyl, 2-aminobiphenyl, and 2-hydroxydiphenylmethane (in order of the relative induction observed). Other compounds and monoaromatics did not yield significant induction. A few compounds even reduced luciferase activities to below the background observed without any effector. These included 2,3-dihydroxybiphenyl, 2,5-dihydroxybiphenyl, 3-methylcatechol, and 2,3-dihydroxybenzaldehyde.
TABLE 4.
HbpR mediated relative luciferase activities in the presence of different compounds
| Compounda | Relative induction (%)b | Compound | Relative induction (%) | |
|---|---|---|---|---|
| Biphenylic compounds | ||||
| Biphenyl | 3.4 ± 0.3 | |||
| 2-Hydroxybiphenyl | 100 ± 16 | |||
| 3-Hydroxybiphenyl | 4.4 ± 0.4 | |||
| 4-Hydroxybiphenyl | 3.5 ± 0.3 | |||
| 2,3-Dihydroxybiphenyl | 1.1 ± 0.2 | |||
| 2,5-Dihydroxybiphenyl | 0.21c ± 0.04 | |||
| 2,2′-Dihydroxybiphenyl | 84 ± 12 | |||
| 4,4′-Dihydroxybiphenyl | 2.0 ± 0.3 | |||
| 2-Aminobiphenyl | 48 ± 7 | |||
| 2-Chlorobiphenyl | 2.2 ± 0.2 | |||
| Polycyclic aromatic hydrocarbons | ||||
| Naphthalene | 2.9 ± 0.3 | |||
| 1-Naphthol | 2.4 ± 0.2 | |||
| Monoaromatic hydrocarbons | ||||
| Toluene | 3.6 ± 0.5 | |||
| Phenol | 3.0 ± 0.4 | |||
| 2-Methylphenol | 4.2 ± 0.4 | |||
| 2-Ethylphenol | 3.7 ± 0.3 | |||
| 2-Propylphenol | 3.8 ± 0.4 | |||
| Salicylic acid | 3.0 ± 0.3 | |||
| Benzoic acid | 3.3 ± 0.3 | |||
| 3-Methylcatechol | 1.0 ± 0.2 | |||
| 2,3-Dihydroxybenzaldehyde | 1.1 ± 0.1 | |||
| 2-Hydroxydiphenylmethane | 32 ± 7 | |||
| Backgroundd | 3.0 ± 0.4 |
Assays were carried out with P. azelaica HBP104. Compounds were added to the assay as 100-fold-concentrated DMSO stock solutions to obtain a final concentration of 0.2 mM.
Luciferase activities were measured after 2 h 15 min and were related to the 2-hydroxybiphenyl-induced light emission of 3.71 × 104 relative light units, which was arbitrarily set to 100%. The values reported here were obtained in two independent experiments, both performed in triplicate. Boldface numbers indicate significant induction compared to background.
The culture turned violet during the assay.
Background luciferase activity was determined by adding only DMSO instead of inducer stock solution to an assay mixture.
Spontaneous mutation in hbpR results in 2-propylphenol utilization.
A previously isolated spontaneous mutant of P. azelaica HBP1, designated strain HBP1 Prp, is capable of using the non-wild-type substrate 2-propylphenol as the sole source of carbon and energy by means of the 2-hydroxybiphenyl pathway enzymes (32) (Fig. 1C). The HbpA, HbpC, and HbpD enzymes in strain HBP1 Prp were expressed not only when grown in the presence of 2-propylphenol but also when grown in the presence of succinate (Table 3), which suggested that the strain carried a regulatory mutation. To analyze the hbpR gene of strain HBP1 Prp, it was retrieved from a partial gene library of strain HBP1 Prp chromosomal DNA in pUC18 by selection for HbpC activity in E. coli. In this way, a 6.8-kb EcoRI fragment, covering the complete hbpRC genes of the Prp mutant, was cloned. Complete double-stranded sequence analysis of the region from the start of hbpC to the 3′-end of hbpR revealed one single transition mutation, T→C, at position 613 from the start of the hbpR gene. This changed codon 205 from TGG to CGG, resulting in a Trp→Arg substitution in the deduced amino acid sequence of HbpR (Fig. 2 and 4). Overexpression of hbpR-T613C in E. coli BL21(DE3)(pLysS) as before for wild-type hbpR resulted in a similar-size protein (63 kDa) (data not shown), indicating that the mutation caused no apparent instability of the protein in E. coli. Strain HBP121 (containing a knockout of the wild-type hbpR gene) could be complemented in trans with the hbpR-T613C gene by mini-Tn5 delivery from plasmid pHYBP129 and conferred on this strain (HBP129) the ability to grow with 2-hydroxybiphenyl (data not shown). This indicated that hbpR-T613C was producing an active HbpR protein. Strain HBP129, in contrast to strains HBP1 (wild-type) and HBP121, could indeed also use 2-propylphenol as the sole source of carbon and energy. The maximum specific growth rate on 2-propylphenol was 0.082 h−1 (td = 8.5 h) for strain HBP129, which was slightly reduced in comparison with the observed μmax on 2-propylphenol for strain HBP1 Prp (μmax = 0.11 h−1, td = 6.4 h). Similar to the findings with strain HBP1 Prp, HbpA, HbpC, and HbpD activities in strain HBP129 were expressed during growth not only on 2-propylphenol but also on succinate (Table 3), although all three enzyme activities were slightly more elevated in HBP1 Prp grown with 2-propylphenol. This showed that HbpR-W205R had the same effects in HBP1 as in the mutant HBP1 Prp and that this mutation (T613C) was solely responsible for the change in activity of HbpR. The same luciferase levels were measured from the PhbpC-luxAB fusion in P. azelaica HBP104 Prp in the absence of inducer (Fig. 5B) as with strain HBP104 in the presence of 2-hydroxybiphenyl (Fig. 5A). This suggests strongly that HbpR-W205R mediates constitutive expression of the hbpC promoter. Luciferase expression in strain HBP104 Prp was even reduced in the presence of 2-hydroxybiphenyl or 2-propylphenol compared to a control reaction with DMSO only (Fig. 5B). This is probably the result of direct inhibition of the luciferase reaction by phenolic compounds (20), since this decrease was not observed from enzyme activity levels of HbpA, HbpC, or HbpD in strain HBP129 (Table 3).
DISCUSSION
In this study we characterized the P. azelaica HBP1 hbpR gene. The nucleotide sequence of the hbpR gene was determined, and the deduced protein product of 570 aa was identified by overproduction in E. coli as a protein with an apparent mass of 63 kDa. The role of the HbpR protein as a key element in the regulation of the hbpCAD genes was inferred from batch growth experiments with strains in which the hbpR gene was disrupted (as in strain HBP121) and complemented again (as in strain HBP127). Induction experiments with E. coli provided evidence that HbpR itself activated expression from the hbpC promoter. Since E. coli cannot degrade 2-hydroxybiphenyl, this also identified 2-hydroxybiphenyl as the actual inducer rather than a metabolite formed during its degradation. Analysis of a spontaneous mutant of P. azelaica HBP1 degrading 2-propylphenol identified one residue located near the C-terminal end of the A domain within HbpR, which might be important for maintaining C-domain repression.
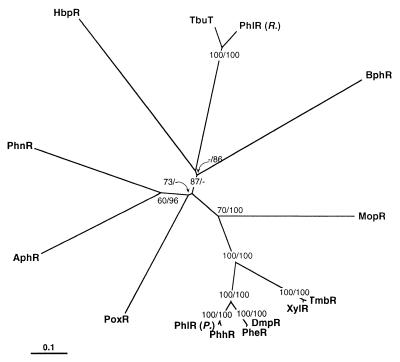
HbpR displayed significant amino acid sequence homology to members of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators. The homology to other members of the NtrC family was restricted to the central C domain. Phylogenetic analysis suggested that the HbpR protein is a distinct member clustering outside most other known XylR/DmpR-type activators (Fig. 6). Nevertheless, HbpR seems to have the same mechanism of direct effector activation as the other members of the XylR/DmpR subclass. The possibility that HbpR is part of a two-component signal transduction system seems very unlikely for two reasons: (i) the A domain of HbpR has no significant homology to the A domains of response regulators within the NtrC family, and (ii) it is very unlikely that E. coli possesses a cognate histidine kinase reacting on 2-hydroxybiphenyl.
FIG. 6.
Single unrooted phylogenetic tree of the HbpR amino acid sequence with 13 members of the XylR/DmpR subgroup. The tree was obtained by protein distance calculations (PROTDIST, Kimura setting) and subsequent neighbor-joining analysis (NEIGHBOR; both programs from the software package Phylip version 3.5 [17]) on a subalignment of 616 aa including gaps. Values at the forks indicate the number of times that particular branching occurred among 100 resamplings of the data set (generated with bootstrapping) as determined by consensus neighbor-joining analysis (first value) or maximum-parsimony analysis (second value). Missing values indicate that particular branching to be absent in the consensus tree. The consensus tree was displayed by using TREEVIEW (45). The bar indicates 0.1 change per site. The origins of the different proteins are as follows: AphR from Comamonas testosteroni TA441 (GenBank accession no. AB006480) (4); BphR encoded on plasmid pNL1 from Sphingomonas aromaticivorans F199 (GenBank accession no. AF079317) (52); DmpR encoded on plasmid pVI150 from Pseudomonas sp. strain CF600 (GenBank accession no. X68033) (59); HbpR from P. azelaica HBP1 (GenBank accession no. U73900) (this study); MopR from Acinetobacter calcoaceticus NCIB8250 (GenBank accession no. Z69251) (56); PheR from P. putida BH (GenBank accession no. D63814); PhhR from P. putida P35X NCIB9869 (GenBank accession no. X79599) (43); PhlR encoded on plasmid pPGH1 from P. putida H (GenBank accession no. X91145) (41) in the figure indicated as PhlR (P.); PhlR from a Ralstonia eutropha JMP134 derivative (GenBank accession no. AF065891), in the figure indicated as PhlR (R.); PhnR from Burkholderia sp. strain RP007 (GenBank accession no. AF061751) (35); PoxR from R. eutropha E2 (GenBank accession no. AF026065) (24); TbuT from R. pickettii PKO1 (GenBank accession no. U72645) (10); TmbR from P. putida TMB (GenBank accession no. U41301) (16); XylR encoded on plasmid pWW0 from P. putida mt-2 (GenBank accession no. M20635) (25).
Most members of the XylR/DmpR type react on a variety of monoaromatic compounds. XylR, for example, recognizes not only the growth substrates toluene, m- and p-xylene, and benzyl alcohol but also several nongrowth compounds like trimethylbenzene, ethyl- and chlorotoluene, and p-chlorobenzaldehyde (1). DmpR reacts not only with the pathway substrates phenol and methylphenols but also with dimethyl-, ethyl-, and chlorophenols, benzyl alcohol, and salicylic acid (60). HbpR is unique in the sense of recognizing compounds with a biphenyl backbone but not monoaromatic structure as effectors. We found only four compounds acting as effectors (Table 4), which all contained a biaromatic ring structure plus a hydroxy or amino group at the ortho ring position, since biphenyl itself and 3-hydroxy, 4-hydroxy-, 4,4′-dihydroxy-, and 2-chlorobiphenyl did not cause activation of HbpR. However, 1-naphthol was not an effector for HbpR activation, indicating that the overall size or orientational flexibility between the two aromatic rings is important.
Interestingly, but not unusually for pathways of aromatic degradation, the substrate spectrum of the 2-hydroxybiphenyl catabolic enzymes is different from the effector spectrum of the cognate regulator HbpR. For example, HbpA, the first enzyme of the hydroxybiphenyl pathway, hydroxylates various molecules with a 2-hydroxyphenyl-R structure, with R being a hydrophobic group (e.g., methyl, ethyl, propyl, sec-butyl, phenyl, or 2-hydroxyphenyl) (30). However, 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl are the only two compounds recognized by HbpR which are also substrates for HbpA. 2-Aminobiphenyl and 2-hydroxydiphenylmethane, on the other hand, cannot serve as substrates for HbpA (57) but are (fortuitous) inducers of the hbpCAD system. A spontaneous mutant of strain HBP1, which seemed to have overcome the regulatory constraints set by wild-type HbpR and which could benefit from the flexible capabilities of the HbpA, HbpC, and HbpD enzymes to using 2-propylphenol as growth substrate as well as 2-hydroxybiphenyl, could be isolated (32). Our results presented here demonstrated that a single mutation in the hbpR regulatory gene was responsible for allowing the mutant strain to grow on 2-propylphenol. However, the mutant HbpR protein did not seem to have acquired any special new recognition specificities. Rather, it seemed to have been locked in a constitutively active form (Table 3 and Fig. 5B). From an evolutionary perspective, this is an effective way for bacteria to deal with potentially new growth substrates. The resulting amino acid substitution (W205R), interestingly, is at a (seemingly) conserved position among activators of the XylR/DmpR type (Fig. 4). For DmpR, one mutant with the same substitution at this position (DmpR-W193R) has been described (42). This mutant was originally selected as a suppressor mutation on DmpR-V276A but does not cause a constitutive phenotype as an isolated mutation. However, since the Trp-205 residue in HbpR is very close to the interdomain hinge or Q-linker region whereas Trp-193 in DmpR is not, substitutions at this position might have different effects on regulator activity.
The DNA sequence of the hbpR gene predicts a similar subdomain (A, B, C, and D) structure for HbpR to that for the other XylR/DmpR-type activators (Fig. 4). The A domains of these regulators are presumed to be the receptor module, directly interacting with the effector molecule and transmitting this signal to the C domain, which carries the ATPase activity (58). The present understanding is that the A domain is an interdomain repressor, keeping the activity of the C domain low when no signal is present (18, 42, 44, 47). Analysis of several mutations, mostly in XylR or DmpR, has illustrated the current activation model (summarized in Fig. 4). For example, mutations which changed the effector specificity of XylR or DmpR were found exclusively in the A domain (Fig. 4). In addition, the isolated A domain of DmpR was shown to bind phenol in vitro (44). Several mutations point to the importance of a small region between residues 140 and 160 (Fig. 4) for effector binding. Since the effector spectrum of HbpR is so different from the other XylR/DmpR-type activators, it would be interesting to determine which amino acid residues allow the unique binding of biaromatic structures into (or onto) the HbpR protein. It is clearly too early to do so, but two features in the A domain attract attention. One of these is the presence of four extra amino acid residues in the HbpR sequence in the region between aa 110 and 130 compared to other XylR/DmpR members. The other feature is the considerably shorter C-terminal end of the A domain directly preceding the B-domain region (Fig. 4). This last feature is shared with the predicted sequence of BphR, a putative regulator which might be involved in regulating the expression of biphenyl degradation (52). Both HbpR and BphR also lack an otherwise conserved Gly residue (at position 199), which often points to structural conservation. The B domain of HbpR itself might still be called a ‘Q’ linker (72), with four Gln residues, whereas those of PhlR (of Ralstonia), TbuT, and PhnR can hardly be named as such (Fig. 4).
There is presently no consensus on which residues within the A domain are functionally important for all XylR/DmpR members, since most studies have concentrated on either DmpR or XylR. For example, the mutant activator DmpR-E135D has semiconstitutive activity (61) whereas the Asp residue at that position in the wild-type XylR protein does not result in semi-constitutive activity (53). XylR-D135E on its turn is a null mutant (53). This suggests that structural information will be needed to understand the subtle differences in signal transmission between the A and C domains of the different XylR/DmpR-type activators.
The specific features of the C domain are much better understood, not least because of its stronger similarity to the NtrC family of transcription activators as a whole. For example, the locations of the seven conserved motifs (C1 through C7) within the C domain (40) can readily be detected on the aligned sequences of the XylR/DmpR members (Fig. 4). The C1 and C4 motifs were shown to be involved in ATP binding and/or hydrolysis (40, 42, 48, 69). The C3 motif might form the site contacting RNAP-ς54, as recently demonstrated for the NtrC-type activator DctD (70), but it also plays a role in ATP hydrolysis (51). For the other motifs, no clear function has been demonstrated, although some (C6 and C7) also seem to be involved in ATP hydrolysis or binding (48, 51). In a number of conserved positions, the XylR/DmpR-type activators differ from the other NtrC family members (Fig. 4). For example, whereas most other NtrC-type proteins carry a His at position 302 within the C3 motif (of the HbpR numbering), most XylR/DmpR members have a Val. The same holds for other residues conserved within the NtrC family (40) but not for the XylR/DmpR members (Fig. 4). The opposite, i.e., conserved residues among XylR/DmpR members but not for NtrC as a whole, is also found (Fig. 4). Some of these substitutions seem to point at protein structure differences (e.g., conserved Pro or Gly residues), and the occurrence of five conserved residues within the XylR/DmpR subclass, found almost next to one another near the C5 motif, is very suggestive for a functional difference (Fig. 4). Such differences might be a useful starting point for future site-directed mutation studies.
The last domain of the XylR/DmpR-type activators is formed by the D domain, which is contacting the DNA at the upstream activating sequences. Preceding the D domain is a region of approximately 40 residues with little conservation among the XylR/DmpR members. Conserved within the D domain is the A(L)-X9-AA-X2-LG motif (40), proposed to correspond to the first ‘helix’ and ‘turn’ of a helix-turn-helix DNA binding motif. The RPXLAYRLXK region directly at the C-terminal part might then form the second recognition helix for XylR/DmpR members. In comparison to other NtrC members, two differences in conserved residues are visible (Fig. 4), which might point to differences in recognition specificity.
In conclusion, the discovery of a novel XylR/DmpR-type transcription activator with completely different effector specificities might open up new avenues to understanding the recognition potential of the A domain and the potential to evolve new recognition specificities. It will also be interesting to study the DNA sequence in this region and find if it is more prone to acquiring small changes.
ACKNOWLEDGMENTS
We thank V. de Lorenzo (Centro Nacional de Biotecnología, CSIC, Madrid, Spain) for kindly providing plasmids pCK218 and pUC18Not and J. Kuhn (Israel Institute of Technology, Haifa, Israel) for providing plasmid pHG171-luxAB. We also thank J. Frey (Institute of Veterinary Bacteriology, Berne, Switzerland) for providing plasmid pHP45Ω and S. E. Lindow (University of Berkeley, Berkeley, Calif.) for providing plasmid pGreenTIR.
The work of M.C.M.J. was supported by grant 5001-044754 from the Swiss Priority Program Environment.
REFERENCES
- 1.Abril M-A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almashanu S, Tuby A, Hadar R, Einy R, Kuhn J. Formation of active bacterial luciferase between interspecific subunits in vivo. J Biolumin Chemilumin. 1995;10:157–167. doi: 10.1002/bio.1170100304. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;144:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 5.Austin S, Buck M, Cannon W, Eydmann T, Dixon R. Purification and in vitro activities of the native nitrogen fixation control proteins NifA and NifL. J Bacteriol. 1994;176:3460–3465. doi: 10.1128/jb.176.12.3460-3465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;130:151–152. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 10.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohn D H, Mileham A J, Simon M I, Nealson K H, Rausch S K, Bonam D, Baldwin T O. Nucleotide sequence of the luxA gene of Vibrio harveyi and the complete amino acid sequence of the α subunit of bacterial luciferase. J Biol Chem. 1985;260:6139–6146. [PubMed] [Google Scholar]
- 12.Delgado A, Ramos J-L. Genetic evidence for activation of the positive transcriptional regulator XylR, a member of the NtrC family of regulators, by effector binding. J Biol Chem. 1994;269:8059–8062. [PubMed] [Google Scholar]
- 13.Delgado A, Salto R, Marqués S, Ramos J L. Single amino acids changes in the signal receptor domain of XylR resulted in mutants that stimulate transcription in the absence of effectors. J Biol Chem. 1995;270:5144–5150. doi: 10.1074/jbc.270.10.5144. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 15.Eckert J W. Control of postharvest diseases. In: Siegel M R, Sisler H D, editors. Antifungal compounds. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1977. pp. 269–352. [Google Scholar]
- 16.Favaro R, Bernasconi C, Passini N, Bertoni G, Bestetti G, Galli E, Dehò G. Organisation of the tmb catabolic operons of Pseudomonas putida TMB and evolutionary relationship with the xyl operons of the TOL plasmid pWW0. Gene. 1996;182:189–193. doi: 10.1016/s0378-1119(96)00552-5. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 18.Fernández S, de Lorenzo V, Pérez-Martin J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 19.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folkert F. Ph.D. thesis. Groningen, The Netherlands: Rijksuniversiteit Groningen; 1996. [Google Scholar]
- 21.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. [Google Scholar]
- 22.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higson F K, Focht D D. Bacterial metabolism of hydroxylated biphenyls. Appl Environ Microbiol. 1989;55:946–952. doi: 10.1128/aem.55.4.946-952.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hino S, Watanabe K, Takahashi N. Phenol hydroxylase cloned from Ralstonia eutropha strain E2 exhibits novel kinetic properties. Microbiology. 1998;144:1765–1772. doi: 10.1099/00221287-144-7-1765. [DOI] [PubMed] [Google Scholar]
- 25.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988;66:301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 26.Izumi Y, Ohshiro T, Ogino H, Hine Y, Shimao M. Selective desulfurization of dibenzothiophene by Rhodococcus erythropolis D-1. Appl Environ Microbiol. 1994;60:223–226. doi: 10.1128/aem.60.1.223-226.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston T C, Thompson R B, Baldwin T O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the β subunit of bacterial luciferase. J Biol Chem. 1986;261:4805–4811. [PubMed] [Google Scholar]
- 28.Kay R, McPherson J. Hybrid pUC vectors for addition of new restriction enzyme sites to the ends of DNA fragments. Nucleic Acids Res. 1987;15:2778. doi: 10.1093/nar/15.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kayser K J, Bielaga-Jones B A, Jackowski K, Odusan O, Kilbane J J., II Utilization of organosulphur compounds by axenic and mixed cultures of Rhodococcus rhodochrous IGTS8. J Gen Microbiol. 1993;139:3123–3129. [Google Scholar]
- 30.Kohler H-P E, Kohler-Staub D, Focht D D. Degradation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1. Appl Environ Microbiol. 1988;54:2683–2688. doi: 10.1128/aem.54.11.2683-2688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler H-P E, Schmid A, van der Maarel M. Metabolism of 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1: production and consumption of 2,2′,3-trihydroxybiphenyl. J Bacteriol. 1993;175:1621–1628. doi: 10.1128/jb.175.6.1621-1628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler H-P E, van der Maarel M J E C, Kohler-Staub D. Selection of Pseudomonas sp. strain HBP1 Prp for metabolism of 2-propylphenol and elucidation of the degradative pathway. Appl Environ Microbiol. 1993;59:860–866. doi: 10.1128/aem.59.3.860-866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 35.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leveau J H J, van der Meer J R. Genetic characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutropha JMP134. Gene. 1997;202:103–114. doi: 10.1016/s0378-1119(97)00460-5. [DOI] [PubMed] [Google Scholar]
- 37.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 39.Miller W G, Lindow S E. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 40.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng L C, O'Neill E, Shingler V. Genetic evidence for interdomain regulation of the phenol-responsive ς54-dependent activator DmpR. J Biol Chem. 1996;271:17281–17286. doi: 10.1074/jbc.271.29.17281. [DOI] [PubMed] [Google Scholar]
- 43.Ng L C, Poh C L, Shingler V. Aromatic effector activation of the NtrC-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J Bacteriol. 1995;177:1485–1490. doi: 10.1128/jb.177.6.1485-1490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill E, Ng L C, Sze C C, Shingler V. Aromatic ligand binding and intramolecular signalling of the phenol-responsive ς54-dependent regulator DmpR. Mol Microbiol. 1998;28:131–141. doi: 10.1046/j.1365-2958.1998.00780.x. [DOI] [PubMed] [Google Scholar]
- 45.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 46.Pavel H, Forsman M, Shingler V. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J Bacteriol. 1994;176:7550–7557. doi: 10.1128/jb.176.24.7550-7557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Martín J, de Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Martín J, de Lorenzo V. In vitro activities of an N-terminal truncated form of XylR, a ς54-dependent transcriptional activator of Pseudomonas putida. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 49.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 50.Ravatn R, Studer S, Zehnder A J B, van der Meer J R. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin Y-K, Kustu S. MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J Bacteriol. 1999;181:4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salto R, Delgado A, Michán C, Marqués S, Ramos J L. Modulation of the function of the signal receptor domain of XylR, a member of a family of prokaryotic enhancer-like positive regulators. J Bacteriol. 1998;180:600–604. doi: 10.1128/jb.180.3.600-604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 56.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid A. Ph.D. thesis. Stuttgart, Germany: University of Stuttgart; 1997. [Google Scholar]
- 58.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 59.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shingler V, Pavel H. Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol Microbiol. 1995;17:505–513. doi: 10.1111/j.1365-2958.1995.mmi_17030505.x. [DOI] [PubMed] [Google Scholar]
- 62.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 63.Sticher P, Jaspers M C M, Stemmler K, Harms H, Zehnder A J B, van der Meer J R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 66.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 67.Suske W A. Ph.D. thesis. Zürich, Switzerland: Swiss Federal Institute of Technology; 1998. [Google Scholar]
- 68.Suske W A, Held M, Schmid A, Fleischmann T, Wubbolts M G, Kohler H-P. Purification and characterization of 2-hydroxybiphenyl 3-monooxygenase, a novel NADH-dependent, FAD-containing aromatic hydroxylase from Pseudomonas azelaica HBP1. J Biol Chem. 1997;272:24257–24265. doi: 10.1074/jbc.272.39.24257. [DOI] [PubMed] [Google Scholar]
- 69.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y-K, Lee J H, Brewer J M, Hoover T R. A conserved region in the ς54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol Microbiol. 1997;26:373–386. doi: 10.1046/j.1365-2958.1997.5851955.x. [DOI] [PubMed] [Google Scholar]
- 71.Wick L Y, Gschwend P M. Source and chemodynamic behavior of diphenyl sulfone and ortho- and para-hydroxybiphenyl in a small lake receiving discharges from an adjacent Superfund site. Environ Sci Technol. 1998;32:1319–1328. [Google Scholar]
- 72.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 73.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 74.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]