Abstract
Drug repurposing is an approach to identify new medical indications of approved drugs. This work presents a graph neural network drug repurposing model, which we refer to as GDRnet, to efficiently screen a large database of approved drugs and predict the possible treatment for novel diseases. We pose drug repurposing as a link prediction problem in a multi-layered heterogeneous network with about 1.4 million edges capturing complex interactions between nearly 42,000 nodes representing drugs, diseases, genes, and human anatomies. GDRnet has an encoder–decoder architecture, which is trained in an end-to-end manner to generate scores for drug–disease pairs under test. We demonstrate the efficacy of the proposed model on real datasets as compared to other state-of-the-art baseline methods. For a majority of the diseases, GDRnet ranks the actual treatment drug in the top 15. Furthermore, we apply GDRnet on a coronavirus disease (COVID-19) dataset and show that many drugs from the predicted list are being studied for their efficacy against the disease.
Keywords: Computational pharmacology, Drug repurposing, Drug repositioning, Graph neural networks, Link prediction
1. Introduction
Drug repurposing involves strategies to identify new medical indications of approved drugs. It includes identifying potential drugs from a large database of clinically approved drugs and monitoring their in vivo efficacy and potency against novel diseases. Drug repurposing is a low-risk strategy as drugs to be screened have already been approved with less unknown harmful adverse effects and requires less financial investment compared to discovering new drugs [1]. Some of the successful examples of repurposed drugs in the past are Sildenafil, which was initially developed as an antihypertensive drug and later proved to be effective also in treating erectile dysfunction [1] and Rituximab that was originally used against cancer was proved to be effective against rheumatoid arthritis [1]. Even during the coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus (SARS-CoV2), which has affected about 450 million people with more than six million deaths worldwide as of February 2022, drug repurposing has been proved very beneficial. Approved drugs like Remdesivir (a drug for treating Ebola virus disease), Ivermectin (anthelmintic drug), Dexamethasone (anti-inflammatory drugs) are being studied for their efficacy against the disease [2], [3], [4].
Experimental and computational approaches are usually considered for identifying the right candidate drugs, which is the most critical step in drug repurposing. To identify the candidate drugs experimentally, a variety of chromatographic and spectroscopic techniques are available for target-based drug discovery. Phenotype screening is used as an alternative to target-based drug discovery when the identity of the specific drug target and its role in the disease is not known [1]. Recently, computational approaches for identifying the candidates for drug repurposing are gaining popularity due to the availability of large biological data. Efficient ways to handle big data have opened up many opportunities in the field of pharmacology. For instance, [5] elaborates several data-driven computational tools using machine learning (ML) and deep learning (DL) techniques to integrate large volumes of heterogeneous data and solve problems in pharmacology such as drug-target interaction prediction and drug–drug interaction prediction [6], to list a few. Drug repurposing has been studied using computational methods such as signature matching methods, molecular docking, matrix factorization-based, and network-based approaches [7], [8], [9], [10], [11], [12], [13]. However, signature matching approaches and molecular docking approaches rely highly on knowing profiles and exact structures of the target genes, that may not be always available. The matrix factorization-based models find new drug–disease interactions by quantifying the similarity between drugs and disease causative viruses using their molecular sequences. However, these approaches are restricted to pairwise similarities and fail to capture the interactions at a global level [13]. The network proximity-based methods predict drugs for a disease by calculating the network proximity scores between the target genes of the drug and the target genes of the disease [9], [10], but these methods cannot easily account for the additional information in the network, such as similarities between drugs or diseases. Recently, representation learning techniques (i.e., machine learning and deep learning) have been gaining attention due to their accelerated and improved benefits for drug repurposing over the traditional non-deep learning methods [14], [15]. Existing deep learning techniques for drug repurposing can be categorized into sequence-based methods and graph-based methods [15]. The sequence-based methods use the molecular structural sequences of drugs and the virus genome sequence of diseases to encode their respective entity-specific information [16]. However, these methods are highly dependent on the availability of the sequence information for each entity. Also, these approaches focus on the consecutive one- or two-dimensional correlation in a sequence, but do not capture the interactions at a global level between different biological entities. On the other hand, the graph-based approaches capture the structural connectivity information between different biological entities and provide more flexible framework for modeling complex biological interactions between the underlying entities [11], [12], [17].
A natural and efficient way to capture complex interactions between different biological entities like drugs, genes, diseases, etc., is to construct a graph with nodes representing entities and edges representing interactions between these entities, e.g., interactions between drugs and genes or between drugs and diseases. The graph-based methods such as the deepwalk-based, or graph neural networks, that are capable of processing such graph structured biological data have been proposed for drug repurposing [11], [12], [17]. The deepwalk-based architecture [17] independently generates the structural information (using the deepwalk algorithm) and the self entity information due to which the entity and the relational correspondence is not well captured. Graph neural networks (GNNs) capture structural information in data by accounting for interactions between various underlying entities while processing data associated with them, thus producing meaningful low-dimensional embeddings for the entities that are useful for downstream machine learning tasks. However, the existing GNN-based models have a considerable computational overhead when processing huge biological networks having interactions of high density. In this work, we address this problem and focus on drug repurposing using computationally-efficient GNNs. We provide a comparative analysis of several graph-based architectures for drug repurposing and showcase the benefits of having a dedicated model through our experiments on real datasets.
1.1. Main results and contributions
We construct a four-layered heterogeneous graph explaining interactions between the four entities, namely, drugs, genes, diseases, and anatomies in each layer. We propose a new dedicated GNN model for drug repurposing, called GDRnet, which has an encoder–decoder architecture. We formulate drug repurposing as a link prediction problem and train GDRnet to predict unknown links between the drug and disease entities, where a link between a drug–disease entity suggests that the drug treats the disease. Specifically, the encoder is based on the scalable inceptive graph neural network (SIGN) architecture [18] for generating the node embeddings of the entities. We propose a learnable quadratic norm scoring function as a decoder to rank the predicted drugs. The proposed norm scorer is particularly designed and tuned for the drug repurposing task that learns correlations between the drug and disease pairs. The main contributions and results are summarized as follows.
-
•
We formulate drug repurposing as a link prediction problem and propose a new dedicated GNN-based drug repurposing model. The trainable encoder of GDRnet precomputes the neighborhood features beforehand, thus, is computationally efficient with reduced training and inference time. The trainable decoder scores a drug–disease pair based on the low-dimensional embeddings obtained from the encoder. The encoder and decoder are trained in an end-to-end manner.
-
•
We validate GDRnet in terms of its link prediction accuracy and how well it ranks the known treatment drug. For a majority of diseases with known treatment in the test set, which were not used while training, GDRnet ranks the approved treatment drugs in the top 15. This suggests the efficacy of the proposed drug repurposing model.
-
•
We perform an ablation study to show the importance of genes and anatomy entities, which model the indirect interactions between the drug and the disease entities.
-
•
We provide a detailed computational runtime analysis of the proposed GDRnet architecture against the existing GNN models. We demonstrate the advantage of using SIGN as an encoder in GDRnet through the performance gain achieved in terms of its training and inference time.
-
•
We apply GDRnet for COVID-19 drug repurposing by including the COVID-19 interactome information from [19] in the dataset. Many of the drugs predicted by GDRnet for COVID-19 are being studied for their efficacy against the disease.
The software to reproduce the results are available in the github repository: https://github.com/siddhant-doshi/GDRnet
2. Multilayered drug repurposing graph
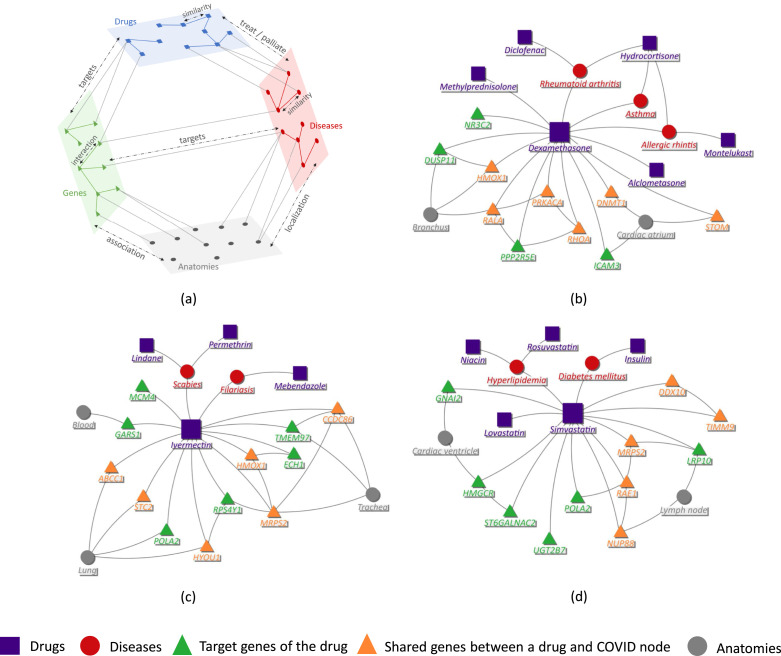
In this section, we model the biological data as a multilayer graph to capture the complex interactions between different biological entities. We consider four entities that are relevant to the drug repurposing task. The four entities are drugs (e.g., Dexamethasone, Sirolimus), diseases (e.g., Scabies, Asthma), anatomies (e.g., Bronchus, Trachea), and genes1 (e.g., DUSP11, PPP2R5E). We form a four-layered heterogeneous graph with these entities as layers; see the illustration in Fig. 1a.
Fig. 1.
Drug repurposing network. (a) Illustration of the four-layered heterogeneous graph with the inter-layer and the intra-layer connections. (b), (c) and (d) Subgraphs centered around the drugs Dexamethasone, Ivermectin and Simvastatin, respectively, illustrate shared target genes between these drugs and COVID-19 disease nodes (see description later on in Section 4.8).
In the multilayer graph, i.e., the interactome there are inter-layered connections between the four layers and intra-layered connections within each layer. The inter-layered connections are of different types. The drug–disease links indicate treatment or palliation, i.e., a drug treats or has a relieving effect on a disease. For example, interaction between Ivermectin-Scabies (as seen in Fig. 1b) and Simvastatin-Hyperlipidemia (as seen in Fig. 1d) are of type treatment, whereas Atropine-Parkinson’s disease is of type palliation. The drug–gene and disease–gene links are the direct gene targets of the compound and the disease, respectively. NR3C2, RHOA, DNMT1 are some of the target genes of the drug Dexamethasone (see Fig. 1b) and PPP1R3D, CAV3 are target genes of the disease Malaria. There are also indirect links between target genes of a drug and a disease, referred to as the shared target genes (see Fig. 1b). For example, genes like ATF3, UPP1, CTSD, are the shared target genes of drug Ivermectin and disease Malaria. The disease–anatomy and gene–anatomy connections indicate how the diseases affect the anatomies and interactions between the genes and anatomies. For example, GNAI2 and HMGCR belong to the cardiac ventricle anatomy (see Fig. 1d); disease Schizophrenia affects multiple anatomies like the central nervous system (CNS) and optic tract.
The intra-layered drug–drug and disease–disease connections show the similarity between a pair of drugs and diseases, respectively. The gene–gene links describe the interaction between genes (e.g., epistasis, complementation) and form the whole gene interactome network. The anatomy information helps by focusing on the local interactions of genes related to the same anatomy as the genes targeted by the new disease. Some examples of the intra-layered connections are Simvastatin-Lovastatin and POLA2-RAE1 as seen in Fig. 1d. This comprehensive network serves as a backbone for our model, which predicts the unknown inter-layered links between drugs and novel diseases by leveraging the multi-layered graph-structured data.
3. Methods and models
Graph neural networks (GNNs) have become very popular for processing and analyzing such graph-structured data in the last few years. Compared to deep learning models such as convolutional neural networks (CNNs), GNNs offer extraordinary performance improvements while dealing with graph-structured data commonly encountered in social networks, biological networks, brain networks, and molecular networks, to name a few. GNN models learn low-dimensional graph representations or node embeddings that capture the nodal connectivity information useful for solving graph analysis tasks like node prediction, graph classification, and link prediction. In this section, we describe the proposed GDRnet architecture for drug repurposing, which is formulated as a link prediction problem.
3.1. Notation
Consider an undirected graph with a set of vertices {} and edges denoting a connection between nodes and . We represent a graph using the adjacency matrix , where the th entry of , denoted by , is if there exists an edge between nodes and , and otherwise. To account for the non-uniformity in the degrees of the nodes, we use the normalized adjacency matrix denoted by , where is the diagonal degree matrix. Each node in the graph has attributes (referred to as input features). Let us denote the input feature vector of node by , which contains attributes of that node.
3.2. Graph neural networks
In most of the existing GNN architectures, the embedding of a node is updated during training by sequentially aggregating information from its 1-hop neighbor nodes, thereby accounting for local interactions in the network. This is also referred to as a GNN layer. Several such GNN layers are cascaded to capture interactions beyond the 1-hop neighborhood. Specifically, by cascading such layers, node features from its -hop neighborhood are captured. For example, in Fig. 1c, the drug Ivermectin is a -hop neighbor of the anatomy Lung and is connected via STC2. Mathematically, the node feature vector updates can be represented by the recursion
| (1) |
where is the embedding for node at the th layer and represents a set of -hop neighbor nodes of node . Local aggregation function combines the neighbor node features (during the training) and transforms it to obtain the updated feature vector. Different choices of the aggregation function and the transformation function lead to different GNN variants like the graph convolutional networks (GCN) [21], GraphSAGE [22], and graph attention networks (GAT) [23], to name a few. However, these GNN models do not scale well on large and dense graphs as their computational cost depends on the number of nodes and edges in the graph. To reduce the runtime computations, a scalable GNN architecture called SIGN [18] has been proposed, where the neighborhood aggregations at various depths (till -hop) are precomputed (before training), and the node embeddings are generated non-iteratively, unlike the GNN models in Eq. (1). As the node features updates are performed beforehand outside the training procedure, these GNN variants easily scale on large graphs, such as the multi-layered drug repurposing graph, as they are independent of the number of edges in the graph. The proposed GDRnet architecture has an encoder–decoder architecture, wherein the encoder is based on the SIGN architecture due to its computational advantages. While SIGN has been used for node classification [18], we utilize it here for link prediction, i.e., to predict links between drugs and diseases. Next, we describe the proposed GDRnet architecture.
3.3. The GDRnet architecture
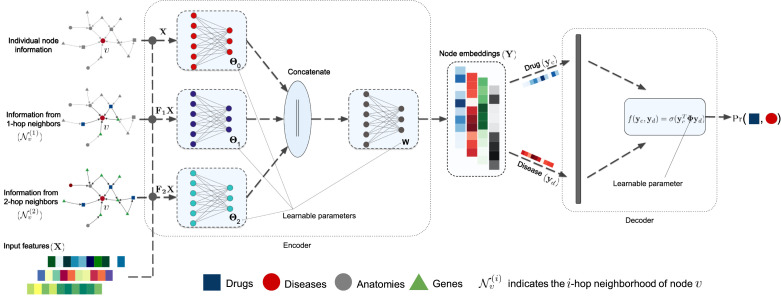
The proposed GNN architecture for drug repurposing has two main components, namely, the encoder and decoder. The encoder generates the node embeddings of all the nodes in the four-layer graph. The decoder scores a drug–disease pair based on the embeddings. The encoder and decoder networks are trained in an end-to-end manner. Next, we describe these two components of the GDRnet architecture, which is illustrated in Fig. 2.
Fig. 2.
The GDRnet architecture.
3.3.1. Encoder
The GDRnet encoder produces low-dimensional node embeddings based on the input features and nodal connectivity information. Recall that the matrix is the normalized adjacency matrix of the four-layered graph . We use graph operators represented using matrices , , to aggregate information in the graph. Here, denotes the th matrix power. By choosing , we aggregate information from the -hop neighborhood. We assume that each node has its own -dimensional feature, which we collect in the matrix to obtain the input feature matrix associated with the nodes of . We can then represent the encoder as
| (2) |
where is the final node embedding matrix for the nodes in the graph and {} are the learnable parameters. Here, represents concatenation, and are the nonlinear tanh and leaky rectified linear unit (leaky ReLU) activation functions, respectively. The matrix aggregates node features from -hop neighbors, which can be related to the neighborhood aggregation performed at the th layer of GNN models that perform sequential neighborhood aggregation as in Eq. (1). Fig. 2 shows the encoder architecture. The main advantage of using SIGN over other models (e.g., GCN, GAT, GraphSAGE) is that the matrix product is independent of the learnable parameters . Thus, this matrix product can be precomputed before training the neural network model. Doing so reduces the computational complexity while incorporating information from the graph structure.
In our experiments, we choose , i.e., the low-dimensional node embeddings have information from 2-hop neighbors. Choosing is found to be not useful for drug repurposing, as we aim to capture the local information of the drug targets such that a drug node embedding should retain information about its target genes and the shared genes in its vicinity. For example, the -hop neighbors of Dexamethasone as shown in Fig. 1b, are the diseases it treats (e.g., Asthma), and the drugs similar to Dexamethasone (e.g., Methylprednisolone) and its target genes (e.g., DUSP11, RHOA). The -hop neighbors are the anatomies of the target genes (e.g., Bronchus), and the drugs that have similar effects on the diseases (e.g., Hydrocortisone and Dexamethasone have similar effects on Asthma). While updating the node for the embedding related to Dexamethasone, it is important to retain this local information for the drug repurposing task.
3.3.2. Decoder
For drug repurposing, we propose a score function based on a general dot-product that takes as input the updated embeddings of drugs and diseases and outputs a score based on which we decide if a certain drug treats the disease. Fig. 2 illustrates the proposed learnable decoder. The columns of the embedding matrix contain the embeddings of all the nodes in the four-layer graph, including the embeddings of the disease and drug nodes. Let us denote the embeddings of the th drug as and the embeddings of the th disease as . The proposed scoring function to infer whether drug is a promising treatment for disease is defined as
| (3) |
where is the nonlinear sigmoid activation function and is a learnable co-efficient matrix. We interpret as the probability that a link exists between drug and disease . The term can be interpreted as a measure of correlation (induced by ) between the disease and drug node embeddings.
3.3.3. Training loss
The model is trained in a mini-batch setting in an end-to-end fashion using stochastic gradient descent to minimize the weighted cross-entropy loss, where the loss function for the sample corresponding to the drug–disease pair is given by
| (4) |
where is the known training label associated with the score for the drug–disease pair , indicates that drug treats or palliates disease , and otherwise. Here, is the weight on the positive samples that we choose to account for the huge class imbalance in the dataset. During training, we include no-drug–disease links, which give us the negative control for learning. For example, there is no link between the drug–disease pair Simvastatin-Scabies, i.e., Simvastatin is not known to treat or suppress the effects of Scabies. The number of no-drug–disease links is almost thirty times the number of positive samples. To handle this class disparity, we explicitly use a weight on the positive samples.
4. Model evaluation and experiments
In this section, we evaluate GDRnet and discuss the choice of various hyper-parameters. The model is evaluated based on two performance measures. Firstly, we report the ability to classify the links correctly, i.e., to predict the known treatments correctly for diseases in the test set. Next, using the list of predicted drugs for the diseases in the test set, we report the model’s ability to rank the actual treatment drug as high as possible (the ranking is obtained by ordering the scores in Eq. (3)). Finally, we also report prediction results for coronavirus related diseases.
4.1. Dataset
We use information from the drug repurposing knowledge graph (DRKG) [24] to form the multi-layered drug repurposing graph. DRKG includes information about six drug databases, namely, Drugbank [25], Hetionet [26], GNBR [27], STRING [28], IntAct [29], and DGIdb [30]. We construct a four-layered graph comprising the drug layer, disease layer, gene layer, and anatomy layer. We extract the details about these entities specifically from the Drugbank, Hetionet, and GNBR databases. We leverage their generic set of low-dimensional embeddings that represent the graph nodes and edges in the Euclidean space for training. The four-layered graph is composed of 8070 drugs, 4166 diseases, 29 848 genes, 400 anatomies, and a total of 1,417,624 links, which include all the inter-layer and intra-layer connections (refer Section 2 for the description of the multi-layered graph). Details about the inter-layered and intra-layered links are given in Table 1.
Table 1.
Multi-layered graph data. The value in each cell represents the number of links between the respective layers. NC represents no connection.
| Drugs | 6486 | |||
| Diseases | 6113 | 543 | ||
| Genes | 76 250 | 123 609 | 474 526 | |
| Anatomies | NC | 3602 | 726 495 | NC |
| Drugs | Diseases | Genes | Anatomies |
4.2. Experimental setup and model parameters
The drug repurposing problem is formulated as a link prediction. It can be viewed as a binary classification problem, wherein a positive class represents the existence of a link between a drug and disease, and otherwise represents a negative class. We have 6113 positive samples (drug–disease links) in our dataset. To account for the negative class samples, we randomly choose 200,000 no-drug–disease links (i.e., those pairs with no link between these drugs and diseases). These links are then divided into the training and testing set with a split. We train the network using mini-batch stochastic gradient descent by grouping the training set in batches of size 512 and train them for nearly 20 epochs. Due to the significant class imbalance, we oversample the drug–disease links while creating batches, thus maintaining the class ratio (ratio of the number of negative samples to the number of positive samples) of 1.5 in each batch. The additional hyperparameters are set as follows. The intermediate embedding dimensions are fixed to 250, the batch size and the learning rate (set to 10−4) are chosen by performing a grid search over the hyperparameter space. Also, we use the leaky rectified linear unit (Leaky-ReLU) as the intermediate activation function. We use the Adam optimizer to perform the back propagation and update the model parameters. The weight on the positive samples (cf. Eq. (4)) is also chosen to be the class imbalance ratio of each batch, i.e., we fix to be 1.5.
4.3. Baselines
We perform experiments on the state-of-the-art network-based drug repurposing methods, the network-proximity based [9], which is based on the Z-scores computed using the permutation test, the HINGRL [17] method based on the autoencoder and deepwalk algorithm, and the Bipartite-GCN method [31], which uses an attention-based GNN layer. In addition, we also provide a comparison with three commonly used GNN encoder architectures, namely, GCN [21], GraphSAGE [22], and GAT [23] for the drug repurposing task, which we treat as a link prediction problem, and compare the classification performance with the GDRnet architecture. Specifically, the encoder in GDRnet is replaced with GCN, GraphSAGE, and GAT to evaluate the model performance. Two blocks of these sequential models are cascaded to maintain consistency with of the GDRnet architecture. We evaluate these models on the test set, which contains known treatments for diseases that are not shown to the model while training. To remain consistent, we use the same initial embeddings for all the experiments.
4.4. Classification performance
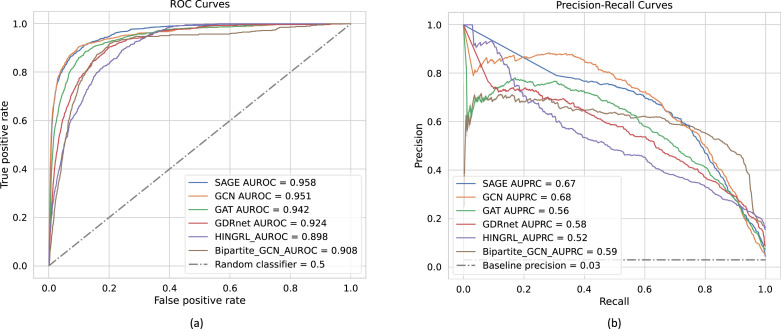
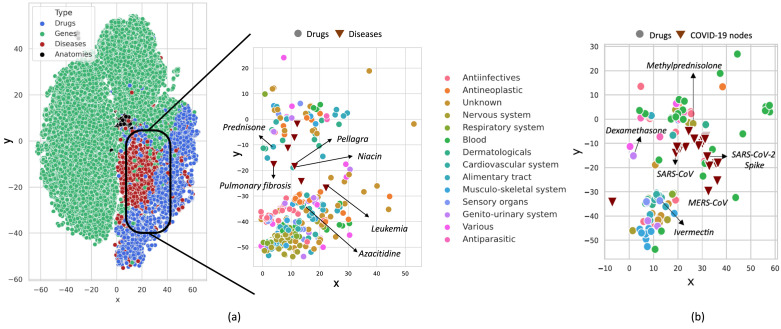
We measure the classification abilities of a model through the receiver operating characteristic (ROC) curve of the true positive rate (TPR) versus the false positive rates (FPR) and the precision–recall (PR) curve of the precision versus the recall. The area under the PR curves (AUPRC) along with the area under the receiver operating characteristics (AUROC), would give a comprehensive view of the performance statistics of the encoders. Fig. 3a shows the ROC curves of different GNN models. We can see that all the models have very similar AUROC values. Also, all the AUPRC values, as shown in Fig. 3b are in a similar range. As compared to the baseline precision of 0.03, which is calculated as the ratio of the minority class in the data, we see a significant gain in the AUPRC values. Fig. 4a provides an illustration of two-dimensional embeddings (from GDRnet), using the t-distributed stochastic neighbor embedding (t-SNE), where we observe that diseases that target certain anatomy or a drug that target certain gene have nearby representations in the embedding space demonstrating the expressive power of GDRnet.
Fig. 3.
Classification performance of GDRnet. (a) and (b) represent the receiver operating curves (ROC) and the precision–recall (PR) curves, respectively, depicting the classification performance of different drug-repurposing models.
Fig. 4.
Embedding visualization. (a) Two-dimensional t-SNE visualization of the high-dimensional embeddings generated by GDRnet for the nodes in the four-layered heterogeneous graph. The left embedding plot shows the representation of all the nodes (around 42 000 nodes), which are colored according to their layer. The right plot focuses on the drugs and diseases used for testing. (b) Embeddings of the COVID-19 disease nodes (27 SARS-CoV-2 proteins and 6 coronavirus related diseases) and the predicted drugs by GDRnet. The drugs in the both the plots (a) and (b) are colored according to their first-level anatomical therapeutic chemical (ATC) categorization.
4.5. Ranking performance
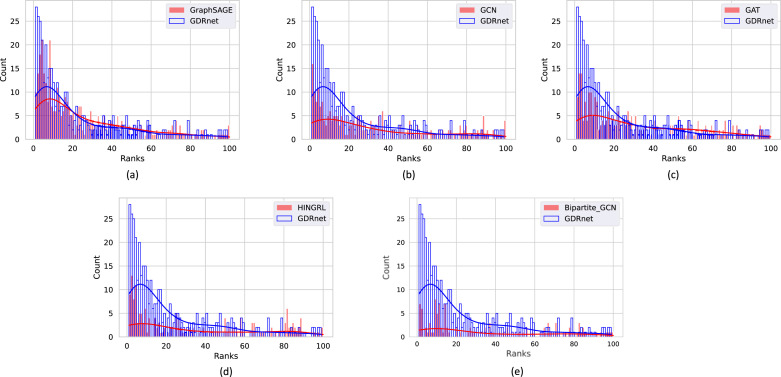
We evaluate GDRnet in terms of ranks of the actual treatment drug in the predicted list for a disease from the testing set, where the rank is computed by rank ordering the scores. Fig. 5 represents the histograms of the ranks of the drug–disease pairs from the testing set for GraphSAGE, GCN, GAT, HINGRL, and Bipartite-GCN compared with GDRnet. To get the histograms, we compute the ranks of the actual treatment drugs for the diseases from the test set and plot the frequencies of those ranks on the vertical axis corresponding to the ranks on the horizontal axis. We see that GDRnet has a higher density of ranks in the top 15 as compared to other models. This clearly illustrates that GDRnet outperforms the other graph-based methods in terms of its ranking abilities. In addition, we compute the network proximity scores [9] and rank order the drugs based on network proximity scores to compare with the GNN-based encoder models. These network proximity scores are a measure of the shortest distance between drugs and diseases through their target genes. They are computed as
| (5) |
where is a proximity score of drug and disease . Here, is the set of target genes of , is the set of target genes of , and is the shortest distance between a gene and a gene in the gene interactome. We convert these into Z-scores using the permutation test , where is the mean proximity score of the pair computed by randomly selecting subsets of genes with the same degree distribution as that of and from the gene interactome, and is the standard deviation of the scores generated in the permutation test of these randomly selected subsets. Table 2 provides the rankings of a few sample drug–disease pairs from the test set that were not shown during the training. We can see that the GDRnet and the other GNN variants result in better ranks on the unseen diseases than the network proximity measure, which is solely based on the gene interactome, by a huge margin. Also, determining the network proximity scores is extremely computationally expensive due to the calculation of Z-scores using the permutation test. For the same reasons we leave off the histogram analysis for the network proximity approach, which evidently through the examples in Table 2, results in poor ranking performance. The diseases on which we evaluate are not confined to a single anatomy (e.g., rectal neoplasms are associated to the rectum anatomy, whereas pulmonary fibrosis is a lung disease), nor do they indicate a similar family of drugs for their treatment (e.g., Fluorouracil is an antineoplastic drug, and Prednisone is an anti-inflammatory corticosteroid). For a majority of the diseases in the test set, GDRnet ranks the treatment drug in the top 15 (as seen in Table 2). In the case of Leukemia, other antineoplastic drugs like Hydroxyurea and Methotrexate are ranked high (in top ) and its known treatment drug Azacitidine is ranked .
Fig. 5.
Ranking histograms. The ranking performance of GDRnet compared with other GNN variants, namely, (a) GraphSAGE (b) GCN and (c) GAT (d) HINGRL (e) Bipartite-GCN.
Table 2.
Ranking. A few examples of the ranks of the actual treatment drugs for the diseases from the testing set. There are no associated genes with some of the disease in our database, which makes it impossible to rank them using the network proximity based method. These are indicated as “Not computable”. The best ranks are highlighted in bold.
| Disease | Treatment drug | Ranks |
||||||
|---|---|---|---|---|---|---|---|---|
| GDRnet | GraphSAGE | GCN | GAT | Network proximity | HINGRL | Bipartite-GCN | ||
| Encephalitis | Acyclovir | 10 | 35 | 35 | 295 | 5462 | 435 | 27 |
| Rectal neoplasms | Fluorouracil | 9 | 421 | 16 | 231 | 2831 | 205 | 117 |
| Pulmonary fibrosis | Prednisone | 5 | 3 | 10 | 9 | 2072 | 2 | 9 |
| Atrioventricular block | Atropine | 6 | 79 | 8 | 14 | 4453 | 26 | 196 |
| Pellagra | Niacin | 2 | 56 | 497 | 484 | Not computable | 460 | 288 |
| Colic | Hyoscyamine | 1 | 1 | 501 | 205 | Not computable | 39 | 101 |
| Leukemia | Azacitidine | 17 | 120 | 31 | 332 | 377 | 527 | 507 |
4.6. Layer ablation study
To gain more insights on the importance of different entities, namely, drugs, disease, genes, and anatomies for drug repurposing, we perform an ablation study on the layers of the constructed graph. We perform link prediction using considered GNN models on the constructed graphs, starting with the only drug–disease two-layered graph, followed by the individual addition of the gene and the anatomy interactome, making it a three-layered graph, and eventually converting it to a four-layered graph by getting all the layers together. We report the corresponding AUROC values in Table 3. We use the degree information as the input features for these experiments to eliminate any biases due to the pre-trained embeddings. As seen in Table 3, the addition of the anatomy and the gene layer shows their importance by giving a significant improvement in the classification performance, demonstrating the significance of the indirect connections provided by the anatomy and the gene layers between the drugs and diseases for drug repurposing. Finally, when all the information from the four layers used together, we see a clear boost in the performance.
Table 3.
Layer ablation study. The AUROC values for a link prediction task compared across different graph layers and different GNN models. Best performances are indicated in bold.
| Graph layers | GDRnet | GraphSAGE | GCN | GAT |
|---|---|---|---|---|
| Drugs, Diseases | ||||
| Drugs, Diseases, Anatomies | ||||
| Drugs, Diseases, Genes | ||||
| Drugs, Diseases, Genes, Anatomies |
In summary, GNNs perform better than the prior network-based approaches in predicting the drugs for a disease. This also signifies the importance of capturing the local interactions in complex biological networks. These interactions are not sufficiently captured by the network proximity methods that restrict their focus only on the target genes of a drug and a disease. The proposed GNN-based GDRnet architecture is computationally attractive and better ranks known treatment drugs for diseases than the popular sequential GNN variants.
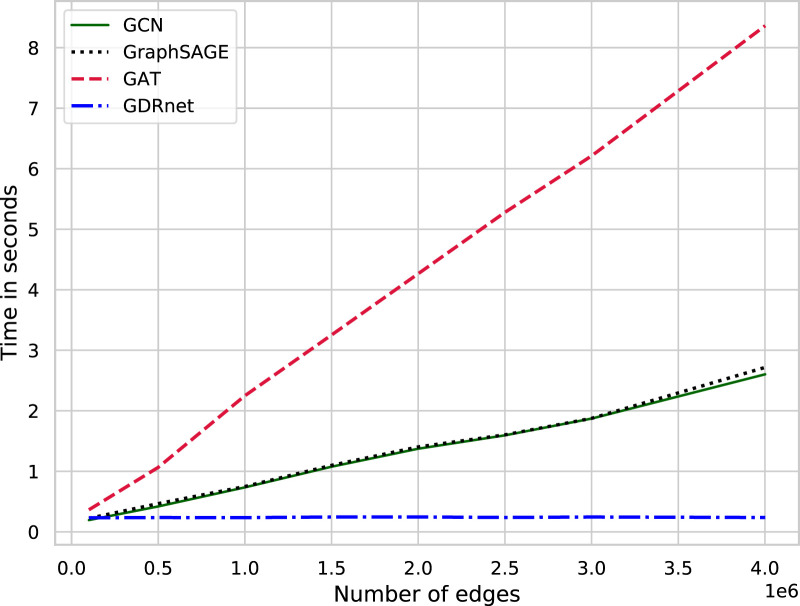
4.7. Computational complexity
The time complexity of GNNs that perform aggregation sequentially like GCN, GraphSAGE, and GAT, is for a graph having nodes and edges with sequential aggregation iterations [32]. The intermediate embedding dimensions are assumed to be . Here, the term corresponds to the feature transformation, and is the additional computations performed to identify the neighborhood for local aggregation during the training. GDRnet benefits itself in terms of the training and inference time due to its parallel framework by precomputing this neighborhood aggregations. This results in the runtime to be independent of the number of edges in the graph, having a time complexity of , where is the number of parallel branches. Fig. 6 illustrates the dependence of GNNs on the number of edges. The time taken for a single epoch (forward pass) on a graph having the same number of nodes as in the constructed multilayered graph in Section 2 (approximately 42 000) are plotted on the vertical axis for varying number of edges on the horizontal axis. GCN, GraphSAGE, and GAT clearly depict their linear dependence on , whereas GDRnet verifies its independence by having a constant time, irrespective of the number of edges. The Bipartite-GCN architecture uses an attention-based graph layer similar to GAT. Thus it has the same complexity as the sequential based GNNs. It is not straightforward to compare the forward pass time complexity incurred by network proximity and HINGRL methods. HINGRL pipeline involves multiple algorithms that are trained independently, like the autoencoder, followed by deepwalk, and finally the random forests, that incur more time complexity as observed during our numerical experiments. For the network proximity method, due to the involvement of the permutation test, it is extremely computationally expensive as well.
Fig. 6.
Computational complexity. Time plot showing the dependence of GNN architectures on the number of edges.
Table 4.
Drugs predicted by GDRnet for COVID-19.
| COVID-19 node | Drugs predicted by GDRnet ranked in top 10 |
|---|---|
| SARS-CoV2-E | Ivermectin, Spironolactone, Sirolimus |
| SARS-CoV2-M | Ivermectin, Cyclosporine, Acyclovir |
| SARS-CoV2-N | Rubella virus vaccine, Sirolimus, Hydralazine |
| SARS-CoV2-spike | Crizanlizumab, Cyclosporine, Cidofovir, Nitazoxanide |
| CoV-NL63 | Dexamethasone, Prednisolone, Celecoxib |
4.8. COVID-19 drug repurposing
Next, we focus on drug repurposing for the four known human coronaviruses (HCoVs), namely, SARS-CoV, MERS-CoV, CoV-229E and CoV-NL63, and two non-human coronaviruses, namely MHV, and IBV. We consider interactions of these disease nodes with human genes. There are 129 known links between these six disease nodes and gene nodes in the dataset [24]. In addition, we consider all the 27 SARS-CoV-2 proteins that include 4 structural proteins, namely, envelope (SARS-CoV2-E), membrane (SARS-CoV2-M), nucleocapsid (SARS-CoV2-N) and surface (SARS-CoV2-spike), 15 non-structural proteins (nsp) and 8 open reading frames (orf), and their 332 links connecting the target human genes [19]. We refer to these 33 nodes (6 disease nodes and 27 SARS-CoV2 proteins) as the COVID-19 nodes. In other words, there are only disease–gene interactions available for these COVID-19 nodes. Some of the genes targeted by the COVID-19 nodes are shown in Fig. 1 (b, c and d), which are also the target genes for the drugs (e.g., Dexamethasone, Ivermectin, Simvastatin).
We individually predict the drugs for all these 33 COVID-19 nodes as each protein in SARS-CoV-2 targets a different set of genes in humans. We select the top 10 ranked predicted drugs out of 8070 clinically approved drugs for each disease entity. Table 4 lists some of the predicted drugs by GDRnet. A complete list of the predicted drugs with their scores and ranks is available in our repository at: https://github.com/siddhant-doshi/GDRnet. Our predictions have corticosteroids like Dexamethasone, Methylprednisolone, antineoplastic drugs like Sirolimus, Anakinra, anti-parasitic drugs like Ivermectin, Nitazoxanide, non-steroidal anti-inflammatory drugs (NSAIDs) like Ibuprofen, Celecoxib, ACE inhibitors and statin drugs like Simvastatin, Atorvastatin, and some of the vaccines discovered previously for other diseases like the Rubella virus vaccine. Fig. 4b gives a two-dimensional t-SNE representation of the embeddings of a few predicted drugs and the COVID-19 disease nodes, where we can see that the representation of the predicted drugs is in the vicinity of the disease nodes in the embedding space.
5. Conclusions and future work
We proposed a GNN model for drug repurposing model, called GDRnet, to predict drugs from a large database of approved drugs for further studies. We leverage a biological network of drugs, diseases, genes, and anatomies and cast the drug repurposing task as a link prediction problem. The proposed GDRnet architecture has a computationally attractive encoder to generate low-dimensional embeddings of the entities and a decoder that scores the drug–disease pairs. Through numerical simulations on real data, we demonstrate the efficacy of the proposed approach for drug repurposing. We also apply GDRnet on COVID-19 data.
This work can be extended along several directions. Considering the availability of substantial biological data, the inclusion of information like individual side effects of drugs, may further improve the predictions. Considering the comorbidities of a patient would help us analyze the biological process and gene interactions in the body specific to an individual and accordingly prescribe the line of treatment. Also, including the edge specific information such as type of drug interactions could help us predicting a synergistic combination of drugs for a disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
S.P. Chepuri is supported in part by the Pratiskha Trust Young Investigator Award, Indian Institute of Science, Bangalore, and the SERB, India grant SRG/2019/000619, and S. Doshi is supported by the Robert Bosch Center for Cyber Physical Systems, Indian Institute of Science, Bangalore , Student Research Grant 2020-M-11. The authors thank the Deep Graph Learning team for making DRKG public at https://github.com/gnn4dr/DRKG.
Footnotes
All the genes are represented using the symbols according to the HUGO gene nomenclature committee (HGNC) [20].
References
- 1.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig T., Latimer J., McNamee C., Norris A. Drug repurposing: progress. and challenges and recommendations. Nat. Rev. Drug Discov. 2018;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 2.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D. Remdesivir for the treatment of Covid-19. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Group T.R. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zitnik M., Nguyen F., Wang B., Leskovec J., Goldenberg A., Hoffman M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zitnik M., Agrawal M., Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34(13):i457–i466. doi: 10.1093/bioinformatics/bty294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaliamurthi S., Selvaraj G., Selvaraj C., Singh S.K., Wei D.Q., Peslherbe G.H. Structure-based virtual screening reveals ibrutinib and zanubrutinib as potential repurposed drugs against COVID-19. Int. J. Mol. Sci. 2021;22(13):7071. doi: 10.3390/ijms22137071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan A., Ali S.S., Khan M.T., Saleem S., Ali A., Suleman M., Babar Z., Shafiq A., Khan M., Wei D.Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J. Biomol. Struct. Dyn. 2021;39(13):4659–4670. doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nature Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gysi D.M., Do Valle I., Zitnik M., Ameli A., Gan X., Varol O., Ghiassian S.D., Patten J.J., Dave R.A., Loscalzo J., Barabási A.L. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. 2021;118(19) doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ioannidis V.N., Zheng D., Karypis G. 2020. Few-shot link prediction via graph neural networks for Covid-19 drug-repurposing. arxiv preprint arxiv:2007.10261. [Google Scholar]
- 13.Su X., Hu L., You Z., Hu P., Wang L., Zhao B. A deep learning method for repurposing antiviral drugs against new viruses via multi-view nonnegative matrix factorization and its application to SARS-CoV-2. Bioinformatics. 2022;23(1) doi: 10.1093/bib/bbab526. [DOI] [PubMed] [Google Scholar]
- 14.Yang F., Zhang Q., Ji X., Zhang Y., Li W., Peng S., Xue F. Machine learning applications in drug repurposing. Interdiscip. Sci.: Comput. Life Sci. 2022:1–7. doi: 10.1007/s12539-021-00487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan X., Lin X., Cao D., Zeng X., Yu P.S., He L., Nussinov R., Cheng F. Deep learning for drug repurposing: Methods. and databases. and and applications. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2022:e1597. [Google Scholar]
- 16.Su X., You Z., Wang L., Hu L., Wong L., Ji B., Zhao B. SANE: a sequence combined attentive network embedding model for COVID-19 drug repositioning. Appl. Soft Comput. 2021;111(107831) doi: 10.1016/j.asoc.2021.107831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B.W., Hu L., You Z.H., Wang L., Su X.R. Hingrl: predicting drug–disease associations with graph representation learning on heterogeneous information networks. Brief. Bioinform. 2022;23(1) doi: 10.1093/bib/bbab515. [DOI] [PubMed] [Google Scholar]
- 18.Frasca F., Rossi E., Eynard D., Chamberlain B., Bronstein M., Monti F. 2020. SIGN: Scalable inception graph neural networks. arxiv preprint arXiv:2004.11198. [Google Scholar]
- 19.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Povey S., Lovering R., Bruford E., Wright M., Lush M., Wain H. The HUGO gene nomenclature committee (HGNC) Hum. Genet. 2001;109(6):678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 21.T.N. Kipf, M. Welling, Semi-supervised classification with graph convolutional networks, in: Proceedings of the International Conference on Learning Representations, Toulon, France, 2017.
- 22.W.L. Hamilton, R. Ying, J. Leskovec, Inductive representation learning on large graphs, in: Advances in Neural Information Processing Systems, California, United States, 2017.
- 23.P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Lio, Y. Bengio, Graph attention networks, in: Proceedings of the International Conference on Learning Representations, Vancouver, Canada, 2018.
- 24.Ioannidis V.N., Song X., Manchanda S., Li M., Pan X., Zheng D., Ning X., Zeng X., Karypis G. 2020. DRKG - drug repurposing knowledge graph for Covid-19. https://github.com/gnn4dr/DRKG/ [Google Scholar]
- 25.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed D., Li C., Sayeeda Z., Assempour N. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2017;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himmelstein D.S., Lizee A., Hessle C., Brueggeman L., Chen S.L., Hadley A., Khankhanian P., Baranzini S.E. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6:e26726. doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Percha B., Altman R.B. A global network of biomedical relationships derived from text. Bioinformatics. 2018;34(15):2614–2624. doi: 10.1093/bioinformatics/bty114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D., Gable A., Lyon D., Junge A., Wyder S., Huerta-Cepas M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J. STRING v11: protein–protein association networks with increased coverage. and supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., Del-Toro N., Duesbury M. The mIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(D1):D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. C.K., H. W.A., Y. F.Y., Kiwala S., Coffman A.C., Spies G., Wollam A., C. S.N., L. G.O., M. G. GIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z., Zhou M., Arnold C. Toward heterogeneous information fusion: bipartite graph convolutional networks for in silico drug repurposing. Bioinformatics. 2020;36(Supplement1):i525–33. doi: 10.1093/bioinformatics/btaa437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z., Pan S., Chen F., Long G., Zhang C., Philip S.Y. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2021;32(1):4–24. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]