Abstract
An understanding of how the heme-deficient gram-positive bacterium Streptococcus pyogenes establishes infections in O2-rich environments requires careful analysis of the gene products important in aerobic metabolism. NADH oxidase (NOXase) is a unique flavoprotein of S. pyogenes and other lactic acid bacteria which directly catalyzes the four-electron reduction of O2 to H2O. To elucidate a putative role for this enzyme in aerobic metabolism, NOXase-deficient mutants were constructed by insertional inactivation of the gene that encodes NOXase. Characterization of the resulting mutants revealed that growth in rich medium under low-O2 conditions was indistinguishable from that of the wild type. However, the mutants were unable to grow under high-O2 conditions and demonstrated enhanced sensitivity to the superoxide-generating agent paraquat. Mutants cultured in liquid medium under conditions of carbohydrate limitation and high O2 tension were characterized by an extended lag phase, a reduction in growth, and a greater accumulation of H2O2 in the growth medium compared to the wild-type strain. All of these mutant phenotypes could be overcome by the addition of glucose. Either the addition of catalase to the culture medium of the mutants or the introduction of a heterologous NADH peroxidase into the mutants eliminated the accumulation of H2O2 and rescued the growth defect of the mutants under high-O2 conditions in carbohydrate-limited liquid medium. Taken together, these data show that NOXase is important for aerobic metabolism and essential in environments high in O2 with carbohydrate limitation.
The gram-positive microorganism Streptococcus pyogenes (group A streptococcus) is the causative agent of numerous infections of the skin and pharynx ranging from superficial diseases including erysipelas, impetigo, and pharyngitis to those characterized by extensive tissue destruction, such as necrotizing fasciitis. The initial stage of all streptococcal infections involves the attachment of the organism to epithelial cells of the nasopharynx or epidermis (49), and considerable evidence suggests that the ability to sense an aerobic environment and survive plays an important role in this process (17, 47, 48). A good example of this is streptococcal fibronectin-binding protein F, which is regulated in response to oxidative stress (16, 48).
The mechanisms and gene products that allow S. pyogenes to survive in aerobic environments remain largely unknown. While S. pyogenes produces a single Mn-containing superoxide dismutase (SOD) that is essential for aerobic streptococcal growth (16), it lacks many of the proteins known to be important for aerobic growth. Since the lactic acid bacteria (including those in the genera Streptococcus, Enterococcus, and Lactococcus) cannot synthesize heme (11), S. pyogenes lacks the catalases and cytochrome oxidases required for oxidative energy-linked metabolism and instead depends on substrate level phosphorylation for growth. In addition, streptococci lack the moderate-to-high levels of intracellular glutathione found in gram-negative bacteria (12). Without such mechanisms for handling oxidative stress, it seems that aerobic conditions should severely restrict streptococcal growth, yet O2 seems to have a positive effect on the growth yields of some other lactic acid bacteria (25, 30). This suggests the existence of other enzymes that are important for aerobic streptococcal growth.
Recently, other lactic acid bacteria have been found to contain unique flavoproteins involved in oxidative metabolism that are very different from the respiratory redox enzymes of cytochrome-containing bacteria like Escherichia coli (8, 20, 41, 42). One such flavoprotein, NADH peroxidase (NPXase), has been characterized extensively in Enterococcus faecalis, where it uses H2O2 as an electron acceptor, thereby providing an enzymatic defense against peroxide stress (41). Another E. faecalis flavoprotein, NADH oxidase (NOXase), catalyzes the direct four-electron reduction of O2 to water and serves as an electron acceptor during active aerobic metabolism in this organism (42). These two flavoproteins have 44% amino acid identity to one another, with the most highly conserved segments containing the nonflavin redox center and the flavin adenine dinucleotide (FAD)- and NADH-binding regions. The nonflavin redox center in each of these enzymes is an unusual stabilized cysteine-sulfenic acid that cycles between oxidized and reduced states (33).
A role for these two flavoproteins in facilitating the aerobic metabolism of lactic acid bacteria may require the regeneration of one NAD+ molecule by NPXase, and the regeneration of two molecules of NAD+ by NOXase would provide oxidized pyridine nucleotides for glycolysis. Furthermore, since NOXase directly reduces O2 to H2O without the formation of harmful reactive O2 intermediates, it may serve to protect group A streptococci against oxidative stress. To address the possibility that either of these two flavoproteins is involved in streptococcal aerobic metabolism, we first examined whether these flavoproteins are present in S. pyogenes. We identified only the H2O-forming NOXase and demonstrated through insertional inactivation of the gene encoding NOXase that this enzyme contributes significantly to aerobic metabolism under conditions of high O2 stress.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains utilized in this study are listed in Table 1. E. coli DH5α was the host for molecular cloning experiments, and HB101 was used in fibronectin-binding assays. E. coli strains were cultured in Luria-Bertani broth (45), and S. pyogenes strains were grown in Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (THY medium) or in C medium, a low-glucose-containing medium (1.5 versus 15 mg/liter for THY medium), as described elsewhere (31). To produce solid media, Bacto Agar (Difco) was added to THY and C media at a final concentration of 1.4%. Streptococci were grown in liquid medium or on solid medium and cultured overnight at 37°C. As in previous studies (16), streptococci were cultured under low-O2 conditions in 10-ml broth cultures tightly sealed in 15-ml conical tubes or on agar plates incubated in an anaerobic gas chamber (GasPak; catalogue no. 70304; BBL). High-O2 conditions were produced when strains were grown in 30-ml broth cultures with vigorous agitation (220 rpm) in 250-ml glass flasks or on agar plates incubated in ambient air. When appropriate, antibiotics were used at the following concentrations: kanamycin at 25 μg ml−1 for E. coli and 500 μg ml−1 for S. pyogenes, chloramphenicol at 20 μg ml−1 for E. coli and 3 μg ml−1 for S. pyogenes. Where indicated, medium was supplemented with various sugars at a final concentration of 1% (wt/vol), pyruvate at 25 mM, catalase at 1 mg/ml, bovine serum albumin at 1 mg/ml, or the intracellular superoxide-generating agent paraquat at 4 mM (Sigma).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Characteristic | Reference or source |
|---|---|---|---|
| Escherichia coli | |||
| DH5α | recA1 endA1 hsdR17 | BRL | |
| HB101 | recA13 proA2 | 3 | |
| Streptococcus pyogenes | |||
| JRS4 | Wild type | 46 | |
| SAM1 | Wild type | Kmr derivative of JRS4 | 17 |
| JNOX1a | JRS4ΩpNOX1 | nox inactivated | This work |
| HSC12 | Wild type | 18 | |
| HNOX1a | HSC12ΩpNOX1 | nox inactivated | This work |
| JCCP1a | JRS4ΩpCcp1 | ccpA inactivated | This work |
| Enterococcus faecalis | |||
| 10C1 | Wild type (ATCC 11700) | 42 | |
| OG1X | Wild type | 23 |
Strain derived through transformation of the designated strain with the indicated integrational plasmid shown.
DNA techniques.
Plasmid DNA was isolated by standard techniques and transformed into E. coli by the method of Kushner (26). S. pyogenes was transformed by electroporation as previously described (6). Restriction endonucleases, ligases, and polymerases were used in accordance with the recommendations of the manufacturers. Chromosomal DNA was purified from S. pyogenes as described previously (6).
Construction of integrational plasmids.
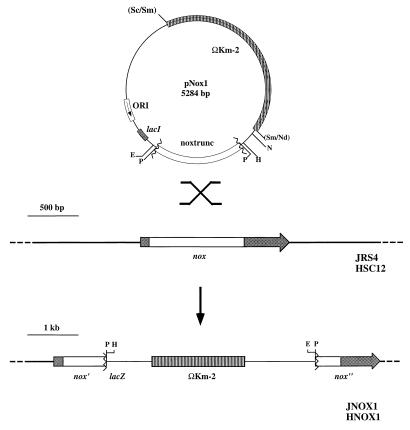
A 1,302-bp internal fragment of the gene encoding NOXase (nox) was obtained by PCR amplification from the S. pyogenes JRS4 chromosomal DNA by using the primers 5 Nox (5′-GTYGTYGTYG GWGCWAAYCA YGCWGGWAC-3′) and 3 Nox (5′-RAWRTGWGGH ARRAARAARA WRTC-3′); R is A/G, W is A/T, and Y is C/T). The PCR product was inserted into a commercial vector (pCRII; Invitrogen) by using a TA tail method to generate pSpNOX. Digestion of pSpNOX with PstI removed 275 bp from the 3′ end of the gene, and then the 1,027 bp PstI fragment containing the truncated nox allele was inserted into the PstI site of pCIV2 (38), generating pNOX1. Introduction of pNOX1 into S. pyogenes targets integration of the plasmid into the nox chromosomal locus by homologous recombination, resulting in the insertional inactivation of nox and the generation of strains JNOX1 and HNOX1 from wild-type strains JRS4 and HSC12, respectively (see Fig. 2). The correct chromosomal structures of JNOX1 and HNOX1 were confirmed by PCR analyses using primers ••• of the appropriate sequences (data not shown).
FIG. 2.
Construction of nox null mutants. Insertional inactivation of nox in two unrelated strains of S. pyogenes was accomplished by first amplifying a region internal to the nox coding region (noxtrunc) and then inserting the fragment into integrational vector pCIV2. The resulting element, pNOX1, contains a kanamycin resistance determinant (striped bar) and the noxtrunc region (empty bar) of JRS4. Recombination between homologous regions of pNOX1 and the S. pyogenes JRS4 and HSC12 chromosomes (indicated by the large X between the plasmid and chromosomal restriction maps) generated JNOX1 and HNOX1, respectively, which have a chromosomal structure that contains two truncated and inactive versions of nox (indicated by the zigzag line). E, EcoRI; H, HindIII; N, NcoI; Nd, NdeI; P, PstI; Sc, ScaI; Sm, SmaI; ORI, origin of replication.
The gene encoding the E. faecalis NADH peroxidase (npr; GenBank accession no. X62755) (41) was amplified from E. faecalis OG1X by PCR using primers NprF (5′-GTGGGGCGTC CCTATCAATC GTATCGGAGA-3′) and NprR (5′-GGTGTTTCCT ATCAACGTGT GGATGAACAA G-3′). The resulting 1,690-bp product was inserted into pCRII as described above, yielding pCMG13. An EcoRI fragment of pCMG13 containing the entire npr coding region was inserted into the EcoRI site of E. coli-streptococcal shuttle vector pLZ12-Km (18, 40) to generate pNPR1, which was then used to introduce a replicating plasmid encoding a functional copy of npr into a streptococcal host.
A 436-bp internal fragment of the gene encoding the gram-positive catabolite repressor protein CcpA was amplified by PCR from the S. pyogenes JRS4 chromosome using primers CcpF1 (5′-GGATCCCAAC CGTTAGTCGT-3′) and CcpR1 (5′-ATAGTCGACG TTGACGCT-3′). The PCR product was inserted into pCRII as described above, yielding pCMG14. A pCMG14 EcoRI fragment containing the ccpA internal region was inserted into the EcoRI site of pCIV2 to generate pCcp1, which was then introduced into the JRS4 chromosomal locus as described above for pNOX1, and the resulting transformant was designated JCCP1.
DNA sequencing.
Sequences of various DNA regions were determined by using fluorescent-dye-labeled nucleotide terminators in accordance with the recommendations of the manufacturer (Big Dye, catalogue no. 4303500; PE Applied Biosystems). Analysis of the resulting sequences was conducted by using the Wisconsin package (Genetics Computer Group), and sequences were compared to the information available through the Oklahoma group A streptococcal genome sequencing project (http://www.genome.ou.edu/strep.html).
NOXase enzyme assays.
S. pyogenes cell extracts were prepared by disruption of bacteria using glass beads (catalogue no. G-4649; Sigma) and agitation in a reciprocating shaking device (model 3110 BX; Biospec Products). NOXase activities were assayed at 25°C in a total volume of 3 ml using an assay buffer (50 mM potassium phosphate [pH 7.0], 0.5 mM EDTA) and conditions that have been previously described (1). The amount of protein in each sample was determined by the method of Bradford (4), and NOXase specific activity is presented as micromoles of NADH oxidized per minute per milligram of protein. Data presented represent the mean and standard deviation of samples analyzed in triplicate and are representative of at least five independent determinations.
RNA techniques.
Relevant S. pyogenes strains were cultured for 14 to 16 h at 37°C under low-O2 conditions in liquid C medium and then diluted 1:100 in 300 ml of fresh medium and grown at 37°C under high-O2 conditions for 2, 4, or 6 h in the presence or absence of added glucose. Streptococcal cells were harvested by centrifugation (2,500 × g, 10 min, 4°C) and resuspended in 200 μl of diethyl pyrocarbonate-treated distilled H2O. Total RNA was isolated by the method of Cheung et al. (7) by using a commercial reagent (FastRNA BLUE; Bio 101) and a high-speed reciprocating shaking device (FP-120; Savant Instruments). RNA samples were then treated with DNase I (GIBCO Bethesda Research Laboratories [BRL], Gaithersburg, Md.) in the presence of the RNase inhibitor RNaseOUT (BRL) in accordance with the manufacturer's instructions to eliminate chromosomal DNA, and the RNA concentrations were determined by measuring A260. To analyze the relative amounts of nox transcripts in comparison to a standard recA transcript, a semiquantitative reverse transcription (RT)-PCR method (35) and a commercial kit (Titan, Boehringer Mannheim) were utilized. Primers RT5Nox (5′-GTTGTTGTTG GTGCAAACCA TGC-3′) and RT3Nox (5′-GTCTTTGGCA CCAAGTGCTG CCA-3′) were used for RT-PCRs of nox, and primers 5RECA1 (5′-CGTCGAAAGC CCGGGATGAT-3′) and 3RECA1 (5′-GCGCATGCCC GGGATCGATA-3′) were used for recA.
Fibronectin-binding assays and SOD activity analysis.
Protein F-dependent fibronectin binding was quantitated by using 125I-labeled fibronectin as described elsewhere (17). The activity of SOD in streptococcal cell lysates was determined by using a native gel assay as described previously (16).
H2O2 measurement.
S. pyogenes was cultured in liquid C medium for 20 h at 37°C under low-O2 conditions and then diluted 1:1,000 in fresh medium and grown at 37°C under high-O2 conditions as indicated in Results. The A600 was measured, and cells were removed by centrifugation. A 180-μl aliquot of each supernatant was added to individual wells of a 96-well microtiter dish. Next, 20 μl of a solution consisting of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS; catalogue no. A-1888; Sigma) at 3 mg/ml and horseradish peroxidase (catalogue no. P-8250; Sigma) at 0.2 mg/ml was prepared in 0.1 M sodium phosphate buffer (pH 7.0) and added to each well. The reaction was allowed to proceed for 20 min at room temperature, and then the A560 was measured. Samples were compared to a standard curve generated by known concentrations of H2O2. Data presented represent the mean and the standard deviation of samples assayed in quintuplicate and are representative of at least three independent experiments.
Estimation of O2 consumption.
An O2 electrode (model 5331; Yellow Springs Instrument Co.) was used to measure O2 uptake by cell extracts as described previously (36).
RESULTS
S. pyogenes contains a single nox homologue but lacks npr.
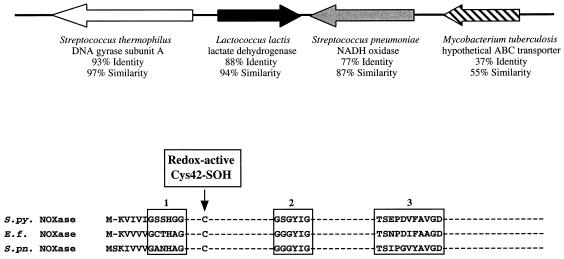
Several methods were used to examine S. pyogenes for the presence of genes that may encode NADH oxidase (nox) or NADH peroxidase (npr). For nox, the DNA sequences of several homologues from other species are available, including a well-characterized gene from E. faecalis (42). Multiple alignment of these sequences revealed highly conserved regions that were then utilized to design primers for PCR (see Materials and Methods). PCR amplification from S. pyogenes JRS4 genomic DNA resulted in a single product, the sequence (GenBank accession no. AF101442) of which was identical to a single open reading frame identified in the S. pyogenes genome sequence database (http://www.genome.ou.edu/strep.html). This open reading frame is highly homologous to nox from E. faecalis (GenBank accession no. X68847) (42). When the identified S. pyogenes open reading frame was compared against the entire GenBank database, it was found to be highly homologous to nox from other lactic acid bacteria and was most homologous to nox from S. pneumoniae (GenBank accession no. AF014458) (77% identical, 87% similar; (Fig. 1). The S. pyogenes nox homologue contains all of the signature residues characteristic of nox from E. faecalis (42), including the cysteine sulfenic acid redox center and the NADH- and FAD-binding regions (Fig. 1).
FIG. 1.
S. pyogenes contains a single nox homologue. (A) The arrows represent the directions of transcription of open reading frames, which are contained within the same chromosomal region as nox. Information presented under the arrows describes the genes to which the open reading frames have the highest homology. A possible factor-independent terminator located 3′ of the ldh homologue was identified by the method of Brendel and Trifonov (5). (B) The S. pyogenes nox homologue (S.py.; GenBank accession no. AF101442) contains all of the signature residues characteristic of E. faecalis nox (E.f., GenBank accession no. X68847) (44) and S. pneumoniae nox (S.pn.; GenBank accession no. AF014458), including the cysteine sulfenic acid redox center, the NADH contact region (box 2), and FAD-binding regions (boxes 1 and 3).
Upon examination of the chromosomal region surrounding the nox homologue, an open reading frame was found that was highly homologous to the lactate dehydrogenase (LDH)-encoding gene (ldh) from Lactococcus lactis (88% identical, 94% similar; Fig. 1). The two genes are 159 bp apart and are oriented so that they are convergently transcribed. Just downstream of the ldh stop codon, there is an inverted repeat of 8 bp that may represent a factor-independent terminator. In addition, sequences with high homology to sites that are bound by the gram-positive catabolite repressor protein CcpA (44), were found upstream of both the nox (86% identity) and ldh (100% identity) homologues. Further examination of the genome revealed that at an unlinked locus, an additional open reading frame with some homology to nox, lies downstream of an alkyl-hydroperoxide reductase C homologue, an arrangement of genes that has been reported in other organisms (37). However, subsequent mutagenesis of this nox-like homologue did not support its identity as an H2O-forming NADH oxidase (J. A. Horenstein and M. G. Caparon, unpublished data).
For npr, examination of the S. pyogenes genome database failed to reveal any genes other than nox that had significant homology to npr from E. faecalis (GenBank accession no. X62755) (41). The lack of an S. pyogenes npr homologue was supported by PCR analysis, which failed to yield a product from S. pyogenes genomic DNA using primers derived from E. faecalis npr. These data are consistent with a previous study that failed to detect NPXase activity in S. pyogenes based on biochemical criteria (A. Claiborne, unpublished data).
Construction of nox null mutant strains.
To verify the identity of the nox homologue and to generate mutants for functional studies, the NOXase coding region was insertionally inactivated in two unrelated strains of S. pyogenes. This was accomplished by insertional mutagenesis in which a region internal to the nox coding sequence is used to target the integration of a plasmid that cannot replicate in streptococci into the chromosomal copy of the gene by homologous recombination (Fig. 2). Disruption of nox in strains JRS4 and HSC12 generated JNOX1 and HNOX1, respectively.
Characterization of nox mutants.
Cultures for all functional assays were conducted in the presence of kanamycin to maintain selection for the integrated plasmid. To ensure that any differences between the wild-type and mutant strains were due to the inactivation of nox and not due to kanamycin, nox mutant JNOX1 was compared to a derivative of the same wild-type strain (JRS4) that has a wild-type nox locus but contains an insertion of the kanamycin resistance determinant into an unrelated chromosomal locus (SAM1 [Table 1]). For clarity, SAM1 will subsequently be referred to as the wild-type strain. In SAM1 cells, NOXase specific activity was readily detectable in cells from 16-h cultures grown under low-O2 conditions (1.6 μmol of NADH oxidized min−1 mg−1). This is approximately one-half of the level of NOXase observed in E. faecalis 10C1 grown under similar conditions (2.8 μmol of NADH oxidized min−1 mg−1). In contrast, JNOX1 had virtually no detectable NOXase activity, even after an additional 6 to 7 h of growth (0.04 μmol of NADH oxidized min−1 mg−1). Similar results were obtained with the mutant derived from the other wild-type strain (HNOX1; data not shown). Since NOXase consumes O2 and produces H2O, these data are in agreement with direct assays of O2 uptake showing that the mutant consumes very little O2 compared to the wild type (0.02 versus 0.31 μmol of O2 min−1 mg−1).
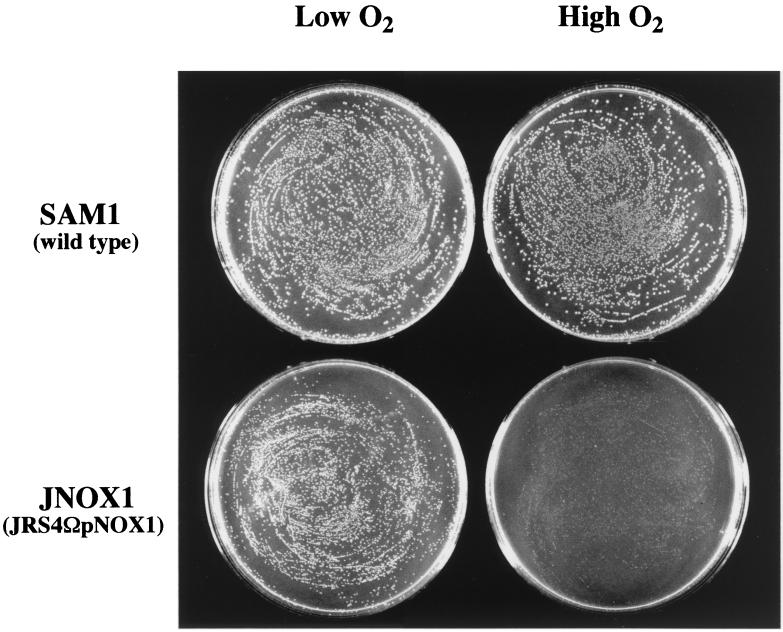
Mutants are defective for aerobic growth.
Next, the capacity of the mutants to grow under conditions of high and low O2 tension in two distinct types of media was evaluated. When bacteria were cultured on solid THY medium under low-O2 conditions, the growth of nox mutant JNOX1 was indistinguishable from that of wild-type SAM1 (Fig. 3). However, when cultured under high-O2 conditions on agar plates, the nox mutant JNOX1 grew poorly and formed barely visible colonies (Fig. 3). Similar results were obtained from growth on C agar plates and with the second nox mutant HNOX1 (data not shown). Since the JNOX1 and HNOX1 mutants exhibited similar phenotypes under all of the conditions tested, only the JNOX1 characterization will be reported in the remainder of this report.
FIG. 3.
Characterization of the nox mutants on solid medium. Both wild-type SAM1 and the nox null mutant JNOX1 were grown under high (ambient atmosphere; see Materials and Methods)- and low-O2 conditions on solid THY medium at 37°C. Wild-type SAM1 was grown for 14 h, and the nox mutant JNOX1 was grown for >20 h.
In liquid medium, the JNOX1 mutant exhibited a complex medium-dependent growth defect in which the mutant grew in THY medium broth cultures but not in C medium broth cultures under high-O2 conditions. This growth defect is referred to as the glucose effect in the remainder of this work. Taken together, these data indicate that the nox mutant has a pronounced growth defect on solid media under high O2 tension and that in liquid media under both high and low O2 tensions, the nox mutant has a growth defect the extent of which is influenced by the type of culture medium.
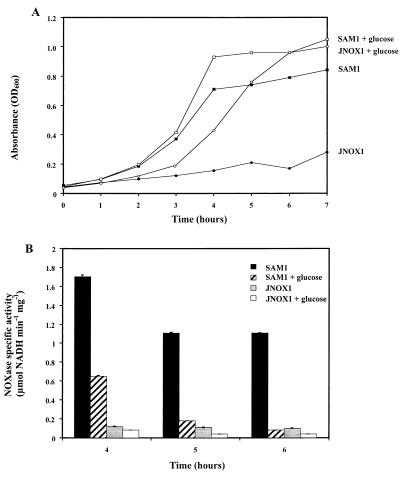
Glucose rescues the nox null mutant phenotype in C medium.
Unlike THY medium, C medium is used to optimize expression of streptococcal genes that are subject to catabolite repression because it contains a minimal concentration of glucose (31). When glucose was added to C medium broth cultures, growth of mutant JNOX1 under low-O2 conditions resembled growth in liquid THY medium, and after a 1-h lag period the bacteria grew to a density equivalent to that of the wild-type strain (Fig. 4A). Other hexose sugars, such as mannose, sucrose, and lactose, were also able to rescue the growth defect of the nox mutant in liquid C medium, as did the addition of pyruvate (data not shown). These data suggest that the concentration of glucose is responsible for the observed medium-dependent growth defects and that under carbohydrate-rich conditions, the NOXase requirement for growth is reduced. To further examine this hypothesis, levels of NOXase from wild-type SAM1 grown in the presence or absence of glucose were analyzed. Consistent with the hypothesis, the highest levels of NOXase were observed during growth in the absence of glucose. Levels of NOXase activity in the presence of glucose were only about 40% of those observed at the first time point analyzed (4 h; Fig. 4B). Furthermore, while the levels of NOXase activity declined only about 30% during culture without added glucose, over the next several hours, the relatively lower NOXase activity observed during growth with added glucose decreased dramatically (Fig. 4B) and NOXase was virtually undetectable when analyzed several hours after the cessation of logarithmic growth (6 h; Fig. 4B). As expected, significant levels of NOXase activity were not detected in the mutant under any growth condition (Fig. 4B).
FIG. 4.
Glucose affects the growth of the JNOX1 mutant and the NOXase activity of the SAM1 wild-type strain. (A) Glucose rescues the JNOX1 mutant growth defect in liquid C medium. Wild-type SAM1 and mutant JNOX1 were cultured for 20 h under low-O2 conditions (static growth) in liquid C medium. Cultures were then diluted to an optical density at 600 nm (OD600) of 0.015 in liquid C medium and grown again under low-O2 conditions in the presence or absence of 1% glucose at 37°C. The OD600 was measured every hour for 7 h. Data presented are representative of at least five independent experiments. (B) Glucose affects NOXase specific activity. At the 4-, 5-, and 6-h time points, aliquots were removed from the cultures described in the legend to panel A and assayed for NOXase specific activity. Data presented represent the mean and standard deviation of samples analyzed in triplicate and are representative of at least five independent experiments.
Analysis of nox transcript levels by semiquantitative RT-PCR (see Materials and Methods) during culture in C medium plus glucose revealed only a twofold decrease in the level of the message at a time point when NOXase activities were undetectable during culture with glucose (6 h; data not shown). This result suggested that the differences in NOXase levels were probably not the result of direct catabolite repression of the nox promoter. This hypothesis was further supported through the analysis of the JCCP1 mutant in which the gene encoding the major catabolite repressor protein of gram-positive bacteria (CcpA) (19, 27, 34, 44) was insertionally inactivated (see Materials and Methods). Expression of NOXase activity in the JCCP1 mutant strain in the presence or absence of glucose was identical to that of the wild type under all of the growth conditions tested (data not shown).
The nox mutants are sensitive to oxidative stress.
To test if the reduced growth of the JNOX1 mutant under high O2 tension is due to increased sensitivity to oxidative stress, the mutant was grown under low-O2 conditions in liquid THY medium in the presence and absence of the oxidative stress-promoting agent paraquat. This comparison revealed that under these conditions, JNOX1 growth was only 23% of that of wild-type SAM1 in the presence of paraquat (data not shown). While this stress-promoting agent is traditionally used to generate O2− stress, the high levels of O2− can rapidly be dismutated to H2O2 in the presence of functional SOD to produce high levels of intracellular H2O2. Examination of SOD activities demonstrated that the mutant was not deficient in the expression of this activity (data not shown), suggesting that either O2− or H2O2 is involved in restricting the growth of the nox mutant. Previous studies have indicated that expression of protein F, a fibronectin-binding surface protein of S. pyogenes, is stimulated by O2− stress but not by H2O2 stress (16, 48). For example, inactivation of the gene which encodes SOD generates an O2− stress that results in the activation of protein F expression under conditions in which it is not normally expressed (static culture in liquid THY medium) (16). However, a similar analysis demonstrated that expression of protein F was not altered in nox mutants (data not shown). These data suggest that some reactive O2 species other than O2− is the source of oxidative stress in the nox mutants.
Higher levels of H2O2 accumulate in the nox null mutant than in the wild type.
Other species of streptococci are known to produce H2O2 when grown in media containing low concentrations of glucose (15). To determine if the same is true of S. pyogenes and if H2O2 is a source of oxidative stress in the nox mutants, the concentration of H2O2 was measured in culture supernatants of the wild-type and mutant strains. Under low-O2 conditions in liquid C medium, neither the wild-type strain nor the mutant strains accumulated any H2O2. However, when bacteria were grown under high-O2 conditions in C medium broth (restrictive growth conditions for the nox mutant), this analysis revealed the accumulation of substantial concentrations of H2O2 in both the wild-type and mutant strains (Table 2). When normalized for cell growth, the mutant JNOX1 accumulated almost three times the level of H2O2 as the wild-type strain under the same condition (liquid C medium with agitation) (Table 2). No accumulation of H2O2 above background levels was detected when either the mutant or the wild-type cultures were supplemented with glucose.
TABLE 2.
Accumulation of H2O2a
| Strain | Plasmid | Low O2
|
High O2
|
||
|---|---|---|---|---|---|
| − Glucose | + Glucoseb | − Glucose | + Glucoseb | ||
| SAM1 | None | <0.010 | <0.010 | 0.580 | <0.010 |
| JNOX1 | None | <0.010 | <0.010 | 1.590 | <0.010 |
| JNOX1 | pNPR1 | <0.010 | <0.010 | <0.010 | <0.010 |
| JNOX1 | pLZ12 | <0.010 | <0.010 | 1.170 | <0.010 |
The amount of H2O2 accumulated in overnight broth cultures is expressed as a ratio of the millimolar of [H2O2] to the units of optical density at 600 nm.
Glucose was added at a final concentration of 1% (wt/vol).
Heterologous peroxidase can rescue the nox mutant growth defect.
The data presented above suggested that H2O2 accumulation is responsible for the growth deficiency of the nox mutants under conditions of increased O2 tension. If this is true, the addition of a heterologous peroxidase or catalase should rescue the growth defect. To further explore this hypothesis, the gene encoding a heterologous NADH peroxidase (npr) from E. faecalis was inserted onto a streptococcal plasmid and the resulting construct (pNPR1 [see Materials and Methods]) was introduced into the nox mutant JNOX1. As expected, JNOX1 containing the vector alone accumulated high levels of H2O2 in the culture supernatant [JNOX1(pLZ12-Km); Table 2] while JNOX1 containing the peroxidase did not accumulate detectable amounts of H2O2 [JNOX1(pNPR1); Table 2], suggesting that the peroxidase is functional in an S. pyogenes background. Analysis of the growth characteristics of the resulting strains revealed that JNOX1 containing the vector alone is unable to grow under high-O2 conditions in liquid C medium or on solid C medium (Table 2). In contrast, the JNOX1 mutant containing the peroxidase is able to grow under high O2 tension in C medium broth, although neither it nor wild-type SAM1 containing the peroxidase is able to grow under high-O2 conditions on agar plates (data not shown). These data were supported by additional studies in which catalase was added to the nox mutant culture medium (data not shown). In the presence of catalase, JNOX1 no longer exhibited any of the mutant phenotypes. For example, similar to the peroxidase results, cultures did not accumulate H2O2 and JNOX1 was able to grow under high-O2 conditions both in liquid medium and on solid medium (data not shown). Taken together, the catalase and peroxidase results suggest that the elimination of excess H2O2 helps alleviate the nox mutant growth defect.
DISCUSSION
Through the inactivation of S. pyogenes nox, we have shown that NOXase is essential for growth in aerobic environments, protects against oxidative stress, and contributes to growth in carbohydrate-limited environments under conditions of intermediate O2 tension. Carbohydrate limitation was associated with a dramatic increase in the production of H2O2 during growth, and the accumulation of H2O2 was enhanced in the absence of NOXase activity. Addition of catalase or introduction of a heterologous peroxidase eliminated the accumulation of H2O2 and relieved the growth defect of the nox mutant under high-O2 conditions. These studies suggest that in the absence of functional NOXase, the accumulation of additional H2O2 contributes to the growth defect.
Because NOXase regenerates two molecules of NAD+ in the reduction of O2 to H2O (42), the loss of NOXase activity likely causes an increase in the levels of NADH that accumulate under aerobic conditions. It has been reported that high levels of NADH are detrimental to cells during exposure to H2O2 (24) because NADH can reduce Fe3+ to Fe2+, which then reacts with H2O2 to form OH· through the Fenton reaction (13, 24). Therefore, one reason why the JNOX1 mutant exhibits a growth defect may be detrimental DNA damage from highly toxic OH· radicals that are generated when the mutant is cultured under aerobic conditions. Furthermore, through production of Fe2+, a high concentration of NADH in the nox mutant may also contribute to increased H2O2 levels because Fe2+ can quickly react with O2 to produce O2− (24), which is rapidly dismutated to H2O2 by SOD.
It is hypothesized that just as a high NADH/NAD+ ratio is harmful to the JNOX1 mutant, a low ratio could also be detrimental to cell growth. If this is true, it may explain why the introduction of NADH peroxidase into the JNOX1 mutant on a multicopy plasmid is not sufficient to allow growth of the mutant on solid media under high-O2 conditions. This hypothesis is further supported by the observation that wild-type SAM1, which normally grows on solid media under high-O2 conditions, is unable to do so when supplemented with the peroxidase. The peroxidase reduces H2O2 and generates one molecule of NAD+; however, since the peroxidase-encoding gene is present on a multicopy plasmid, its overexpression would produce a low NADH/NAD+ ratio, which could affect cellular metabolism and growth. Therefore, a balanced NADH/NAD+ ratio is probably crucial for proper streptococcal cellular metabolism; any gross alteration of the ratio likely effects the ability of the bacteria to grow under high-O2 conditions.
In the presence of glucose, NOXase specific activity was decreased, indicating that in this environment some other enzyme can compensate for the loss of NOXase function. One candidate is LDH, which catalyzes the conversion of pyruvate to lactic acid and regenerates one molecule of NAD+ in the process. LDH is allosterically activated by the glycolytic intermediate fructose-1,6-diphosphate (9); therefore, in the presence of excess glucose, LDH is activated. Since LDH regenerates one NAD+ molecule and NOXase regenerates two NAD+ molecules, activation of one or both enzymes will ultimately affect cellular metabolism through an increase or a decrease in the NADH/NAD+ ratio (14). Support of this comes from the observation that overexpression of NOXase in L. lactis results in a low NADH/NAD+ ratio, which in turn diverts pyruvate to other pathways instead of its conversion to lactic acid by LDH (29). As reported here, an ldh homologue lies just downstream of nox in S. pyogenes. The significance of this observation is unknown; however, it is not unusual for genes encoding enzymes with interdependent activities to lie in close proximity to one another in a genome (28).
The hypothesis that under certain environmental conditions the expression of some other enzyme allows growth of the JNOX1 mutant under otherwise inhibitory conditions is further supported by the observation that upon repeated culture of JNOX1 under high-O2 conditions on agar plates, a large-colony variant arose. In contrast to the wild type or the JNOX1 mutant, this variant does not accumulate any H2O2, suggesting that it has acquired a compensating mutation in a locus which simultaneously allows aerobic growth and eliminates the accumulation of H2O2. These data imply the existence of a separate oxidative protective response that may or may not involve specific defenses against H2O2 and other O2 intermediates.
The source of the H2O2 that accumulates under conditions of high O2 and carbohydrate limitation is unknown, but this H2O2 accumulation is most likely due to many different factors, such as an inability to destroy H2O2, the presence of H2O2-producing enzymes, or both. Other lactic acid bacteria have been reported to accumulate H2O2 in the culture medium (15, 32). This accumulation has been attributed to various H2O2-producing enzymes, including pyruvate oxidase (25, 43), H2O2-producing NADH oxidase (2, 21, 22), lactate oxidase (50), and α-glycerophosphate oxidase (39). Previous studies did not detect a pyruvate oxidase activity in S. pyogenes (50), and examination of the streptococcal genome did not reveal a pyruvate oxidase homologue. However, consistent with previous reports (50), open reading frames with significant homology to the Streptococcus iniae lactate oxidase (GenBank accession no. Y07622; 84% identical, 93% similar), the Enterococcus casseliflavus α-glycerophosphate oxidase (GenBank accession no. U57498; 66% identical, 78% similar), and an H2O2-producing NADH oxidase (see above) were discovered upon examination of the S. pyogenes genome database. The E. casseliflavus α-glycerophosphate oxidase is regulated by catabolite repression and is aerobically active (39). In some organisms NAD-independent LDHs have been shown to be aerobically active, subject to catabolite repression, and sensitive to the NADH/NAD+ ratio (10). Lactate oxidase is an H2O2-producing enzyme, and if it is regulated like the NAD-independent LDHs and is subject to catabolite repression, this could explain why the addition of glucose to carbohydrate-limited medium eliminates H2O2 accumulation. However, this could also be due to increased intracellular levels of pyruvate, which is a known scavenger of H2O2 (20). This hypothesis is supported by the observation that upon addition of pyruvate to the JNOX1 mutant cultured in carbohydrate-limited liquid medium, the mutant grew to wild-type densities.
The studies presented here demonstrate that NOXase function is necessary for S. pyogenes aerobic metabolism, as well as growth under conditions of intermediate O2 tension and carbohydrate limitation. This is the first report of a targeted mutation in the gene encoding NOXase and demonstrates a dual metabolic and protective function for this enzyme in S. pyogenes. The continued characterization of gene products required for aerobic survival should provide a better understanding of streptococcal pathogenesis at the earliest stages of infection.
ACKNOWLEDGMENTS
We thank Danny Kohl for the use of his O2 electrode and Arne Olsén for his recipe for C medium. We also thank the University of Oklahoma Genome Center for their gracious public release of genome data prior to completion of the project.
Public Health Service grants AI38273 (M.G.C.) and GM35394 (A.C.) from the National Institutes of Health supported this work. M.G.C. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Ahmed S A, Claiborne A. The streptococcal flavoprotein NADH oxidase. J Biol Chem. 1989;264:19856–19863. [PubMed] [Google Scholar]
- 2.Anders R F, Hogg D M, Jago G R. Formation of hydrogen peroxide by group N streptococci and its effect on their growth and metabolism. Appl Microbiol. 1970;19:608–612. doi: 10.1128/am.19.4.608-612.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon M G, Scott J R. Genetic manipulation of the pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 7.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 8.Claiborne A, Ross R P, Parsonage D. Flavin-linked peroxide reductases: protein-sulfenic acids and the oxidative stress response. Trends Biochem Sci. 1992;17:183–186. doi: 10.1016/0968-0004(92)90263-9. [DOI] [PubMed] [Google Scholar]
- 9.Crow V L, Pritchard G G. Fructose 1,6-diphosphate-activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J Bacteriol. 1977;131:82–91. doi: 10.1128/jb.131.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Da Cunha M V, Foster M A. Sugar-glycerol cofermentations in lactobacilli: the fate of lactate. J Bacteriol. 1992;174:1013–1019. doi: 10.1128/jb.174.3.1013-1019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolin M I. Cytochrome-independent electron transport enzymes of bacteria. In: Gunsalus I C, Stanier R Y, editors. The bacteria. III. New York, N.Y: Academic Press, Inc.; 1961. pp. 425–460. [Google Scholar]
- 12.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fordyce A M, Crow V L, Thomas T D. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl Environ Microbiol. 1984;48:332–337. doi: 10.1128/aem.48.2.332-337.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Mendoza A, Liébana J, Castillo A M, De La Higuera A, Piédrola G. Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol. 1993;39:434–439. doi: 10.1099/00222615-39-6-434. [DOI] [PubMed] [Google Scholar]
- 16.Gibson C M, Caparon M G. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J Bacteriol. 1996;178:4688–4695. doi: 10.1128/jb.178.15.4688-4695.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanski E, Caparon M G. Protein F, the fibronectin-binding protein, is an adhesin of the group A streptococcus, Streptococcus pyogenes. Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanski E, Horwitz P A, Caparon M G. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect Immun. 1992;60:5119–5125. doi: 10.1128/iai.60.12.5119-5125.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi M. Reduced nicotinamide adenine dinucleotide oxidase involvement in defense against oxygen toxicity of Streptococcus mutans. Oral Microbiol Immunol. 1992;7:309–314. doi: 10.1111/j.1399-302x.1992.tb00594.x. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol. 1993;139:2343–2351. doi: 10.1099/00221287-139-10-2343. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi M, Shimada M, Matsumoto J, Yamamoto Y, Rhaman A, Kamio Y. Molecular cloning and sequence analysis of the gene encoding the H2O2-forming NADH oxidase from Streptococcus mutans. Biosci Biotech Biochem. 1994;58:1603–1607. doi: 10.1271/bbb.58.1603. [DOI] [PubMed] [Google Scholar]
- 23.Ike Y, Craig R A, White B A, Yagi Y, Clewell D B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci USA. 1983;80:5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 25.Kodama T, Fukui K, Shimamoto T, Ohta H, Kokeguchi S, Kato K. Effects of oxygen on glucose-limited growth of Streptococcus mutans. Infect Immun. 1987;55:169–173. doi: 10.1128/iai.55.1.169-173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kushner S R. An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Boyer H W, Micosia S, editors. Genetic engineering. New York, N.Y: Elsevier/North-Holland Biomedical Press; 1978. pp. 17–23. [Google Scholar]
- 27.Küster E, Luesink E J, de Vos W M, Hillen W. Immunological crossreactivity to the catabolite control protein CcpA from Bacillus megaterium is found in many gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 28.Llanos R M, Harris C J, Hillier A J, Davidson B E. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J Bacteriol. 1993;175:2541–2551. doi: 10.1128/jb.175.9.2541-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez de Felipe F, Kleerebezem M, De Vos W M, Hugenholtz J. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol. 1998;180:3804–3808. doi: 10.1128/jb.180.15.3804-3808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucey C A, Condon S. Active role of oxygen and NADH oxidase in growth and energy metabolism of Leuconostoc. J Gen Microbiol. 1986;132:1789–1796. [Google Scholar]
- 31.Lyon L R, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine protease of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malke H, Starke R, Jacob H E, Köhler W. Bacteriocine-like activity of group A streptococci due to the production of peroxide. J Med Microbiol. 1974;7:367–374. doi: 10.1099/00222615-7-3-367. [DOI] [PubMed] [Google Scholar]
- 33.Mallett T C, Parsonage D, Claiborne A. Equilibrium analyses of the active-site asymmetry in enterococcal NADH oxidase: role of the cysteine-sulfenic acid redox center. Biochemistry. 1999;38:3000–3011. doi: 10.1021/bi9817717. [DOI] [PubMed] [Google Scholar]
- 34.Monedero V, Gosalbes M J, Pérez-Martínez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy E, Hieny S, Sher A, O'Garra A. Detection of in vivo expression of interleukin-10 using a semi-quantitative polymerase chain reaction method in Schistosoma mansoni infected mice. J Immunol Methods. 1993;162:211–223. doi: 10.1016/0022-1759(93)90386-l. [DOI] [PubMed] [Google Scholar]
- 36.Murphy M G, Condon S. Comparison of aerobic and anaerobic growth of Lactobacillus plantarum in a glucose medium. Arch Microbiol. 1984;138:49–53. [Google Scholar]
- 37.Niimura Y, Poole L B, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J Biol Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 38.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 39.Parsonage D, Luba J, Mallett T C, Claiborne A. The soluble alpha-glycerophosphate oxidase from Enterococcus casseliflavus. Sequence homology with the membrane-associated dehydrogenase and kinetic analysis of the recombinant enzyme. J Biol Chem. 1998;273:23812–23822. doi: 10.1074/jbc.273.37.23812. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross R P, Claiborne A. Cloning, sequence and overexpression of NADH peroxidase from Streptococcus faecalis 10C1. J Mol Biol. 1991;221:857–871. doi: 10.1016/0022-2836(91)80180-3. [DOI] [PubMed] [Google Scholar]
- 42.Ross R P, Claiborne A. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1. J Mol Biol. 1992;227:658–671. doi: 10.1016/0022-2836(92)90215-6. [DOI] [PubMed] [Google Scholar]
- 43.Ryan C S, Kleinberg I. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch Oral Biol. 1995;40:753–763. doi: 10.1016/0003-9969(95)00029-o. [DOI] [PubMed] [Google Scholar]
- 44.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 45.Scott J R. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology. 1972;62:344–349. doi: 10.1016/0042-6822(74)90397-3. [DOI] [PubMed] [Google Scholar]
- 46.Scott J R, Guenther P C, Malone L M, Fischetti V A. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson W A, Courtney H S, Ofek I. Interactions of fibronectin with streptococci: the role of fibronectin as a receptor for Streptococcus pyogenes. Rev Infect Dis. 1987;9:S351–S359. doi: 10.1093/clinids/9.supplement_4.s351. [DOI] [PubMed] [Google Scholar]
- 48.VanHeyningen T, Fogg G C, Yates D, Hanski E, Caparon M G. Adherence and fibronectin-binding are environmentally regulated in the group A streptococci. Mol Microbiol. 1993;9:1213–1222. doi: 10.1111/j.1365-2958.1993.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 49.Wannamaker L W. Differences between streptococcal infection of the throat and skin. N Engl J Med. 1970;282:23–30. doi: 10.1056/NEJM197001012820106. [DOI] [PubMed] [Google Scholar]
- 50.Zitzelsberger W, Götz F, Schleifer K H. Distribution of superoxide dismutases, oxidases, and NADH peroxidase in various streptococci. FEMS Microbiol Lett. 1984;21:243–246. [Google Scholar]