Abstract
Differential gene expression during Bacillus subtilis sporulation is controlled by sigma factors and other regulatory effectors. The first compartmentalized sigma factor, ςF, is active specifically in the prespore compartment. During our screening for new chromosome segregation mutants using a ςF-dependent gpr-lacZ reporter as a probe, we identified a new gene (ywfN) required for maximal expression of the reporter and named it rsfA. The product of rsfA has features of gene regulatory proteins, and the protein colocalizes with DNA. The expression of rsfA is under the control of both ςF and ςG. Null mutations in rsfA have different effects on the expression of ςF-dependent genes, suggesting that the RsfA protein is a regulator of transcription that fine-tunes gene expression in the prespore.
During sporulation in Bacillus subtilis, differential gene expression in the prespore and the mother cell compartments is governed by four different, sporulation-specific sigma factors, whose activities are tightly regulated both temporally and spatially (9, 11, 18, 30). Two sigma factors are active specifically in the prespore—first ςF and then later ςG—while ςE and then ςK control transcription in the mother cell.
In the mother cell, the expression of both the ςE and ςK regulons has been shown to be subject to extra levels of control by small DNA-binding transcriptional regulators, SpoIIID and GerE (reviewed recently in reference 18). Similarly, in the prespore, the SpoVT protein has been identified to control ςG-dependent gene expression (3). A number of genes are now known to be regulated by ςF, including various genes required for sporulation (gpr [33], spoIIIG [32], spoIIR [16, 23], and spoIIQ [22]), a catalase (katX [2]), a dd-carboxypeptidase (dacF [28]), and several genes of unknown function (6), but so far, no factor capable of modulating ςF-dependent transcription has been found. Here we report the identification and characterization of a new prespore-specific regulatory gene, rsfA, which codes for a protein with similarity to regulatory leucine zipper proteins. We show that the expression of rsfA is dependent on both ςF and ςG. Disruption of rsfA had different effects on the expression of ςF-dependent genes. Consistent with its role as a transcriptional regulator, an RsfA-green fluorescent protein (GFP) fusion protein was shown to colocalize with chromosomal DNA. Limited experiments with a paralogue of rsfA, called ylbO, suggest that it may play an analogous role to rsfA in the control of mother cell gene expression.
MATERIALS AND METHODS
Bacterial strains.
The B. subtilis strains and plasmids used in this study are described in Table 1. The Escherichia coli strain used was DH5α [F− endA1 hsdR17 (rK− mK+) supE44 λ− thi-1 recA1 gyrA96 (Nalr) relA1 Δ(lacZYA-argF)U169 φ80dlacΔ(lacZ)M15; Gibco-BRL].
TABLE 1.
B. subtilis strains and plasmids used
| Strain or plasmid | Relevant genotypea | Construction, source, or referenceb |
|---|---|---|
| Strains | ||
| 168 | trpC2 | Laboratory stock |
| 36.3 | trpC2 spoIIIE36 | 12 |
| BFA1335 | trpC2 rsfA::pMUTIN4-′rsfA′ | Gene Function Analysis Strain Collection |
| BFA3241 | trpC2 ylbO::pMUTIN4-′ylbO′ | Gene Function Analysis Strain Collection |
| BS5:JH642 | trpC2 pheA1 amyE::spoIIQ-lacZ cat | 22 |
| IB427 | katX::katX-lacZ cat | 2 |
| MO2029 | trpC2 pheA1 amyE::spoIIR-lacZ cat | 23 |
| PS943 | spoIIIGΔ1 amyE::spoIIIG-lacZ cat | P. Setlow, unpublished data |
| PS1287 | amyE::sspE-2G-lacZ cat | 31 |
| 1.5 | trpC2 spoIIAC1 | 10 |
| 168 (pDG298) | trpC2 (pDG298) Pspac-spoIIIG kan | 32 |
| 735 | trpC2 spoIIAC::pSG635 (lacI Pspac-spoIIAC kan) | S. R. Partridge, unpublished data |
| 901 | trpC2 spoIIGA::aphA3 | 35 |
| 1224 | trpC2 Δ(ywhD-rsfA)::(′neo tet) | Materials and Methods |
| 1225 | trpC2 ΔrsfA::tet | Materials and Methods |
| 1226 | trpC2 rsfA::pMUTIN4-′rsfA′ spoIIAC1.5 | BFA1335 (chr)→1.5 |
| 1227 | trpC2 rsfA::pMUTIN4-′rsfA′ spoIIGA::aphA3 | BFA1335 (chr)→901 |
| 1228 | trpC2 rsfA::pMUTIN4-′rsfA′ spoIIIG::aphA3 | BFA1335 (chr)→1248 |
| 1229 | trpC2 rsfA::pMUTIN4-′rsfA′ spoIIAC::pSG635 (lacI Pspac-spoIIAC kan) | 735 (chr)→BFA1335 |
| 1230 | trpC2 rsfA::pMUTIN4-′rsfA′ (pDG298) Pspac-spoIIIG kan | 168 (pDG298)→BFA1335 |
| 1231 | trpC2 ΔrsfA::tet spoIIIG::aphA3 | 1248 (chr)→1225 |
| 1232 | trpC2 ΔrsfA::tet spoIIIG::aphA3 amyE::spoIIR-lacZ cat | MO2029 (chr)→1231 |
| 1233 | trpC2 ΔrsfA::tet spoIIIG::aphA3 amyE::spoIIQ-lacZ cat | BS5:JH642 (chr)→1231 |
| 1234 | trpC2 ΔrsfA::tet spoIIIG::aphA3 katX::katX-lacZ cat | IB427 (chr)→1231 |
| 1235 | trpC2 ΔrsfA::tet spoIIIG::aphA3 amyE::spoIIIG-lacZ cat | PS943 (chr)→1231 |
| 1236 | trpC2 ΔrsfA::tet spoIIIG::aphA3 amyE::sspE-2G-lacZ cat | PS1287 (chr)→1231 |
| 1237 | trpC2 ΔrsfA::tet amyE::(rsfA+ neo) | Materials and Methods |
| 1239 | trpC2 rsfA::pMUTIN4-′rsfA′ amyE::(rsfA+ neo) | 1237 (chr)→BFA1335 |
| 1241 | trpC2 amyE::Pxyl-rsfA cat | pSG4907→168 |
| 1242 | trpC2 rsfA::pSG4904 (rsfA-gfp cat) | pSG4904→168 |
| 1243 | trpC2 spoIIIG::aphA3 amyE::spoIIR-lacZ cat | MO2029 (chr)→1248 |
| 1244 | trpC2 spoIIIG::aphA3 amyE::spoIIQ-lacZ cat | BS5:JH642 (chr)→1248 |
| 1245 | trpC2 spoIIIG::aphA3 katX::katX-lacZ cat | IB427 (chr)→1248 |
| 1246 | trpC2 spoIIIG::aphA3 amyE::spoIIIG-lacZ cat | PS943 (chr)→1248 |
| 1247 | trpC2 spoIIIG::aphA3 amyE::sspE-2G-lacZ cat | PS1287 (chr)→1248 |
| 1248 | trpC2 spoIIIG::aphA3 | D. Foulger, unpublished data |
| 1249 | trpC2 amyE::gpr-lacZ cat | pPS1326→168 |
| 1250 | trpC2 amyE::gpr-lacZ cat ΔrsfA::tet | 1225 (chr)→1249 |
| 1251 | trpC2 amyE::Pgpr-gfp cat | pSG4908→168 |
| 1252 | trpC2 amyE::Pgpr-gfp cat spoIIAC::pSG635 (lacI Pspac-spoIIAC kan) | 735 (chr)→1251 |
| 1253 | trpC2 rsfA::pSG4904 (rsfA-gfp cat) spoIIAC::pSG635 (lacI Pspac-spoIIAC kan) | 735 (chr)→1242 |
| 1254 | trpC2 spoIIIE36 rsfA::pSG4904 (rsfA-gfp cat) | pSG4904→36.3 |
| 1255 | trpC2 ylbO::pMUTIN4-′ylbO′ spoIIAC1 | BFA3241 (chr)→1.5 |
| 1256 | trpC2 ylbO::pMUTIN4-′ylbO′ spoIIGA::aphA3 | BFA3241 (chr)→901 |
| Plasmids | ||
| pBEST309 | bla tet | 13 |
| pBEST502 | bla neo | 14 |
| pJPR1 | bla amyE′ cat Pxyl ′amyE | J. Rawlins, unpublished data |
| pMLK83 | bla amyE′ neo gusA ′amyE | 17 |
| pPS1326 | gpr-lacZ in ptrpBG1 | 31 |
| pRD154 | bla amyE′ ′amyE | R. Daniel, unpublished data |
| pSG635 | bla kan lacI Pspac-spoIIAC | 26 |
| pSG4903 | bla amyE′ neo ′amyE | neo from pMLK83 (XbalI+NotI)→pRD154 (SpeI+NotI) |
| pSG1151 | bla gfp cat | 20 |
| PSG1154 | bla amyE′ spc Pxyl-gfp ′amyE | 20 |
| pSG4904 | bla ′rsfA-gfp cat | ′rsfA (PCR, XhoI+EcoRI)→pSG1151 (XhoI+EcoRI) |
| pSG4905 | bla ′neo | pBEST502 (Bsp120I+NotI) religated |
| pSG4906 | bla 'neo tet | tet from pBEST309 (NotI)→pSG4905 (NotI) |
| pSG4907 | bla amyE′ cat Pxyl-rsfA ′amyE | rsfA (PCR, EcoRI+SpeI)→pJPR1 (EcoRI+SpeI) |
| pSG4908 | bla amyE′ Pgpr-gfp cat ′amyE | cat Pgpr of pPS1326 (SalI+HindIII)→pSG1154 (SalI+HindIII) |
′X′, internal segment of gene X; ′X or X′, 5′ or 3′ truncation, respectively, of gene X; aphA3, kanamycin resistance gene; cat, chloramphenicol resistance gene; tet, tetracycline resistance gene; neo, neomycin resistance gene; Pxyl, xylose-inducible promoter; Pspac, IPTG-inducible promoter; gfp, gene for the mut1 derivative of GFP (4).
For strains constructed by transformation, the source of the DNA used in the transformation is given first, and the recipient strain is indicated after the arrow. chr, genomic DNA.
General methods.
B. subtilis cells were made competent for transformation with DNA by the method of Anagnostopoulos and Spizizen (1) as modified by Jenkinson (15). B. subtilis chromosomal DNA was prepared by a scaled-down method based on the one described by Errington (8). DNA manipulations and E. coli transformations were carried out by standard methods (27).
The solid medium used for growing B. subtilis was nutrient agar (Oxoid). Chloramphenicol (5 μg ml−1), kanamycin (5 μg ml−1), tetracycline (12 μg ml−1), erythromycin (1 μg ml−1) and lincomycin (25 μg ml−1), and 0.01% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were added as required. The media used for growing E. coli were 2× TY (tryptone-yeast extract) (27) and nutrient agar (Oxoid) supplemented with ampicillin (100 μg ml−1) as required.
PCR primers.
Detailed information on the primers used is available upon request.
Construction of the deletion strains 1224 [Δ(ywhD-rsfA)] and 1225 (ΔrsfA) by double-crossover homologous recombination.
The 1,513-bp DNA fragment containing the C-terminal coding region of ywhE and the 1,454-bp DNA fragment containing ywfM and part of ywfL were amplified by PCR with Pfu DNA polymerase with primers ywhE-F and ywhE-R and ywfM-F and ywfL-R, respectively. The two PCR products were then digested with SalI, ligated to an XhoI-XhoI fragment containing the tetracycline resistance gene (isolated from plasmid pSG4906). The ligation reaction was transformed into a wild-type B. subtilis strain, 168, with selection for tetracycline resistance. One of the transformants was examined by PCR to confirm that it had contained the deletion, and the strain was designated 1224. The deleted region was 10 kb long and contained genes ywhD-A, thrZ, mmr, ywgBA, ywfO, ywzC, and rsfA.
Strain 1225 (ΔrsfA) was constructed in a similar way, with primers ywfO-F2 and ywfN-R5 and ywfN-F2 and ywfL-R. The PCR products were digested with SstI and PstI, respectively, and ligated to the tetracycline resistance gene isolated from pBEST309 (digested with SstI and PstI). The ligation reaction was transformed into strain 168 directly, and one of the transformants was examined by PCR to confirm the deletion.
Identification of the gene responsible for the phenotype of the deletion in strain 1224.
DNA fragments containing the ywhD-to-mmr (6 kb) and mmr-to-rsfA (6.9 kb) regions were amplified by long-range PCR with primers ywhD-F and mmr-R and thrZ-F and ywfM-R, respectively. The PCR products were then digested with XhoI, ligated to SalI-digested pPS1326, and transformed directly into strain 1224. The transformants were examined by long-range PCR to confirm the structure of the insertions at the amyE locus.
Complementation analysis with small overlapping DNA fragments (of approximately 1 to 1.2 kb) in the mmr-rsfA region was performed in the same way with different primers.
Insertion of rsfA at amyE.
A 1,240-bp DNA fragment containing the rsfA gene (and its putative promoter) was amplified by PCR with primers ywzC-F and ywfM-R and Pfu DNA polymerase. The PCR product was digested with restriction enzyme SalI, ligated to SalI-digested pSG4903, and then transformed directly into strain 1225, with selection for kanamycin resistance. Several Spo+-appearing transformants were first checked for an Amy− phenotype and then were examined by PCR and with restriction enzyme digestions to confirm the site of the insertion and its orientation relative to amyE. Strain 1237 had rsfA inserted in the opposite orientation relative to amyE.
Induction of sporulation.
B. subtilis cells were grown in hydrolyzed casein growth medium (CH) at 37°C and induced to sporulate by the resuspension method of Sterlini and Mandelstam (29), as specified by Partridge and Errington (25). Times (minutes) after resuspension of cells in the starvation medium were denoted t0, t60, etc. Spore heat resistance (85°C, 15 min) was measured 9 h after the initiation of sporulation and expressed as the percentage of viable cells.
Induction of sigma factors during vegetative growth.
Overnight cultures grown at 30°C were diluted in fresh warm medium to an optical density at 570 nm (OD570) of 0.03 and grown at 37°C. For each strain, the culture was divided into two portions at an OD570 of 0.2, and IPTG (isopropyl-β-d-thiogalactopyranoside [1 mM, final concentration]) was added to one of the two cultures. The incubation of both cultures was continued and samples were removed at intervals for monitoring β-galactosidase and OD570.
Assay for β-galactosidase activity.
β-Galactosidase activity was measured either with ONPG (o-nitrophenyl-β-d-galactopyranoside) and expressed as Miller units as described by Daniel et al. (5) or with MUG (4-methylumbelliferyl-β-d-galactopyranoside) and expressed as MUG units by the method of Errington and Mandelstam (10). One MUG unit is the amount of enzyme that catalyzes the production of 1 nmol of 4-methylumbelliferone per min.
Live cell fluorescence microscopy.
Live cell examination of GFP and DAPI (4,6-diamidino-2-phenylindole) fluorescence and image capture were done as described previously (36).
RESULTS
Identification of the rsfA gene.
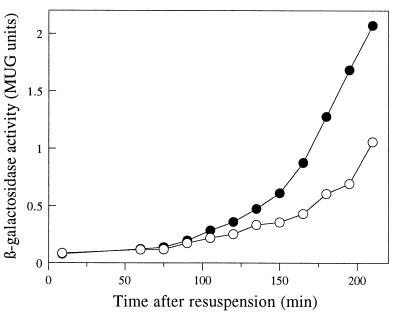
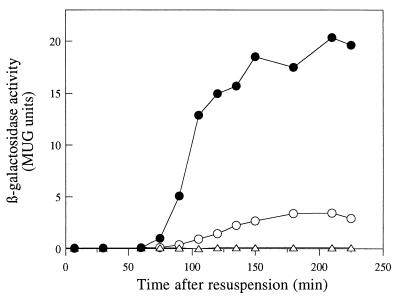
In the course of conducting a screen for chromosome segregation mutants, we noticed that deletion of a 10-kb fragment from the chromosome (3,851 to 3,861 kb), containing genes ywhD to ywfN, reduced the level of expression of a prespore-specific reporter gene, gpr-lacZ (data not shown). When introduced into wild-type strain 168, the deletion caused a reduction in sporulation frequency of about 30% (strain 1224). To identify the gene(s) or DNA sequence(s) responsible for this phenotype, we reintroduced into strain 1224 segments of DNA from the deleted region at the amyE locus, in two overlapping fragments of about 6 kb each (ywhD to mmr and mmr to ywfN). Only the fragment containing genes mmr to ywfN complemented the Spo phenotype of strain 1224. We then divided this region into seven overlapping fragments and again introduced the individual fragments into strain 1224 at the amyE locus. Only one of the fragments, the one containing the intact ywfN gene, showed complementation of the Spo phenotype. We next constructed a ΔywfN::tet null mutant (strain 1225) and confirmed that the mutant showed a decrease in gpr-lacZ expression (Fig. 1) and reduction in frequency of heat resistance similar to that of strain 1224. We renamed the ywfN gene rsfA (for regulator of sigma F-dependent gene expression [see below]).
FIG. 1.
Disruption of rsfA reduces the expression of a ςF-dependent reporter gene, gpr-lacZ. Wild-type (●) and the ΔrsfA::tet mutant (○) strains containing a gpr-lacZ fusion were induced to sporulate, and samples were taken at intervals for assay of β-galactosidase activity. Time zero is the time of initiation of sporulation.
In the course of these experiments, an rsfA (ywfN) mutant became available via the European Community-sponsored Functional Analysis Project on B. subtilis. The BFA1335 mutant contains an insertion of the nonreplicating pMUTIN4 plasmid (34), which disrupts rsfA and fuses its promoter to lacZ. Again, this mutant showed a reduced sporulation frequency similar to that of strain 1224.
The rsfA gene encodes a protein 258 amino acids long with an acidic isoelectric point (5.1). The deduced amino acid sequence showed some features of eukaryotic gene regulatory proteins, including a stretch of basic amino acids and a possible leucine zipper motif, suggesting that the protein forms dimers or multimers by means of a coiled-coil structure (Fig. 2).
FIG. 2.
The deduced amino acid sequence of RsfA. The stretch of basic amino acids is underlined. The heptad repeat of leucine residues characteristic of “leucine zippers” is indicated by underlined boldface type.
The expression of rsfA is prespore specific and is dependent on both ςF and ςG.
To monitor the expression of the rsfA gene, strain BFA1335 was grown in CH medium and then induced to sporulate, and samples were taken for analysis of β-galactosidase expression during both vegetative growth and sporulation. No expression of rsfA-lacZ could be detected during vegetative growth (data not shown). During sporulation, expression was induced from about t75 (75 min after the initiation of sporulation) (Fig. 3). Expression was abolished in a spoIIAC mutant (in which ςF is not formed), and it was approximately doubled in a spoIIGA mutant (in which ςE is not activated) (Fig. 3A), which is characteristic of several ςF-dependent genes (16, 21–23).
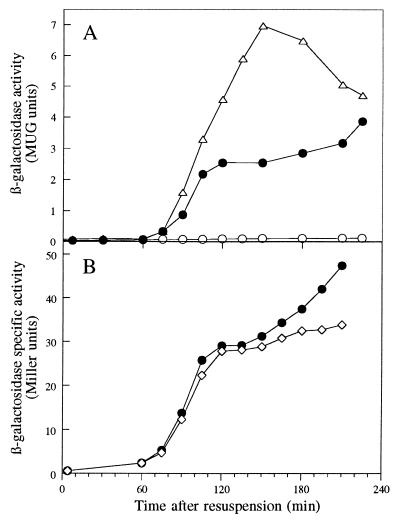
FIG. 3.
Effects of mutations in genes encoding sporulation-specific sigma factors on expression of rsfA-lacZ. Strains containing an rsfA-lacZ fusion in a wild-type background (● [strain BFA1335]) or in the presence of mutations in spoIIAC (○ [strain 1226]) and spoIIGA (▵ [strain 1227]) (A) or spoIIIG (◊ [strain 1228]) (B) were induced to sporulate at time zero, and samples were taken at intervals for assay of β-galactosidase activity.
The expression of rsfA-lacZ was reduced slightly at later times in a spoIIIG mutant (Fig. 3B), suggesting that expression of rsfA is driven not only by EςF, but also by EςG. This was confirmed by the strong induction of rsfA-lacZ during exponential growth in cells engineered to synthesize either ςF or ςG (Fig. 4).
FIG. 4.
Induction of rsfA-lacZ in vegetative cells producing ςF or ςG. Strains 1229 (▵, ▴ [producing ςF under Pspac control]), 1230 (□, ■ [producing ςG under Pspac control]), and BFA1335 (○, ● [an isogenic strain without the Pspac construct]) were grown in 2× TY, and samples from cultures with (solid symbols) or without (open symbols) IPTG were taken for assay of β-galactosidase activity.
Effects of an rsfA null mutation on prespore-specific gene expression.
The structural similarity of RsfA to leucine zipper proteins and the existence of the basic amino acid stretch suggested that the protein might function as a transcriptional regulator. Since rsfA was expressed long before ςG becomes active, it was likely that RsfA mainly regulates ςF-dependent transcription. To test this, we examined the expression of lacZ fusions to several ςF-dependent genes in the ΔrsfA::tet null mutant. To eliminate possible secondary effects on ςG (because the recognition specificities of ςF and ςG overlap [31, 32]), the strains used also carried a spoIIIG deletion. The rsfA mutation had small but reproducible effects on the levels of transcription from all of the promoters tested, except that of spoIIQ, which was unaffected (typical results are shown in Fig. 5). Expression of fusions to the katX, spoIIIG, and sspE-2G promoters was, like that of gpr, reduced in an rsfA mutant background compared with that of the wild type. Interestingly, in contrast, the spoIIR gene showed increased expression, suggesting that it is negatively regulated by RsfA (Fig. 5D). Similar results were obtained in a spoIIIG+ background (data not shown), in accordance with the idea that RsfA mainly affects transcription directed by EςF.
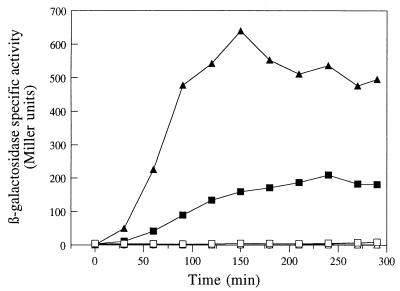
FIG. 5.
Contrasting effects of an rsfA null mutation on expression of lacZ fusions to various prespore-specific genes. Isogenic strains containing a spoIIIG mutation and lacZ fusions to katX (A [strains 1234 and 1245]), spoIIIG (B [strains 1235 and 1246]), sspE-2G (C [strains 1236 and 1247]), spoIIR (D [strains 1232 and 1243]), spoIIQ (E [strains 1233 and 1244]), or rsfA (F [see below]) were induced to sporulate at time zero, and samples were taken at intervals for assay of β-galactosidase activity. In panels A to E, results are compared for the rsfA+ strain (solid symbols) and the isogenic rsfA::tet derivative (open symbols). In panel F, the open symbols correspond to strain BFA1335 (rsfA::pMUTIN4 rsfA-lacZ), and the solid symbols correspond to a derivative carrying a wild-type copy of rsfA inserted into the amyE locus (strain 1239).
Negative autoregulation of rsfA expression.
To test whether rsfA was autoregulated, we introduced a wild-type copy of rsfA at the amyE locus into the strain BFA1335. The expression of rsfA-lacZ in the resulting strain, 1239, was then examined and compared with that of BFA1335. As shown in Fig. 5F, the presence of a wild-type copy of rsfA reduced the expression of rsfA-lacZ at later time points. It thus, appeared that rsfA exerts negative regulation on its own promoter, as on the spoIIR promoter.
An RsfA-GFP fusion colocalizes with chromosomal DNA.
The different effects of the rsfA null mutation on the expression of prespore-specific genes suggested that the protein acts directly on DNA, rather than by affecting the stability or activity of ςF. As one approach to testing whether the protein associates with DNA in vivo, we examined the subcellular localization of RsfA by making a fusion of the GFP from Aequorea victoria to the C terminus of RsfA. The strain, 1242, containing the rsfA-gfp fusion construct (and a truncated rsfA gene) produced an amount of heat-resistant spores similar to that of wild-type strain 168 (not shown), suggesting that the fusion protein is functional. As shown in Fig. 6A, the RsfA-GFP fusion protein was localized to the prespore in wild-type strain 1242, as expected for a ςF-dependent gene. To see more clearly the relative locations of the fusion protein and DNA, we used a spoIIIE36 mutant, in which only 30% of the prespore chromosome is present in the prespore compartment (35). Figure 6B shows that in the spoIIIE36 mutant, the RsfA-GFP fusion protein colocalized with the prespore chromosome.
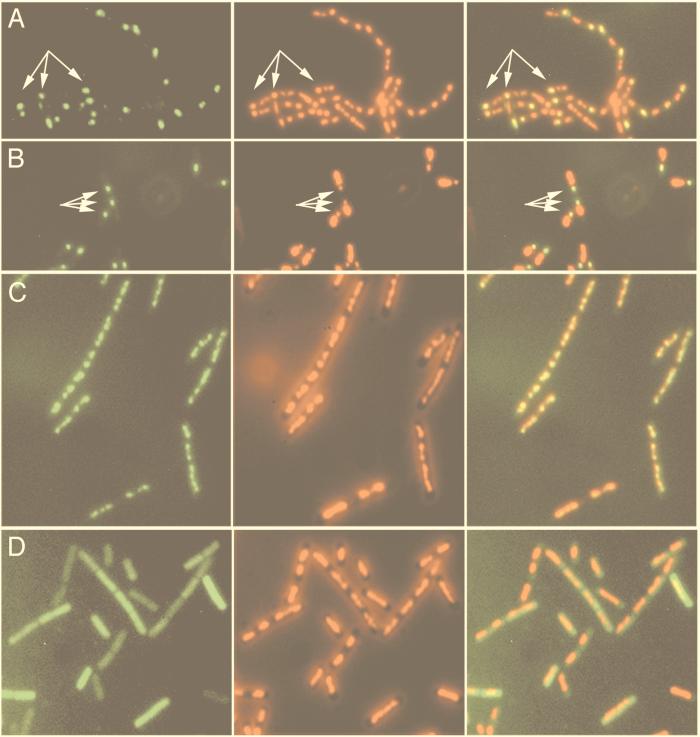
FIG. 6.
Fluorescence microscopic localization of RsfA-GFP in sporulating cells or in vegetative cells engineered to produce ςF. (A and B) Sporulating cells (t180) of the wild type (A [strain 1242]) or spoIIIE36 mutant (B [strain 1254]) containing the rsfA-gfp fusion. (C and D). Cells of strains containing an IPTG-inducible spoIIAC gene allowing ectopic synthesis of ςF in vegetative cells and either the rsfA-gfp fusion (C [strain 1253]) or a Pgpr-gfp fusion (D [strain 1252]) 120 min after the addition of IPTG. (Left panels) GFP signals showing the localization of the GFP fusions. (Middle panels) DAPI signals showing the nucleoids. (Right panels) Overlays of the GFP and corresponding DAPI images. Arrows point to some of the prespore compartments.
The affinity of RsfA for DNA was further demonstrated by the colocalization of the RsfA-GFP fusion to chromosomal DNA when the expression of rsfA-gfp was induced during vegetative growth by the synthesis of ςF, as shown in Fig. 6C. Under the same conditions, the control Pgpr-GFP fusion (also ςF-dependent) showed a uniformly distributed GFP signal throughout the whole cell (Fig. 6D), confirming that the localization pattern of RsfA-GFP was not caused by the GFP moiety of the fusion protein.
Interestingly, when rsfA was placed under the control of a xylose-inducible Pxyl promoter and strain 1241 was induced to synthesize RsfA during vegetative growth, the cells showed impaired chromosome distribution and segregation and eventually lysed (data not shown). The molecular basis for this effect is not clear, but it could be consistent with the notion that RsfA acts directly on DNA.
The ylbO gene encodes a protein with a high degree of similarity to RsfA.
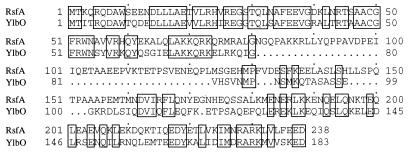
A search of the B. subtilis genome DNA sequence revealed that the ylbO gene located at 1,575.5 kb on the chromosome encodes a 193-amino-acid protein with high similarity to RsfA, particularly over the N- and C-terminal regions of the proteins (Fig. 7).
FIG. 7.
Alignment of the deduced amino acid sequences of RsfA and YlbO. Identical residues are boxed.
To test whether the function of ylbO was related to that of rsfA, we examined the phenotype of strain BFA3241, in which ylbO has been disrupted and its promoter fused to lacZ. This strain showed no obvious phenotype, and it produced amounts of heat-resistant spores similar to those produced by wild-type strain 168 (not shown). Nevertheless, expression of the ylbO-lacZ fusion was induced to a rather high level during sporulation (Fig. 8). In contrast to rsfA, the expression of ylbO-lacZ was completely abolished by a mutation in the spoIIGA gene required for activation of ςE (Fig. 8). In view of this dependence and the relatively early timing of expression during sporulation, ylbO is almost certainly a mother cell-specific gene under the control of ςE (9), but its function remains unknown.
FIG. 8.
Expression of ylbO-lacZ during sporulation. Isogenic strains containing a ylbO-lacZ fusion were induced to sporulate at time zero, and samples were taken at intervals for assay of β-galactosidase activity. The strains used were otherwise wild type (● [strain BFA3241]) or carried mutations in the spoIIAC (○ [strain 1255]) or spoIIGA (▵ [strain 1256]) gene.
DISCUSSION
We have discovered the first accessory regulator for ςF-dependent transcription, RsfA. Its gene was initially identified as being required for maximal expression of the prespore-specific gpr-lacZ fusion in a spoIIIE36 mutant. We then found that rsfA disruption affects the expression of most genes in the ςF regulon, suggesting that it is a global regulator of transcription. Comparison of previous published results on expression of genes in the ςF regulon reveals a range of subtle differences in their levels of expression and kinetics, and this is well illustrated by the time courses shown in Fig. 1 and 5. Some of the differences in kinetics are likely to be due to differential dependence of the promoters on EςF and EςG. However, RsfA can now also be implicated in these effects. Inspection of the data shown in Fig. 1 and 5 reveals that the genes that are expressed independently of RsfA (spoIIQ) or are negatively regulated by it (spoIIR and rsfA itself) show relatively early and steep expression kinetics (at least as judged by the lacZ fusion data in Fig. 5D to F). This would be consistent with their promoters being recognizable by EςF independent of accessory factors. In contrast, those genes that are positively regulated by RsfA (gpr, katX, spoIIIG, and sspE-2G) all show later kinetics, and their rate of rise to maximal expression is slower. This could reflect the need for the RsfA product to accumulate in the cell before these genes become fully expressed, as we suggested previously for the action of SpoIIID on some genes in the ςE regulon (12). Thus, although RsfA is not essential for sporulation, it appears to improve the efficiency of sporulation by fine-tuning the expression of genes in the ςF regulon, particularly the timing of their expression. It is not yet clear whether RsfA also modulates the expression of ςG-dependent genes. However, since rsfA can also be transcribed by EςG, as for many other ςF-dependent genes (e.g., gpr and spoIIIG), it would not be surprising if RsfA also played a role in the regulation of some ςG-dependent genes. The deleterious effects of expression of rsfA in vegetative cells were surprising. It seems possible that one role of this protein could be to shut down expression of some genes that are needed for vegetative growth, but not in the developing prespore. However, it is also possible that this effect is an artifact due to overproduction of RsfA.
The RsfA protein has a leucine zipper motif (Fig. 2), which is present in many eukaryotic gene regulatory proteins, such as the Jun/AP1 family of transcription factors (24) and the yeast general control protein GCN4 (7). It has been proposed that the leucine side chains extending from one α-helix interact with those from a similar α-helix of a second polypeptide, forming a coiled-coil which facilitates formation of dimers or multimers. RsfA also contains a stretch of basic amino acids (Fig. 2), which could serve to bind to phosphate groups on DNA. As one way of testing for an association with DNA, we examined the localization of an RsfA-GFP fusion. As shown in Fig. 6, the protein was closely associated with DNA, both in sporulating cells and when ectopically expressed during vegetative growth. Alignment of the promoter regions of genes affected by rsfA did not reveal common sequence motifs that might represent a recognition site for binding of the protein. In some respects, its modest, though global, effects on gene expression are reminiscent of proteins such as H-NS of E. coli, which bind fairly nonspecifically to the nucleoid and regulate the expression of many genes. Biochemical studies are now needed to establish whether RsfA binds directly to DNA to bring about its regulatory effects, whether the DNA binding is sequence specific, and how the positive and negative effects are achieved.
RsfA had a clear paralogue, in the form of the ylbO gene, identified in the complete genome sequence (19). Although a ylbO disruption mutant had no obvious phenotypic effect, the gene turned out to be expressed specifically during sporulation and at quite a high level. The limited experiments we have done (time course and dependence on spoIIGA) suggest that it is expressed within the ςE regulon. It will be interesting to determine whether ylbO has a role in the regulation of gene expression in the mother cell, paralleling that of RsfA in the prespore.
ACKNOWLEDGMENTS
We thank M. Arnaud, R. Daniel, D. Foulger, N. Fransden, A. Moir, J. P. Rawlins, P. Setlow, and P. Stragier for strains and plasmids.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagyan I, Casillas-Martinez L, Setlow P. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by ςF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J Bacteriol. 1998;180:2057–2062. doi: 10.1128/jb.180.8.2057-2062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.Daniel R A, Williams A M, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decatur A, Losick R. Identification of additional genes under the control of the transcription factor ςF of Bacillus subtilis. J Bacteriol. 1996;178:5039–5041. doi: 10.1128/jb.178.16.5039-5041.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 8.Errington J. Efficient Bacillus subtilis cloning system using bacteriophage vector φ105J9. J Gen Microbiol. 1984;130:2615–2628. doi: 10.1099/00221287-130-10-2615. [DOI] [PubMed] [Google Scholar]
- 9.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington J, Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986;132:2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- 11.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illing N, Errington J. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the ςE form of RNA polymerase. Mol Microbiol. 1991;5:1927–1940. doi: 10.1111/j.1365-2958.1991.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 13.Itaya M. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci Biotechnol Biochem. 1992;56:685–686. doi: 10.1271/bbb.56.685. [DOI] [PubMed] [Google Scholar]
- 14.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson H F. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol. 1983;129:1945–1958. doi: 10.1099/00221287-129-6-1945. [DOI] [PubMed] [Google Scholar]
- 16.Karow M L, Glaser P, Piggot P J. Identification of a gene, spoIIR, that links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karow M L, Piggot P. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 18.Kroos L, Zhang B, Ichikawa H, Yu Y-T N. Control of ς factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 19.Kunst F, et al. The complete genome sequence of the Gram positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 20.Lewis P J, Marston A L. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene. 1999;227:101–109. doi: 10.1016/s0378-1119(98)00580-0. [DOI] [PubMed] [Google Scholar]
- 21.Lewis P J, Partridge S R, Errington J. ς factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londoño-Vallejo J-A, Frehel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 23.Londoño-Vallejo J-A, Stragier P. Cell-cell signalling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 24.Neuberg M, Adamkiewicz J, Hunter J B, Muller R. A Fos protein containing the Jun leucine zipper forms a homodimer which binds to the AP1 binding site. Nature. 1989;341:243–245. doi: 10.1038/341243a0. [DOI] [PubMed] [Google Scholar]
- 25.Partridge S R, Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 26.Partridge S R, Foulger D, Errington J. The role of ςF in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991;5:757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schuch R, Piggot P J. The dacF-spoIIA operon of Bacillus subtilis, encoding ςF, is autoregulated. J Bacteriol. 1994;176:4104–4110. doi: 10.1128/jb.176.13.4104-4110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, Fajardo-Cavazos P, Sussman M D, Tovar-Rojo F, Cabrera-Martinez R-M, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EςF: identification of features of good EςF-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun D, Stragier P, Setlow P. Identification of a new ς-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 1989;3:141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- 33.Sussman M D, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J Bacteriol. 1991;173:291–300. doi: 10.1128/jb.173.1.291-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 35.Wu L J, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 36.Wu L J, Feucht A, Errington J. Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum. Genes Dev. 1998;12:1371–1380. doi: 10.1101/gad.12.9.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]