Abstract
The diphtheria toxin repressor (DtxR) of Corynebacterium diphtheriae uses Fe2+ as a corepressor. Holo-DtxR inhibits transcription from the iron-regulated promoters (IRPs) designated IRP1 through IRP5 as well as from the promoters for the tox and hmuO genes. DtxR binds to 19-bp operators with the consensus sequence 5′-TTAGGTTAGCCTAACCTAA-3′, a perfect 9-bp palindrome interrupted by a single C · G base pair. Among the seven known DtxR-specific operators, IRP3 exhibits the weakest binding to DtxR. The message (sense) strand of the IRP3 operator (5′-TTAGGTGAGACGCACCCAT-3′ [nonconsensus nucleotides underlined]) overlaps by 2 nucleotides at its 5′ end with the putative −10 sequence of the IRP3 promoter. The underlined C at position +7 from the center of the IRP3 operator [C(+7)] is unique, because T is conserved at that position in other DtxR-specific operators. The present study examined the effects of nucleotide substitutions at position +7 or −7 in the IRP3 operator. In gel mobility shift assays, only the change of C(+7) to the consensus nucleotide T caused a dramatic increase in the binding of DtxR, whereas other nucleotide substitutions for C(+7) or replacements for A(−7) had only small positive or negative effects on DtxR binding. All substitutions for C(+7) or A(−7) except for A(−7)C dramatically decreased IRP3 promoter strength. In contrast, the A(−7)C variant caused increased promoter strength at the cost of nearly eliminating repressibility by DtxR. The message (sense) strand of the IRP1 operator (5′-TTAGGTTAGCCAAACCTTT-3′) includes the −35 region of the IRP3 promoter. A T(+7)C variant of the IRP1 operator was also constructed, and it was shown to exhibit decreased binding to DtxR, decreased repressibility by DtxR, and increased promoter strength. The nucleotides at positions +7 and −7 in DtxR-specific operators are therefore important determinants of DtxR binding and repressibility of transcription by DtxR, and they also have significant effects on promoter activity for IRP3 and IRP1.
Corynebacterium diphtheriae is the causative agent of diphtheria, a local infection that most often involves the respiratory tract. Diphtheria toxin is the most important virulence determinant of C. diphtheriae, and it is responsible for the most serious systemic manifestations of diphtheria, which include myocarditis and polyneuropathy (26). Diphtheria toxin is synthesized and secreted by toxinogenic strains of C. diphtheriae that are lysogenic for tox+ corynebacteriophages, such as phage β, that carry the gene for diphtheria toxin (tox) (21, 26, 45). The chromosomally encoded diphtheria toxin repressor (DtxR) and iron negatively regulate expression of the tox gene (3, 36). DtxR homologs are present in several other bacterial species, including Mycobacterium tuberculosis and Mycobacterium smegmatis (designated IdeR) (6, 38), Streptomyces lividans and Streptomyces pilosus (11), Brevibacterium lactofermentum (23), Staphylococcus epidermidis and Staphylococcus aureus (SirR) (15), and Treponema pallidum (TroR) (12).
DtxR functions as an iron-dependent global regulatory protein, in a manner similar to the ferric uptake regulator (Fur) protein in gram-negative bacteria (1, 3, 36, 41). DtxR-regulated loci contain operators that overlap with proven or putative promoters and contain an interrupted 9-bp inverted repeat within a 19-bp sequence (18, 37, 39, 43). Molecular footprinting techniques demonstrated that DtxR binding protects a region surrounding the dyad axis of the corresponding operator (37, 40, 42), in a manner resembling that reported for several other well-characterized bacterial repressors (13, 14, 24, 25). Recent crystallographic findings demonstrated that two dimeric DtxR holorepressor molecules bind simultaneously to DtxR-specific operators on opposite faces of the DNA helix (28, 48).
At least seven promoters in C. diphtheriae are negatively regulated by DtxR and iron (18, 34, 35, 37, 39, 41). These include the iron-regulated promoters (IRPs) designated IRP1 through IRP5, as well as the promoters for the tox and hmuO genes. The tox gene encodes diphtheria toxin (45); hmuO encodes a heme oxygenase that is essential for the acquisition of iron by C. diphtheriae from heme and hemoglobin (34, 35); and the deduced products of the genes downstream from IRP1 and IRP3 are predicted to be a ferric siderophore receptor and a transcriptional regulator homolog in the AraC family, respectively (18, 34). The functions of the gene products regulated by IRP2, IRP4, and IRP5 have not yet been established (18, 37).
Each of the seven DtxR-regulated promoters described above has been tested for its ability to drive expression of a β-galactosidase reporter gene, under high-iron and low-iron growth conditions, in Escherichia coli strains in which DtxR is constitutively expressed at a low level from pDSK29 (18, 35, 37). Among them, IRP3 is least repressible by DtxR and iron, i.e., it exhibits both the lowest repression ratio (β-galactosidase activity under low-iron [derepressed] growth conditions/β-galactosidase activity under high-iron [repressed] growth conditions) and the highest level of β-galactosidase activity under high-iron (repressed) growth conditions (18, 35, 37). In gel shift assays and DNase I footprinting assays with DNA fragments containing IRP3, a higher concentration of DtxR is needed to demonstrate protein-DNA binding than in assays with DNA fragments containing other DtxR-specific operators (18). These findings provide strong evidence that DtxR has lower affinity for the IRP3 operator than for other operators that are regulated more stringently by DtxR.
In the present study we used site-directed mutagenesis to make substitutions for the nonconsensus nucleotide C at position +7 [C(+7)] in the IRP3 operator as well as for the consensus nucleotides A(−7) in the IRP3 operator and T(+7) in the IRP1 operator. We determined the effects of these substitutions on the binding of DtxR, transcriptional repressibility by DtxR, and promoter strength. These studies extend the available data on the relationships between the structure and function of DtxR-regulated promoter/operators.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli K-12 DH5α [F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1] (Bethesda Research Laboratories, Gaithersburg, Md.) was used for all purposes in this study, except that E. coli CJ236 (dut ung thi relA pCJ105 Cmr) (Bio-Rad, Hercules, Calif.) was used to generate uracil-containing single-strand DNA (ssDNA) as a template for site-directed mutagenesis. Strains were routinely cultured in Luria-Bertani broth (LB) (32) or terrific broth (TB) (44). Antibiotics and chromogenic substrates, when required, were included in the culture medium or plates at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 150 μg/ml; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40 μg/ml. In order to create iron-limiting growth conditions, the iron chelator ethylenediamine-di(o-hydroxyphenyl) acetic acid (EDDA) was added at 500 μg/ml to LB cultures and at 50 μg/ml to LB agar medium. Clones of DtxR-regulated promoter/operators in the promoter probe vector pQF50 (7) were used for measuring promoter activity and repressibility in E. coli hosts, as described previously (37). Plasmid pDSK29, an RSF1010 derivative carrying a 5-kb fragment containing the dtxR gene, was used for testing iron-dependent regulation of the pQF50 clones by DtxR (37). Plasmid pIRP3-1 (18) was used as the source of a 0.2-kb HindIII-NotI fragment carrying the IRP3 promoter/operator, and pIRP1-1 (37) was used as the source of a 0.18-kb AluI-MspI fragment carrying the IRP1 promoter/operator. Each of these fragments was cloned into plasmid pBluescript II KS(+) (Stratagene, La Jolla, Calif.) to generate ssDNA for site-directed mutagenesis, and the fragments containing the desired mutations were then recloned into pQF50 for subsequent testing.
DNA preparation, cloning, and sequencing.
Restriction enzymes and other DNA-modifying enzymes were used according to the instructions of the manufacturer (Life Technologies, Gaithersburg, Md.). DNA fragments were separated by electrophoresis in low-melting-point agarose gels, excised, and purified by using a gel extraction kit (Qiagen Inc., Chatsworth, Calif.). Recombinant DNA was introduced into E. coli strains by electroporation (Bio-Rad). Wizard miniprep kits (Promega, Madison, Wis.) were used to prepare plasmid DNA for subcloning and sequencing. Nucleotide sequence analysis of DNA fragments cloned into pBluescript II KS(+) was performed by an automated sequencing facility (Department of Biochemistry, Colorado State University, Fort Collins).
Site-directed mutagenesis.
Twenty-one-mer oligonucleotides designed on the basis of the IRP3 and IRP1 sequences [GAG ACG CAC C(A/C/G)A TCG GAA TGC for the +7 nucleotide in IRP3; GCA GTC TAT TG(C/G/T) GTG AGA CGC for the −7 nucleotide in IRP3; and TAG CCA AAC CCT TGT TGG TGT for the +7 nucleotide in IRP1] were purchased from Life Technologies. Mutagenesis was performed as described in the Bio-Rad Muta-Gene Manual. A uracil-containing ssDNA template was prepared from pBluescript II KS(+) containing the IRP3 and IRP1 region in E. coli CJ236 by using helper phage M13K07 (Pharmacia, Uppsala, Sweden). The products of oligonucleotide-primed DNA synthesis reactions were transformed into E. coli DH5α, and the mutations were confirmed by DNA sequencing.
Gel mobility shift assays and footprinting assays.
The Klenow fragment of DNA polymerase I was used for [α-32P]dCTP labeling of 220-bp NotI-BamHI fragments carrying alleles of the IRP3 operator and 180-bp KpnI-SpeI fragments carrying alleles of the IRP1 operator. The end-labeled DNA fragments at approximately 0.5 nM were incubated with various concentrations (0 to 2 μM) of purified DtxR in 10-μl reaction volumes. CoSO4 was present at 300 μM in all experiments. Other conditions were as described in a previous report from our laboratory (18).
β-Galactosidase assays.
E. coli DH5α(pDSK29) containing pIRP3, pIRP1, or one of the derivative plasmids was grown overnight in LB medium with either 500 μg of EDDA per ml (low-iron conditions) or no added EDDA (high-iron conditions). Units of β-galactosidase activity were calculated according to the method of Miller (20). Data presented are means and standard deviations from assays of three independent cultures grown under each set of specified conditions.
RESULTS
Construction of site-directed mutations in the IRP3 and IRP1 operators.
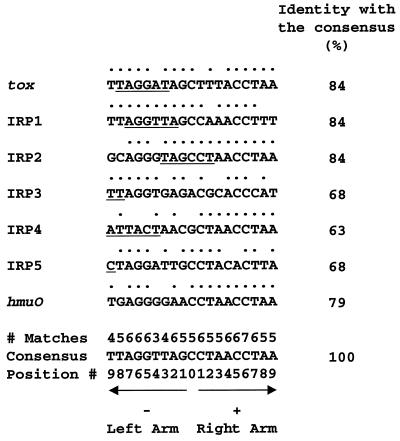
The nucleotide sequences of the reported DtxR-specific operators and the relationship of each operator to the demonstrated or putative promoter that it regulates are shown in Fig. 1. The tox, IRP1, and hmuO promoters were identified by primer extension or RNase protection experiments (19, 35, 39), and the putative IRP2, IRP3, IRP4, and IRP5 promoters were deduced by DNA sequence analysis (18, 37). The putative IRP3 promoter has the −35 sequence 5′-ATGATT-3′ separated by 17 nucleotides from the −10 sequence 5′-TCTATT-3′, and the 2 nucleotides at the 3′ end of the −10 promoter region overlap with the 5′ end of the operator. The IRP1 promoter has the −35 sequence 5′-AGGTTA-3′ separated by 18 nucleotides from the −10 sequence 5′-TATATT-3′ (39), and the entire −35 region is located within the operator. As noted previously, IRP3 is the least repressible of the known DtxR-regulated promoter/operators, whereas IRP1 is strongly repressible by DtxR and iron.
FIG. 1.
DtxR-specific operators. Sequences of the message (sense) strands of seven DtxR-specific operators are shown with the 5′-to-3′ orientation from left to right (18, 35, 37). A dot above a nucleotide indicates that it is identical with the corresponding nucleotide in the consensus sequence. The numbers above the consensus sequence indicate how many times matching nucleotides are found at the corresponding positions in these DtxR-specific operators. The left and right arms of the interrupted palindrome are indicated by the leftward- and rightward-pointing arrows, respectively. The central nucleotide is numbered 0, and the nucleotides in the left and right arms of the palindrome are numbered from −1 to −9 and from +1 to 9, respectively, with increasing distance from the center of the operator. Underlining shows the locations of proven or putative promoter regions either within the operators or overlapping them. For the tox, IRP2, IRP3, IRP4, and IRP5 operators, the underlined sequences identify complete or partial −10 promoter regions (18, 19, 37). For the IRP1 operator, the underlined sequence identifies the −35 promoter region (39). For hmuO, the −35 promoter region is immediately contiguous with the 5′ end of the operator, and the −10 promoter region is adjacent to the 3′ end of the operator but separated from it by one intervening nucleotide (35).
The sequence of the 19-bp operator in IRP3 differs from the consensus sequence of DtxR-regulated promoters at 6 positions (Fig. 1). The nonconsensus residue C at position +7 in IRP3 is unique, since all of the other DtxR-regulated promoter/operators have T at that position. The nonconsensus residues A at position 0 and C at position +3 in IRP3 are also unusual, since G or C is found at position 0 and A or T is found at position +3 in the other DtxR-regulated promoter/operators. The nonconsensus residues G at position −3, C at position +1, and T at position +9 in IRP3 are each present in the core sequence of at least one other promoter/operator that is more tightly regulated by DtxR than is IRP3. The primary purpose of the present study was to test the hypothesis that the unique nonconsensus residue C at position +7 in the IRP3 core sequence plays a major role in the poor repressibility of IRP3 by DtxR and its weak binding to DtxR. Toward this end we constructed variants of the IRP3 operator containing all possible single-nucleotide substitutions at position +7 and at the symmetrically located position −7, and we assessed the effects of these substitutions on binding to DtxR, repressibility by DtxR, and promoter activity. To extend these studies with IRP3, we also constructed a C(+7)T substitution in the operator sequence of the highly DtxR-repressible promoter/operator IRP1.
Analysis of site-directed mutations at positions +7 and −7 in IRP3.
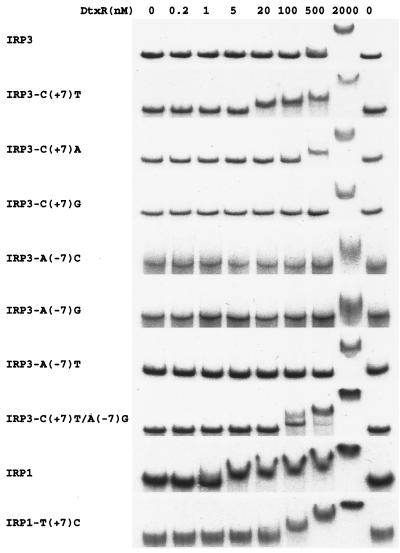
To analyze the relative binding of the sequence variants of IRP3 to DtxR, gel mobility shift assays were performed with each variant at several different DtxR concentrations ranging from 0 to 2,000 nM in the presence of 300 μM Co2+ (Fig. 2). The 0.22-kb DNA fragments containing the wild-type and mutant IRP3 core sequences were purified and end labeled with [α-32P]dCTP. The fragment containing the wild-type IRP3 operator sequence exhibited an easily detectable mobility shift only in the presence of 2,000 nM DtxR. In contrast, the fragment containing the IRP3 C(+7)T substitution exhibited a detectable mobility shift in the presence of as little as 20 nM DtxR. The substitution of T for C at position +7 in IRP3, therefore, caused a dramatic increase in the binding of DtxR to the DNA fragment. Changing the C at position +7 in IRP3 to A caused a slight increase in the binding of DtxR, manifested by the appearance of a detectable mobility shift at 500 nM DtxR. In contrast, changing C to G at position +7 caused a slight decrease in the binding of DtxR, resulting in a mobility shift of smaller magnitude at 2,000 nM DtxR than that seen with wild-type IRP3.
FIG. 2.
Gel mobility shift assays. Each of the DNA fragments of pIRP3 and its variants was 220 bp long, and the DNA fragments of pIRP1 and its variant were 180 bp. The fragments were end labeled with [α-32P]dCTP and incubated in the presence of Co2+ (300 μM) and various concentrations of DtxR (0 to 2,000 nM) as indicated.
To analyze the effect of the nucleotide at the reciprocal position −7 on the binding of DtxR to IRP3, A(−7) in wild-type IRP3 was changed systematically to all other nucleotides, and fragments containing each of the −7 variants of IRP3 were also subjected to gel mobility shift assays (Fig. 2). C or G substitutions caused a decrease in DtxR binding and resulted in a mobility shift of smaller magnitude in the presence of 2,000 nM DtxR, whereas the A(−7)T substitution did not significantly change the magnitude of the shift at 2,000 nM DtxR.
A double mutant of the IRP3 operator was constructed with the A-to-G replacement at position −7, which had the strongest negative effect on DtxR binding, and the C-to-T replacement at position +7, which had the strongest positive effect on DtxR binding. In gel shift assays the DNA fragment containing this double mutant exhibited a detectable mobility shift at DtxR concentrations as low as 100 nM, indicating that the binding of the double mutant fragment to DtxR was significantly greater than that of wild-type IRP3. Therefore, the substitution of T for C at position +7 in IRP3 had a greater effect on binding to DtxR than any other single substitution at position +7 or −7, and the negative effect of the A(−7)G substitution did not completely counteract the stronger positive effect of the C(+7)T substitution on DtxR binding to the IRP3 variant containing both substitutions.
Promoter strength and repressibility were examined by cloning each variant of IRP3 into pQF50, which has a promoterless lacZ gene, transforming each clone into E. coli DH5α with and without the compatible dtxR-containing plasmid pDSK29, and measuring the β-galactosidase activities of each transformant under high-iron and low-iron growth conditions (Table 1). All variants of IRP3 except A(−7)C showed very low or undetectable β-galactosidase activity, demonstrating that most of the nucleotide substitutions at position −7 or +7 in IRP3 caused markedly decreased promoter activity, to the extent that repressibility could no longer be measured accurately. In contrast, the A(−7)C substitution increased promoter activity approximately 1.7-fold but simultaneously decreased the repression ratio from the value of approximately 9 for wild-type IRP3 to approximately 1.3.
TABLE 1.
Characterization of wild-type and mutant IRP3 and IRP1 operators
| Plasmid | Sequence of operatora | Iron | β-Galactosidase activity (Miller units)b
|
DtxR bindingc to operator | |
|---|---|---|---|---|---|
| dtxR+ | DtxR− | ||||
| Consensus sequenced | TTAGGTTAGCCTAACCTAA | ||||
| pIRP3-1 | TTAGGTGAGACGCACCCAT | + | 5.2 ± 1.3 | 45.3 ± 2.3 | + |
| − | 46.7 ± 3.8 | 43.5 ± 1.7 | |||
| pIRP3 C(+7)T | TTAGGTGAGACGCACC*TAT | + | 0.2 ± 0.2 | 0.2 ± 0.1 | ++++ |
| − | 0.3 ± 0.3 | 0.4 ± 0.2 | |||
| pIRP3 C(+7)A | TTAGGTGAGACGCACC*AAT | + | 1.7 ± 0.5 | 6.2 ± 1.7 | ++ |
| − | 4.8 ± 1.2 | 7.4 ± 1.5 | |||
| pIRP3 C(+7)G | TTAGGTGAGACGCACC*GAT | + | 1.0 ± 0.1 | 3.1 ± 0.4 | ± |
| − | 2.8 ± 0.3 | 3.7 ± 0.9 | |||
| pIRP3 A(−7)C | TT*CGGTGAGACGCACCCAT | + | 56.4 ± 5.5 | 75.1 ± 6.4 | ± |
| − | 75.5 ± 4.1 | 79.3 ± 5.3 | |||
| pIRP3 A(−7)G | TT*GGGTGAGACGCACCCAT | + | 0.6 ± 0.3 | 1.7 ± 0.9 | ± |
| − | 2.5 ± 1.2 | 1.5 ± 0.5 | |||
| pIRP3 A(−7)T | TT*TGGTGAGACGCACCCAT | + | 1.0 ± 0.2 | 4.5 ± 0.9 | + |
| − | 3.6 ± 0.7 | 4.1 ± 1.1 | |||
| pIRP3 C(+7)T/A(−7)G | TT*GGGTGAGACGCACC*TAT | + | 0.1 ± 0.1 | 0.3 ± 0.2 | +++ |
| − | 0.3 ± 0.2 | 0.2 ± 0.1 | |||
| pIRP1-1 | TTAGGTTAGCCAAACCTTT | + | 1.1 ± 0.2 | 19.5 ± 1.8 | +++++ |
| − | 20.3 ± 2.1 | 22.3 ± 2.6 | |||
| pIRP1 T(+7)C | TTAGGTTAGCCAAACC*CTT | + | 12.7 ± 0.6 | 95.1 ± 3.8 | +++ |
| − | 87.0 ± 4.4 | 91.5 ± 4.2 | |||
| pQF50 | + | 0.2 ± 0.1 | 0.2 ± 0.1 | ||
| − | 0.1 ± 0.1 | 0.2 ± 0.2 | |||
Asterisks above operator sequences identify locations of nucleotide substitutions.
Average of at least three determinations for E. coli DH5α ± standard deviation.
Relative binding of DtxR to the operators from IRP3, IRP1, and their mutants, based an gel mobility shift data from Fig. 2 (pIRP3 was assigned an arbitrary value of +, and pIRP1 was assigned an arbitrary value of +++++).
19-bp consensus sequence for DtxR-specific operators (see text).
Analysis of a T-to-C substitution at the +7 position in IRP1.
The above findings indicated that the nucleotide at position +7 in the operator is important for the binding of IRP3 to DtxR and for the transcriptional activity of IRP3. To determine whether this was also true for IRP1, which is tightly regulated by DtxR, a T-to-C substitution at position +7 in the IRP1 operator was generated by site-directed mutagenesis. The binding of DtxR to a DNA fragment containing this T(+7)C variant of IRP1 was analyzed by gel mobility shift assays (Fig. 2). For the fragment containing wild-type IRP1, a small but distinct shift in mobility was detectable at 5 nM DtxR. In contrast, for the fragment with the T(+7)C variant of IRP1, the lowest concentration of DtxR at which an unambiguous shift in mobility was visible was 100 nM.
Expression of the β-galactosidase gene under the control of wild-type IRP1 and the T(+7)C variant of IRP1 was also compared in E. coli DH5α containing the dtxR+ plasmid pDSK29 under high-iron and low-iron conditions as described above (Table 1). The repression ratio decreased from approximately 18-fold for wild-type IRP1 to approximately 6.8-fold for the T(+7)C variant, and the most striking difference was a much higher level of β-galactosidase production from the T(+7)C variant than from wild-type IRP1 under high-iron (repressing) growth conditions (12.7 versus 1.1 β-galactosidase units). Therefore, both in IRP3 and in IRP1, the presence of C instead of T at position +7 was associated with decreased binding of holo-DtxR to the operator and with decreased repression of the promoter/operator by DtxR in vivo under high-iron conditions. In IRP1, the T(+7)C substitution caused an increase of approximately fourfold in promoter activity (from 20.3 to 87 β-galactosidase units under low-iron conditions [Table 1]). In contrast, the reciprocal C(+7)T substitution in IRP3 abolished promoter activity (from 46.7 to 0.3 β-galactosidase unit under low-iron conditions).
Footprinting analysis of selected IRP3 and IRP1 variants.
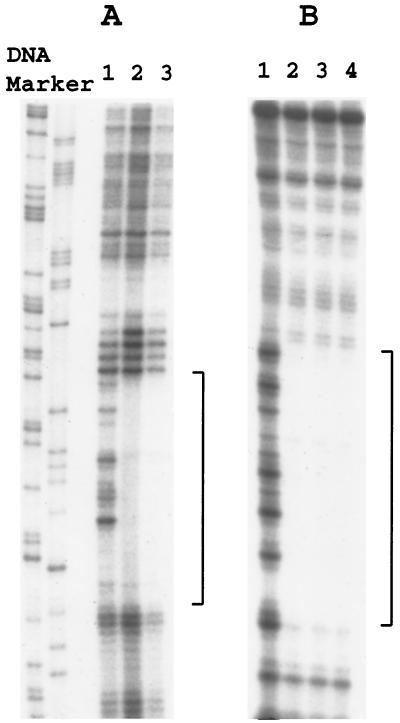
DNase I footprinting was performed to confirm that DtxR binds to the same sequences in wild-type and mutant alleles of IRP3 and IRP1 (Fig. 3). These experiments demonstrated that the DNase I footprints were similar for the wild type, the C(+7)T single mutant, and the A(−7)G/C(+7)T double mutant of IRP3. Similarly, the DNase I footprints for the wild type and T(+7)C variants of IRP1 were indistinguishable. Therefore, the altered affinity of holo-DtxR to these mutant alleles of IRP3 and IRP1 in the gel shift experiments described above was not caused by inactivation of a primary DtxR binding site and utilization of a weaker secondary DtxR binding site at a different location. These data demonstrate that the susceptibility of DtxR-regulated promoter/operators to repression by DtxR in vivo is directly related to the strength of their binding to holo-DtxR in vitro.
FIG. 3.
DNase I footprinting assays. All DNA fragments were 3′ end labeled with [α-32P]dCTP on one strand and were incubated in the presence of Co2+ (300 μM) and DtxR (1000 nM for pIRP3 and its variants; 200 nM for pIRP1 and its variant). (A) Lanes: 1, pIRP1 with no DtxR; 2, pIRP1 with DtxR; 3, pIRP1 T(+7)C with DtxR. (B) Lanes: 1, pIRP3 with no DtxR; 2, pIRP3 with DtxR; 3, pIRP3 C(+7)T with DtxR; 4, pIRP3 C(+7)T/A(−7)G with DtxR. Brackets indicate sequences protected by DtxR from DNase I digestion.
DISCUSSION
DtxR is an iron-activated global regulatory protein that represses the synthesis of diphtheria toxin and several other proteins of C. diphtheriae. The seven DtxR-regulated operators characterized so far either overlap with the −10 region or the −35 region of the associated promoter or are located between them (Fig. 1). Regulatory proteins that bind to ς70-like promoters at regions centered downstream from position −30 almost always function as repressors (10). The locations of the known DtxR-specific operators in relation to the proven or putative promoters that they regulate (Fig. 1) are fully consistent with the finding that DtxR functions as a repressor at multiple promoter/operators from C. diphtheriae but has not yet been shown to act as a transcriptional activator. Determining the molecular basis for sequence-specific DNA binding by holo-DtxR is critical for understanding its biological function. How changes in the structure of DtxR affect its function has been discussed elsewhere (8, 9, 16, 27, 28, 48). Here we examined how variations in the nucleotide sequence of DtxR-regulated promoter/operators affect their function.
The consensus sequence for DtxR-specific operators is 19 bp long and contains a perfect 9-bp AT-rich palindrome interrupted by a single base pair (Fig. 1). This consensus sequence was established by comparing the sequences both of wild-type IRPs from C. diphtheriae (18, 37) and of selected tox operator variants with partially randomized sequences that exhibited high-affinity binding to holo-DtxR in vitro (43). The DNA on either side of this 19-bp region in DtxR-specific operators from C. diphtheriae is highly variable both in nucleotide sequence and in AT content (18, 37). A 19-bp segment is sufficient, therefore, to identify operators that are recognized by DtxR.
Variations from the consensus sequence that do not abolish operator function will be considered first (Fig. 1). Identity between the consensus sequence and the message (sense) strand for each of these seven operators varies from 12 to 16 bp. Nucleotide C(+6) is invariant, and 15 other nucleotides are conserved in either five or six of these operators. Nucleotides T(−3), T(−4), and T(−9) are least conserved and occur in four, three, and four of these operators, respectively. All four possible nucleotides are found only at position −9, but three different nucleotides occur at positions −8, −4, −3, −1, 0, and +3. In addition, among the 21 tox operator variants mentioned previously that exhibit high-affinity binding to DtxR (43), all four possible nucleotides occur at positions −3, −4, and +4. In summary, although some nucleotide substitutions can occur at almost every position in the operator without abolishing its function, the greatest variability is found at positions −9, −8, −4, −3, −1, 0, +3, and +4.
Mutations that alter the function of DtxR-regulated promoter/operators will be considered next. Several corynephage β mutants with partially operator-constitutive tox phenotypes have been characterized (17, 22, 46, 47). C. diphtheriae lysogens carrying such mutants as prophages produce more diphtheria toxin under repressing high-iron growth conditions than do strains carrying wild-type β prophage, but they are not totally resistant to inhibition of toxin production by high concentrations of iron in the culture medium. The tox-201 and tox-202 alleles, which exhibit the strongest operator-constitutive phenotypes, have single G(−5)A and G(−6)A substitutions in the message (sense) strand of the tox operator (17), suggesting important roles of the highly conserved nucleotides G(−5) and G(−6) for the binding of DtxR. Nevertheless, the wild-type IRP4 promoter/operator, which is highly repressible by DtxR (18), also has A at position (−6) in the message (sense) strand of its operator (Fig. 1). Therefore, a specific nucleotide at a particular position, such as A(−6), is not an absolute determinant of operator function, because its effect is influenced by the local DNA sequence.
The present study is the first to analyze the effects of single-nucleotide changes at positions +7 and −7 in DtxR-specific operators. At C(+7) and A(−7) in IRP3 (Fig. 1), substitution of each other possible nucleotide causes changes in binding to DtxR (Fig. 2), repression by DtxR (Table 1), and promoter activity (Table 1), in various combinations. There appears to be strong selective pressure for C at position +7 in the wild-type IRP3 operator, because any other nucleotide at that position interferes dramatically with promoter activity (Table 1). In contrast, the A(−7)C substitution in IRP3 causes increased promoter activity, but at the expense of markedly decreased operator function (Table 1). It is not surprising that single-nucleotide substitutions can affect both operator and promoter functions, because the operator and promoter sequences in DtxR-regulated promoter/operators usually overlap (Fig. 1) (2, 18, 19, 35, 37, 40). Historically, the tox-201 allele mentioned above was the first example of decreased operator function and increased promoter activity shown to be caused by a single-nucleotide substitution [G(−5)A] in a DtxR-regulated promoter/operator (17, 46).
Structures of wild-type and/or mutant forms of apo-DtxR, holo-DtxR, and holo-DtxR in complex with DNA provide important additional information about mechanisms for DtxR activation and DtxR binding to its cognate operators (5, 27–31, 33, 48). Two independent DtxR dimers bind on opposite faces of the DNA to symmetrically disposed regions that are separated by 5 nucleotides (28, 48). The DNA helical axis is distorted slightly from that of linear canonical B-form DNA, and the recognition helix of the helix-turn-helix motif from each DtxR monomer inserts into the major groove. Only the side chains of Gln43 in the recognition helices interact directly with bases. Gln43 from one monomer in each DtxR dimer interacts with the central CG base pair and possibly with an adjacent base (28, 46). The Gln43 residues of the second monomers in the two DtxR dimers interact, respectively, with C(+5) and with the complement of G(−5) in the opposite DNA strand (28). Disruption of the latter interaction by a G(−5)A substitution provides a likely explanation for the operator-constitutive phenotype of phage βtox-201 described above (17, 46, 47). In contrast to the limited direct interactions of DtxR with bases, 9 residues from each helix-turn-helix motif are reported to contact ligands in the DNA backbone (28, 48). Although interactions with ligands in the DNA backbone are known to complement interactions with bases in determining the sequence-specific binding of repressor proteins to DNA (13, 14, 28, 48), the dramatic preponderance of binding to ligands in the DNA backbone versus ligands in the bases reported for DtxR is a striking aspect of its sequence-specific DNA-binding activity.
Although a Gln43 residue from each DtxR dimer interacts directly with the central CG base pair in the tox operator, G or C at position 0 is not required for binding of DtxR to a cognate operator (Fig. 1). The exception is IRP3, which has A at position 0 in the operator (Fig. 1). It is not yet known, however, whether A(0) contributes to the weaker affinity of IRP3 for DtxR and the poorer repressibility of IRP3 by DtxR, in comparison with several other DtxR-regulated operator/promoters (Fig. 2; Table 1) (18, 37).
If the structures described above for the complexes of holo-DtxR with DNA are representative of all DtxR-operator complexes, then the striking effects of nucleotide substitutions at positions +7 and −7 in the IRP3 operator reported here are not caused by disrupting direct interactions between DtxR and bases in the major groove. However, these nucleotide substitutions could cause changes in local DNA flexibility, which is determined by nucleotide sequence and is believed to provide an “indirect readout” of sequence-specific information in DNA (4, 14). Local flexibility is important in determining whether a short segment in DNA can adopt the confirmation needed for it to interact with a sequence-specific DNA-binding protein such as a repressor. Unfortunately, rules that can accurately predict local DNA conformations from DNA sequences are not yet available (4). The results of the studies presented here are fully consistent with the hypothesis that local DNA flexibility makes an important contribution to the interaction of DtxR with its cognate operators.
Some DtxR-specific operators exhibit high homology with the consensus sequence in only one arm of the palindrome. The most striking example is IRP4, which is identical with the consensus sequence in the right arm but has only 3 of 9 matching nucleotides in the left arm (Fig. 1). The stepwise patterns in gel mobility shifts seen with increasing DtxR concentrations for some DtxR-specific operators, particularly for wild-type IRP1 and IRP3 C(+7)T in Fig. 2, suggest that those operators may contain both high-affinity and low-affinity DtxR-binding sites. Although X-ray crystallography reveals that two DtxR dimers can bind to the tox operator (28, 48), it is not yet established whether binding of both DtxR dimers is required for the repression of transcription in vivo.
In summary, although rapid progress has been made in the last several years, much remains to be learned about the molecular basis for sequence-specific binding of DtxR to its cognate operators. Additional genetic, biochemical, and structural studies are required to determine whether there are significant differences in the molecular bases of interaction of DtxR with the various operators that it can recognize and to refine current models of DtxR-operator interactions. Such studies should provide new insights about this process, which plays a central role in the DtxR-dependent global regulation of gene expression by iron in C. diphtheriae. Such studies should also contribute to an improved general understanding of sequence-specific protein-DNA interactions, which have fundamental importance for all living cells.
ACKNOWLEDGMENTS
This research was supported in part by Public Health Service grant R01 AI14107.
We thank Michael D. Feese, Joanne Goranson-Siekierke, Wim G. J. Hol, Michael G. Jobling, Diana M. Marra, and Yilei Qian for constructive comments and criticism during the preparation of this article.
REFERENCES
- 1.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd J, Murphy J R. Analysis of the diphtheria tox promoter by site-directed mutagenesis. J Bacteriol. 1988;170:5949–5952. doi: 10.1128/jb.170.12.5949-5952.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd J, Oza M N, Murphy J R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickerson R E. Sequence-dependent B-DNA conformation in crystals and in protein complexes. In: Ramaswamy H S, Sarma M H, editors. Structure, motion, interaction and expression of biological macromolecules. Albany, N.Y: Proceedings of the Tenth Conversation, State University of New York. Adenine Press; 1998. pp. 17–36. [Google Scholar]
- 5.Ding X, Zeng H, Schiering N, Ringe D, Murphy J R. Identification of the primary metal ion-activation sites of the diphtheria tox repressor by X-ray crystallography and site-directed mutational analysis. Nat Struct Biol. 1996;3:382–387. doi: 10.1038/nsb0496-382. [DOI] [PubMed] [Google Scholar]
- 6.Doukhan L, Predich M, Nair G, Dussurget O, Mandic-Mulec I, Cole S T, Smith D R, Smith I. Genomic organization of the mycobacterial sigma gene cluster. Gene. 1995;165:67–70. doi: 10.1016/0378-1119(95)00427-8. [DOI] [PubMed] [Google Scholar]
- 7.Farinha M A, Kropinski A M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goranson-Siekierke J, Holmes R K. Regulation of diphtheria toxin production: characterization of the role of iron and the diphtheria toxin repressor. In: Alouf J E, Freer J H, editors. The comprehensive sourcebook of bacterial protein toxins. London, United Kingdom: Academic Press; 1999. pp. 94–103. [Google Scholar]
- 9.Goranson-Siekierke J, Pohl E, Hol W G J, Holmes R K. Anion-coordinating residues at binding site 1 are essential for the biological activity of the diphtheria toxin repressor. Infect Immun. 1999;67:1806–1800. doi: 10.1128/iai.67.4.1806-1811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Resnikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 11.Gunter-Seeboth K, Schupp T. Cloning and sequence analysis of the Corynebacterium diphtheriae dtxR homologue from Streptomyces lividans and S. pilosus encoding a putative iron repressor protein. Gene. 1995;166:117–119. doi: 10.1016/0378-1119(95)00628-7. [DOI] [PubMed] [Google Scholar]
- 12.Hardham J M, Stamm L V, Porcella S F, Frye J G, Barnes N Y, Howell J K, Mueller S L, Radolf J D, Weinstock G M, Norris S J. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene. 1997;197:47–64. doi: 10.1016/s0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 13.Harrison S C. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 14.Harrison S C, Aggarwal A K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 15.Hill P J, Cockayne A, Landers P, Morrissey J A, Sims C M, Williams P. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect Immun. 1998;66:4123–4129. doi: 10.1128/iai.66.9.4123-4129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, R. K. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. (Suppl.), in press. [DOI] [PubMed]
- 17.Krafft A E, Tai S P, Coker C, Holmes R K. Transcription analysis and nucleotide sequence of tox promoter/operator mutants of corynebacteriophage beta. Microb Pathog. 1992;13:85–92. doi: 10.1016/0882-4010(92)90069-z. [DOI] [PubMed] [Google Scholar]
- 18.Lee J H, Wang T, Ault K, Liu J, Schmitt M P, Holmes R K. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:4273–4280. doi: 10.1128/iai.65.10.4273-4280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leong D, Murphy J R. Characterization of the diphtheria tox transcript in Corynebacterium diphtheriae and Escherichia coli. J Bacteriol. 1985;163:1114–1119. doi: 10.1128/jb.163.3.1114-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Murphy J R, Pappenheimer A M, Jr, de Borms S T. Synthesis of diphtheria tox-gene products in Escherichia coli extracts. Proc Natl Acad Sci USA. 1974;71:11–15. doi: 10.1073/pnas.71.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy J R, Skiver J, McBride G. Isolation and partial characterization of a corynebacteriophage beta, tox operator constitutive-like mutant lysogen of Corynebacterium diphtheriae. J Virol. 1976;18:235–244. doi: 10.1128/jvi.18.1.235-244.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oguiza J A, Tao X, Marcos A T, Martin J F, Murphy J R. Molecular cloning, DNA sequence analysis, and characterization of the Corynebacterium diphtheriae dtxR homolog from Brevibacterium lactofermentum. J Bacteriol. 1995;177:465–467. doi: 10.1128/jb.177.2.465-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 25.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 26.Pappenheimer A M., Jr Diphtheria toxin. Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 27.Pohl E, Holmes R K, Hol W G J. Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR. J Biol Chem. 1998;273:22420–22427. doi: 10.1074/jbc.273.35.22420. [DOI] [PubMed] [Google Scholar]
- 28.Pohl E, Holmes R K, Hol W G J. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J Mol Biol. 1999;292:653–667. doi: 10.1006/jmbi.1999.3073. [DOI] [PubMed] [Google Scholar]
- 29.Pohl E, Qui X, Must L M, Holmes R K, Hol W G J. Comparison of high-resolution structures of the diphtheria toxin repressor in complex with cobalt and zinc at the cation-anion binding site. Protein Sci. 1997;6:1114–1118. doi: 10.1002/pro.5560060519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu X, Pohl E, Holmes R K, Hol W G J. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry. 1996;35:12292–12302. doi: 10.1021/bi960861d. [DOI] [PubMed] [Google Scholar]
- 31.Qiu X, Verlinde C L, Zhang S, Schmitt M P, Holmes R K, Hol W G J. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure. 1995;3:87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schiering N, Tao X, Zeng H, Murphy J R, Petsko G A, Ringe D. Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1995;92:9843–9850. doi: 10.1073/pnas.92.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt M P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt M P. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect Immun. 1997;65:4634–4641. doi: 10.1128/iai.65.11.4634-4641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt M P, Holmes R K. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt M P, Holmes R K. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J Bacteriol. 1994;176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt M P, Predich M, Doukhan L, Smith I, Holmes R K. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect Immun. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. (Erratum, 64:681, 1996.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt M P, Talley B G, Holmes R K. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:5364–5367. doi: 10.1128/iai.65.12.5364-5367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt M P, Twiddy E M, Holmes R K. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci USA. 1992;89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tai S P, Krafft A E, Nootheti P, Holmes R K. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb Pathog. 1990;9:267–273. doi: 10.1016/0882-4010(90)90015-i. [DOI] [PubMed] [Google Scholar]
- 42.Tao X, Murphy J R. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J Biol Chem. 1992;267:21761–21764. [PubMed] [Google Scholar]
- 43.Tao X, Murphy J R. Determination of the minimal essential nucleotide sequence for diphtheria tox repressor binding by in vitro affinity selection. Proc Natl Acad Sci USA. 1994;91:9646–9650. doi: 10.1073/pnas.91.20.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tartof K D, Hobbs C A. Improved media for growing plasmid and cosmid clones. Focus. 1987;9:12. [Google Scholar]
- 45.Uchida T, Gill D M, Pappenheimer A M., Jr Mutation in the structural gene for diphtheria toxin carried by temperate phage. Nat New Biol. 1971;233:8–11. doi: 10.1038/newbio233008a0. [DOI] [PubMed] [Google Scholar]
- 46.Welkos S L, Holmes R K. Regulation of toxinogenesis in Corynebacterium diphtheriae. I. Mutations in bacteriophage beta that alter the effects of iron on toxin production. J Virol. 1981;37:936–945. doi: 10.1128/jvi.37.3.936-945.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welkos S L, Holmes R K. Regulation of toxinogenesis in Corynebacterium diphtheriae. II. Genetic mapping of a tox regulatory mutation in bacteriophage beta. J Virol. 1981;37:946–954. doi: 10.1128/jvi.37.3.946-954.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White A, Ding X, vanderSpek J C, Murphy J R, Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998;394:502–506. doi: 10.1038/28893. [DOI] [PubMed] [Google Scholar]