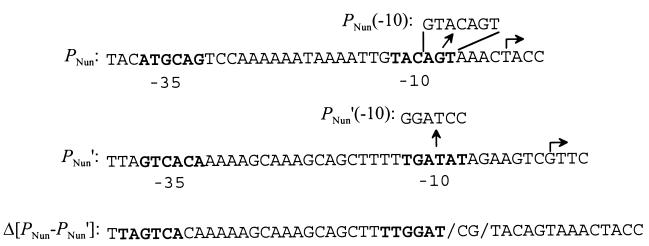Abstract
Lysogens of phage HK022 are resistant to infection by phage λ. Lambda resistance is caused by the action of the HK022 Nun protein, which prematurely terminates early λ transcripts. We report here that transcription of the nun gene initiates at a constitutive prophage promoter, PNun, located just upstream of the protein coding sequence. The 5′ end of the transcript was determined by primer extension analysis of RNA isolated from HK022 lysogens or RNA made in vitro by transcribing a template containing the promoter with purified Escherichia coli RNA polymerase. Inactivation of PNun by mutation greatly reduced Nun activity and Nun antigen in an HK022 lysogen. However, a low level of residual activity was detected, suggesting that a secondary promoter also contributes to nun expression. We found one possible secondary promoter, PNun′, just upstream of PNun. Neither promoter is likely to increase the expression of other phage genes in a lysogen because their transcripts should be terminated downstream of nun. We estimate that HK022 lysogens in stationary phase contain several hundred molecules of Nun per cell and that cells in exponential phase probably contain fewer.
Only a few prophage genes are expressed in lysogens. Some, such as the genes encoding prophage repressors, prevent the expression of genes that lead to cell death and prophage loss. Others, such as phage λ lom, bor, rexA, and rexB (2, 6), phage P22 sieA, sieB, and mnt (30, 31), phage P1 res and mod (35), phage P2 old and tin (9, 18), and prophage genes encoding virulence factors in a variety of pathogenic bacteria (see reference 32), have diverse functions. It is believed that the products of most of these genes confer a selective advantage on the lysogen, or, like repressors, prevent prophage curing and lytic phage growth. In some cases, for example λ rexA (16), the expressed genes are cotranscribed with the repressor gene. In others, for example, P2 old (9), transcription initiates at dedicated promoters. In both cases downstream transcription terminators can prevent cotranscription of other prophage genes that might harm the host or destabilize the prophage. In phages of the λ family, transcription antitermination mechanisms that function during lytic growth allow full expression of genes located downstream of the terminators only when they are needed for phage production (see reference 34).
Lysogens of temperate phage HK022 produce Nun, a protein that confers resistance to (or excludes) the related phage λ (24). Although the nun gene is located immediately downstream of pL, a major HK022 early promoter, this promoter can be inactivated by mutation without preventing nun expression (3). A second promoter, pRM, is located upstream of pL (Fig. 1). Transcripts initiating at pRM in a lysogen direct the synthesis of prophage repressor, which is encoded by the cI gene. Previous results showed that at least some pRM-directed transcripts extend through the repressed pL promoter, which suggested that nun is cotranscribed with cI (3).
FIG. 1.
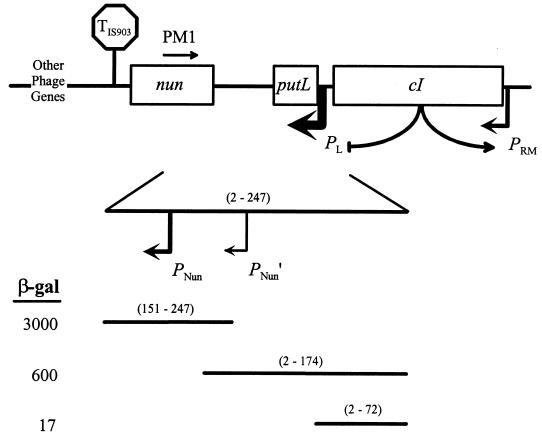
Genetic map of the nun-cI region of HK022 (not to scale). Promoters and the direction of transcription are indicated by bent arrows. TIS903 is a transcription termination site within an insertion element found in our original isolate of HK022 (21). In a lysogen, pL is repressed and pRM is activated by the product of the cI gene. The map underneath is an expanded view of the region that contains two new promoters identified in this study (PNun and PNun′). DNA fragments from positions 2 to 72, positions 2 to 174, and positions 151 to 247, relative to the HK022 pL transcription start point, were fused to a promoterless lacZ gene. The corresponding β-galactosidase (β-gal) activities (numbers at left) were measured in exponentially growing cells carrying multicopy fusions. The relative strengths of PNun, PNun′, and derepressed pL (but not pRM) are suggested by the thickness of the corresponding arrows. The binding site for oligonucleotide PM1, which was used to map the PNun and PNun′ transcription start sites, is shown.
A potential problem with this idea is that nun transcripts initiating at pRM will read through putL, a cis-acting transcription antitermination site that lies between pRM and the beginning of nun (Fig. 1). Nascent putL transcripts modify elongating RNA polymerase molecules so that they read through downstream transcription termination sites (14). Indeed, the activity of putL is required for pL-directed expression of phage genes located downstream of nun during lytic phage growth (20). Therefore, if nun were transcribed from pRM in a lysogen, antitermination could lead to transcription of these downstream genes. This is likely to be harmful because some of the resulting proteins can reduce the fitness of the lysogen or destabilize the prophage (see Discussion). Accordingly, we looked for nun promoters located downstream of putL. In this article, we report that nun is transcribed from a dedicated promoter located between putL and the coding sequence, and that transcription from pRM contributes little if anything to nun expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophages.
Bacterial strains, plasmids, and bacteriophages are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and bacteriophages
| Strain, plasmid, or bacteriophage | Relevant genotype or descriptiona | Comment and/or source or reference |
|---|---|---|
| Strains | ||
| CF881 | rna | Lacks RNase I (36) |
| JO350 | MC1000 (HK022 nun::Tn10)1 | 21 |
| LM86 | MC1000 (HK022::kan PNun′[−10])1 | This work |
| LM87 | MC1000 (HK022::kan)1 | The prophage is strain O295 (this work) |
| LM89 | MC1000 (HK022::kan Δ[PNun-PNun′])1 | This work |
| MC1000 | lacΔX74 | Host for HK022 prophages (27) |
| N5947 | lacZΔ21 (λ int2 xis1 Δ[S-X] Nam7 Nam53 cI857 ΔH1[cro-bio-uvrB]) | Used as a host for pL(λ)-lacZ fusions (22) |
| RK422 | MC1000 (HK022::kan PNun[−10])1 | This work |
| RK444 | MC1000 (λRS88 PNun-lacZ)1 | This work |
| RK445 | MC1000 (λRS88 PNun[−10]-lacZ)1 | This work |
| RW4055 | W3110 lacZΔM15 lacIq/pNUN | pNUN carries a PLac-nun fusion (1) |
| RW4072 | (argF-lac)ΔU169 recA56 (λ imm-HK022 cIts12 nun::lacZ sam7)n | |
| RW3177 | N5947 (λ imm21 pL[λ]-lacZ) (HK022 nun::Tn10) | This work |
| RW3181 | N5947 (λ imm21 pL[λ]-nutL-lacZ) (HK022 nun::Tn10) | This work |
| RW4089 | N5947 (λ imm21 pL[λ]-lacZ) (HK022::kan PNun′[−10])1 | This work |
| RW4091 | N5947 (λ imm21 pL[λ]-lacZ) (HK022::kan)1 | This work |
| RW4093 | N5947 (λ imm21 pL[λ]-lacZ) (HK022::kan Δ[PNun-PNun′])1 | This work |
| RW4095 | N5947 (λ imm21 pL[λ]-lacZ) (HK022::kan PNun[−10])1 | This work |
| RW4097 | N5947 (λ imm21 pL[λ]-nutL-lacZ) (HK022::kan PNun′[−10])1 | This work |
| RW4099 | N5947 (λ imm21 pL[λ]-nutL-lacZ) (HK022::kan)1 | This work |
| RW4101 | N5947 (λ imm21 pL[λ]-nutL-lacZ) (HK022::kan Δ[PNun-PNun′])1 | This work |
| RW4103 | N5947 (λ imm21 pL[λ]-nutL-lacZ) (HK022::kan PNun[−10])1 | This work |
| SK38 | pK1 in XL1 | 3 |
| SK37 | pJO9.18 in XL1 | 3 |
| SK11 | CF881 (HK022) | 3 |
| XL1 | recA | Host for plasmids (Stratagene) |
| Plasmids | ||
| pJO9.18 | pEMBL-18 derivative that carries the pR-cI-pL-nun segment of HK022 DNA | 21; see reference 7 for a description of pEMBL-18 |
| pK1 | pJO9.18 with a mutation inactivating pL | 3 |
| pLM11 | pJO9.18 PNun′(−10) | This work |
| pLM12 | pJO9.18 Δ(PNun-PNun′) | This work |
| pMOC192 | pRS415 containing HK022 sequence from positions +2 to +174 relative to the start site of pL transcription fused to lacZ | M. Clerget, personal communication |
| pMOC194 | pRS415 containing HK022 sequence from positions +2 to +72 relative to the start site of pL transcription fused to lacZ | M. Clerget, personal communication |
| pNGC25 | pGB2 containing the HK022 cI and nun genes | J. Little, personal communication |
| pRK395 | pRS415 with a PNun-lacZ fusion | This work |
| pRK412 | pJO9.18 with the PNun(−10) mutation | This work |
| pRK438 | pRS415 with a PNun(−10)-lacZ fusion | This work |
| pRS415 | Promoterless lacZ expression vector | 28 |
| Bacteriophages | ||
| O295 | HK022 cIts12 with a kanamycin resistance determinant inserted in the pL operon | 3 |
| RS88 | λ imm434 cIind− | Parent of single-copy PNun-lacZ fusions (28) |
| W30 | λ b2 cI− | NIH collectionb |
Subscripts indicate the number of tandem prophages, as follows: 1, single lysogen; and n, polylysogen. pL(λ)-lacZ (fusion HA25) and pL(λ)-nutL-lacZ (fusion HA22) are located in the b2 region of a modified λ imm21 prophage (24). Fusion HA25, which has 22 bp less of the λ pL operon than does fusion HA22, lacks the boxB element of nutL.
NIH, National Institutes of Health.
Phage and bacterial growth.
Bacteria were grown in tryptone or Luria-Bertani (LB) broth and supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) where appropriate. HK022 was grown and assayed as described in reference 21.
Cloning of fragments with promoter activity.
Oligonucleotides were purchased from BioServe Biotechnologies (Laurel, Md.). Plasmid DNAs were isolated by using a Qiagen spin miniprep kit. Restriction enzymes and ligase were purchased from New England Biolabs (NEB). A 97-bp DNA fragment that immediately precedes the nun gene and contains the PNun promoter was amplified from plasmid pNGC25 by PCR with primers RK99 (5′-GGCGGATCCAATAAGCACCGTACGG-3′) and RK100 (5′-CAGCGAATTCAAGTATTTATTGCAAAGATTC-3′). The amplified DNA was digested with EcoRI and BamHI (underlined sequences) and cloned into the promoterless lacZ vector pRS415.
Primer extension.
Total RNA was isolated with an RNA isolation kit (Totally RNA; Ambion) from log-phase cultures of the following strains: SK37, SK38, and SK11 (Table 1). Strains SK37 and SK38 carry plasmids with inserts of HK022 DNA, and strain SK11 is an HK022 lysogen. Seven milliliters of culture was added directly to 10 ml of lysing buffer and then processed according to the Ambion protocol. RNA was suspended in diethyl pyrocarbonate-treated water supplemented with 1 mM EDTA. The purity and integrity of the RNA was analyzed on a 1.2% agarose gel, followed by staining with ethidium bromide. Primer extension reactions (20-μl volumes) were performed by using the Promega reverse transcription system. Oligonucleotide PM1 (5′-CTCGAGATGTAAGACCTC-3′) is complementary to the nun mRNA and hybridizes approximately 90 bp downstream of the PNun start site. PM1 was 5′ end-labeled in a 30-μl reaction mixture containing 50 μCi of [γ-32P]ATP (Redivue [3,000 Ci/mmol]; Amersham) 10 U of T4 polynucleotide kinase (NEB) and 1× kinase buffer (NEB). Reaction mixtures were incubated for 30 min at 37°C. Primer extension reaction mixtures were incubated at 42°C for 1 h, and the reactions were stopped by adding 10 μl of Stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.02% xylene cyanol). Reaction mixtures were heated at 94°C for 3 min, chilled immediately on ice, and then loaded onto an 8% denaturing sequencing gel. Sequencing reactions were performed with the Perkin-Elmer Amplicycle sequencing kit. Sequence was obtained directly from purified pK1 plasmid and PCR-amplified template from wild-type PNun and PNun(−10) mutant clones by using 5′ end-labeled oligonucleotide PM1 as the primer.
In vitro transcription.
Multiple round in vitro transcription reactions were performed on templates amplified from PNun+ and PNun(−10) mutant clones. Reaction mixtures consisted of approximately 50 nM PCR template, 100 nM RNA polymerase holoenzyme (Epicentre, Madison, Wis.), 1 mM concentrations of each nucleoside triphosphate (NTP), 20 mM Tris-glutamate, 50 mM K-glutamate, 10 mM Mg-glutamate, and 0.001 μg of bovine serum albumin per ml. The reaction mixtures were incubated for approximately 1 h at 37°C. After ethanol precipitation and washing, the pelleted material was suspended in diethyl pyrocarbonate-treated water supplemented with 1 mM EDTA. The RNA products were then used directly in a primer extension reaction.
PNun and PNun′ mutations.
To inactivate the PNun promoter, plasmid pJO9.18 was digested with BsrGI, and the overhangs were filled in with Klenow fragment. The resulting blunt-ended plasmid was ligated and electroporated into MC1000 cells. Transformants were screened by PCR with primers PM1 and RK100, and the products were tested for their susceptibility to BsrGI digestion. The PNun′ promoter was inactivated by replacing the sequence in the predicted −10 hexamer with a BamHI restriction site. The replacement was accomplished by overlap extension PCR with primers PM2 (5′-GCGGATAACAATTTCACACAGG-3′), PM3 (5′-CTTCTATTTTTTCGAACGACTTCTGGATCCAAAAGCTGCTTTGCTTTTTGTGAC-3′), PM4 (5′-CACAAAAAGCAAAGCAGCTTTTTGGATCCAGAAGTCGTTCGAAAAAATAG-3′), and PM6 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′). The BamHI site that replaces the predicted −10 sequence is underlined in the relevant oligonucleotide sequences. Amplifications were performed with Vent polymerase with pJO9.18 used as the template. The final amplified product was digested with HindIII and EcoRI and ligated into pJO9.18. The desired clones were identified by restriction analysis of purified plasmid DNA. The mutation was transferred onto HK022 by recombination, and the presence of the BamHI site was confirmed by PCR amplification with primers PM1 and PM5 (5′-CCTCATTAGGCAGTCAATCG-3′) followed by digestion of the products with BamHI. Inactivation of both the PNun′ and PNun promoters was accomplished by digesting the PNun′ mutant construct with BamHI and BsrGI. The DNA ends were filled in by Klenow fragment, ligated, and electroporated into MC1000 cells. The desired deletion mutant was identified by analyzing the size of DNA amplified by PCR with primers PM1 and PM5.
Crossing PNun and PNun′ mutations onto HK022 phage.
Cells carrying pJO9.18 or various mutant derivatives were infected with HK022::Tn10(kan) cIts12 (phage O295), and the resulting lysate was used to infect MC1000 cells. This plasmid carries the wild-type HK022 cI gene but, for unknown reasons, does not prevent HK022 growth (21). The Tn10(kan) insertion is in the pL operon, downstream from nun. Phage recombinants carrying the plasmid cI+ gene were selected by infection of a sensitive host and isolation of Kmr colonies at 42°C, the nonpermissive temperature for cIts12. The presence of the mutation on the prophage was confirmed by the size of PCR-amplified products and the ability to digest the products with the enzymes BsrGI or BamHI. Lysates of recombinant phage were made by induction of the lysogens with UV light. These lysates were used to lysogenize cells containing λpL-nutL-lacZ fusions (24).
Crossing lacZ fusions onto λRS88.
Promoter fusions in the lacZ reporter vector, pRS415, were transferred onto λRS88 by recombination as described previously (14, 28). Lambda prophage copy number was determined as described previously (23).
Beta-galactosidase activities.
Strains containing thermosensitive λ repressor and a pL(λ)-lacZ fusion were grown overnight at 32°C in tryptone broth. The overnight cultures were diluted 1:50 in LB broth, grown to approximately 1 × 108 to 2 × 108 cells/ml, and shifted to 42°C for 60 min to allow derepression of the λ pL promoter. Beta-galactosidase activity was measured as described previously (17). It was important to use toluene to permeabilize the cells because β-galactosidase produced by these fusions was destabilized by the alternative treatment with chloroform and detergent. Cells containing single-copy lacZ fusions were grown overnight at 37°C in tryptone broth. The overnight cultures were diluted 1:1,000 in LB broth and grown at 37°C to an optical density at 650 nm of approximately 0.5 × 108 to 1 × 108 cells/ml. Thereafter, aliquots were removed and placed on ice at various intervals and the β-galactosidase activities were determined as described above.
Determination of HK022 prophage copy number.
The copy number of HK022 prophages was determined essentially as described in reference 23 by using primers specific for the HK022 int and attP and the host attB sites, as follows: RK111 (int oligonucleotide [5′-AGCAATGCAGGGAGGCCAGC-3′]), RK112 (attP oligonucleotide [5′-AATAATCTTGCGGTAGCCAGAC-3′]), and RK113 (attB oligonucleotide [5′-GGAGATCTCCTGCTCCTGTTG-3′]). Single-copy lysogens were identified by the amplification of an approximately 747-bp DNA generated from the attB and int primers. Polylysogens produced the 747-bp fragment in addition to an approximately 613-bp fragment.
Immunoblots (10).
Cells were grown in LB broth at 37°C, chilled, concentrated, diluted into loading buffer, boiled for 5 min, and frozen at −70°C. Aliquots were loaded directly onto sodium dodecyl sulfate Tris-tricine–16.5% acrylamide gels (Bio-Rad), electrophoresed, and electroblotted to Immobilon polyvinylidene difluoride membranes. We found that Nun comigrated with prestained marker proteins (Bio-Rad) in the 14- to 18-kDa range. The membranes were blocked with 5% milk powder and allowed to react overnight with antiserum against partially purified Nun raised in rabbits (antiserum kindly supplied to us by David Friedman [11]). We treated the antiserum before use with an acetone powder prepared from strain MC1000 to remove antibodies to bacterial proteins (10). It was diluted 1:50 into a solution of 5% milk powder for use. Bands were visualized by treating the membranes with peroxidase-coupled anti-rabbit serum for 1 h, allowing them to react with ECL Western blotting detecting reagent (Amersham), and placing them in contact with X-ray film. To quantitatively estimate the amount of Nun, we mixed serial dilutions of purified Nun (a gift of Randy Watnick) with extracts of strain MC1000 and fractionated the mixtures in separate lanes of a gel (see Fig. 3).
FIG. 3.
Sequence of predicted promoters and promoter mutants. The bent arrows indicate the observed 5′ ends of PNun and PNun′ transcripts (top and middle lines, respectively), as indicated by primer extension analysis (Fig. 2). The predicted −10 and −35 hexamers for these promoters are shown in bold type. The E. coli ς70 consensus hexamers are TATAAT and TTGACA, respectively. The straight arrows indicate the sequence changes in the PNun(−10) and PNun′(−10) mutants (see Materials and Methods). The sequence Δ[PNun-PNun′] is shown in the third line (see Materials and Methods). The slashes indicate the fusion points between PNun′(−10) sequence (left) and PNun sequence (right). Possible alternative −10 and −35 hexamers are shown in bold type.
RESULTS
Identification of promoters located between putL and nun.
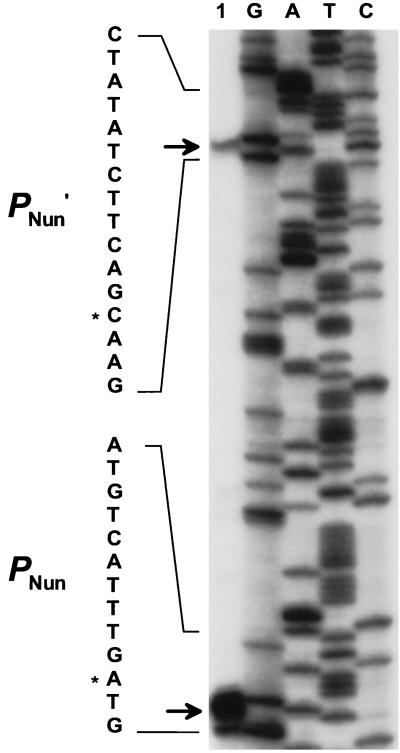
We have discovered two promoters located between putL and nun. These were found by extending an oligonucleotide primer that was hybridized to RNA isolated from cells that carry plasmid clones of HK022 DNA. Extension of the primer, which is complementary to the beginning of the nun coding sequence, produced a strong signal corresponding to a 5′ RNA end close to the start of the nun coding region and a weak signal corresponding to a 5′ RNA end further upstream, but still downstream of putL (Fig. 2). We also saw the strong primer extension signal with RNA isolated from an HK022 lysogen, although the amount of product was much less in this case (data not shown) (we did not see the weak signal, as would be expected if its intensity were correspondingly reduced). We call the putative promoter corresponding to the strong downstream signal PNun, and we called the putative promoter corresponding to the weak upstream signal PNun′.
FIG. 2.
Mapping of the transcription start sites of the PNun and PNun′ promoters. Oligonucleotide PM1, complementary to the nun mRNA, was used for reverse transcriptase primer extension and sequencing. The DNA sequence of the promoter −10 regions is shown on the left. The arrows mark the position of the extension products (lane 1). The predominant initiation sites are marked by asterisks beside the DNA sequence (lanes G, A, T, and C).
Short DNA segments containing either of these putative promoters had promoter activity when fused to a promoterless lacZ reporter gene in a multicopy plasmid (Fig. 1). A 97-bp DNA fragment containing the presumed start point of PNun-directed transcription accumulated about 3,000 U of β-galactosidase during exponential growth of a culture carrying the plasmid-borne fusion, suggesting that this interval contains a functional promoter (plasmid pRK395, the segment from positions +151 to +247 relative to the pL start). A 173-bp DNA fragment containing the presumed start point of PNun′-directed transcription accumulated about 600 U of β-galactosidase under the same conditions (plasmid pMOC192, the segment from positions +2 to +174 relative to the pL start). A similar plasmid clone lacking the PNun′ start accumulated about 17 U of β-galactosidase (pMOC194, the segment from positions +2 to +72 relative to the pL start), suggesting that the segment from positions +73 to +174 contains at least part of a promoter.
A computer-assisted search (19) identified a candidate promoter in the PNun region and a second one in the PNun′ region. The sequences, predicted −10 and −35 hexamers, and observed start sites are shown in Fig. 3. Although the match to E. coli consensus ς70 promoters is poor, the location of the predicted promoters is completely consistent with the functional analysis described in the preceding paragraph.
PNun and PNun′ mutations.
To see if these promoters contribute to Nun production in an HK022 lysogen, we mutated them individually or together and measured the activity and intracellular concentration of Nun. In mutant PNun(−10), the predicted −10 hexamer of PNun was changed, in mutant PNun′(−10), the predicted −10 hexamer of PNun′ was changed, and in mutant Δ[PNun-PNun′], the DNA segment extending from the (altered) −10 hexamer of PNun′(−10) to the −10 hexamer of PNun was deleted (Fig. 3). We showed that PNun(−10) had substantially reduced promoter activity: the steady-state level of β-galactosidase activity in cells carrying a single copy of a PNun(−10)-lacZ fusion was 1 to 3% of that of a comparable PNun-lacZ fusion (strains RK445 and RK444, respectively) (data not shown). The PNun′(−10) and Δ[PNun-PNun′] mutations were not checked in this way, and we have no experimental evidence that they inactivate the respective promoters. Indeed, inspection of the sequence revealed Δ[PNun-PNun′] could have created a new promoter (Fig. 3). The three mutations were crossed from the plasmids in which they were constructed onto HK022 as described in Materials and Methods, and single lysogens of the mutant phages were isolated. The Nun phenotype of these lysogens was checked in several ways.
Plaque formation by λ on lawns of HK022 lysogens (Table 2).
TABLE 2.
Phenotypes of PNun and PNun′ mutations in single-copy HK022 lysogensa
| Phenotype
|
λ Plaque formation (EOP)b | β-Galactosidase activity
|
|||
|---|---|---|---|---|---|
| PNun | PNun′ | nun | pL(λ)-nutL-lacZc | pL(λ)-lacZd | |
| + | + | + | <10−8 | 20 | 1,349 |
| + | (−10) | + | <10−8 | 37 | 1,690 |
| (−10) | + | + | ∼10−2e | 2,069 | 1,445 |
| Δ | Δ | + | ∼10−3e | 408 | 1,543 |
| + | + | Tn10 | 1.0 | 1,735 | 1,281 |
The nun::Tn10 insertion mutation and the two pL(λ)-lacZ fusions have been described previously (21, 24). The phage used for measuring the efficiency of plating (EOP) was λ b2 c (strain W30), and efficiencies are relative to that on lawns of MC1000, the nonlysogenic host. Beta-galactosidase activity was measured 1 h after thermal derepression of the λ pL promoter, and values are averages obtained from at least two independent determinations. The standard error of the mean was ≤14%.
The bacterial strains used were (from top to bottom) LM87, LM86, RK422, LM89, and JO350.
The bacterial strains used were (from top to bottom) RW4099, RW4097, RW4103, RW4101, and RW3181.
The bacterial strains used were (from top to bottom) RW4091, RW4089, RW4095, RW4093, and RW3177.
There was a background of many tiny plaques, which were not included in the estimate of plating efficiency.
A wild-type HK022 prophage reduced the number of plaques formed by superinfecting λ by more than 8 orders of magnitude. No λ mutants able to form plaques were found, probably because they cannot arise in a single step (25). However, the HK022 PNun(−10) mutant prophage excluded λ much less efficiently than did its parent. We observed a large number of tiny, uncountable plaques at a frequency of ≥10−2 plaques per added λ phage. These are probably formed by limited growth of wild-type λ on this host. We also found plaques of normal or near-normal size at a frequency of about 10−2 plaques per added phage. Most or all of these were formed by mutants that probably arose during limited λ growth on the lawn (see below). The residual λ exclusion by the PNun(−10) lysogen is clearly due to Nun activity, since a lysogen of a nun::Tn10 insertion mutant plated λ with unit efficiency, and the plaques were of near-normal size. The phenotype of a Δ[PNun-PNun′] lysogen was similar to that of the PNun single mutant, although, surprisingly, the frequency of normal-sized (i.e., mutant; see below) λ plaques was somewhat lower for the former (ca. 10−3 plaques per added phage). These results suggest that PNun makes a major but not the only contribution to nun transcription in a lysogen. The quantitative difference in λ exclusion between PNun(−10) and Δ[PNun-PNun′] can be accounted for by assuming that the deletion mutation creates a new promoter that is weaker than PNun but stronger than PNun(−10) (see above). The PNun′(−10) lysogen excluded λ as efficiently as did its wild-type parent, but this result cannot be unambiguously interpreted because we have no experimental evidence that the mutation inactivated the promoter.
Most or all of the normal-sized λ plaques that arose on lawns of HK022 PNun(−10) and Δ[PNun-PNun′] lysogens were formed by mutants. Phage from two such plaques recovered from lawns of each lysogen were subjected to several cycles of single-plaque purification on lawns of a nonlysogenic host and then amplified on the same host. We found that each of these phage lines formed plaques with unit efficiency on lawns of both the PNun(−10) and the Δ[PNun-PNun′] lysogens. However, they did not form plaques on wild-type or PNun′(−10) lysogens (efficiency of plating, ≤10−6), and they have not been further characterized.
Effect of promoter mutations on the expression of a pL(λ)-nutL-lacZ fusion.
Nun blocks λ growth by binding to the boxB element of nascent nut transcripts and prematurely terminating early λ transcription (5, 24) (see Discussion). Thus, measurement of β-galactosidase production in a cell containing a nut site fused to a reporter gene gives a direct and quantitative estimate of Nun activity. As shown in Table 2, the wild-type Nun concentration greatly reduced β-galactosidase accumulation by the pL(λ)-nutL-lacZ fusion but had no effect on a similar fusion that lacks a complete boxB. This agrees with previously published results (24). The PNun(−10) mutation restored a high level of β-galactosidase activity, similar to that observed in a nun insertion mutant. These results confirm that PNun makes a major contribution to nun expression. In contrast to measurement of phage exclusion, we saw no evidence of any residual Nun activity in this mutant. We propose an explanation for this difference in the Discussion. The Δ[PNun-PNun′] mutation gave an intermediate level of lacZ expression compared to the wild-type and the PNun mutant lysogens, a result that is qualitatively consistent with its intermediate effect on λ exclusion. The PNun′(−10) mutation caused a small but reproducible increase in the expression of the nutL-lacZ fusion.
Effect of promoter mutations on the level of Nun antigen.
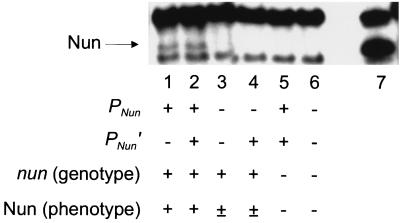
We measured the effects of our promoter mutants on the intracellular concentration of Nun protein in HK022 lysogens by immunoblots. We found that the PNun(−10) and the Δ[PNun-PNun′] mutations reduced Nun protein concentrations to undetectable levels (Fig. 4). We estimate that we would have been able to see as little as 25% of the level of Nun found in a wild-type lysogen. By contrast, the PNun′(−10) mutation had no detectable effect on Nun level. The simplest conclusion from these results is that PNun is the major promoter directing nun transcription in a lysogen. We consider the source of the residual phage exclusion activity by the PNun(−10) prophage in the Discussion.
FIG. 4.
Detection of Nun in HK022 lysogens by immunoblotting. Cell extracts of strains lysogenic for wild-type or mutant HK022 prophages, of a nonlysogen, or of a strain carrying an induced PLac-nun fusion (as indicated below the gel image) were fractionated by electrophoresis in a denaturing gel. The Nun phenotype refers to the λ exclusion data of Table 1. Nun was detected with antibody as described in Materials and Methods. The position of Nun is indicated by the arrow. Its mobility relative to protein standards is consistent with a previous report (21), and the identification is confirmed by the large amount of protein with identical mobility extracted from a Nun-overproducing strain (lane 7). Lanes 1 to 6 each contained 0.02 to 0.025 mg of protein, and lane 7 contained about half that amount. Lane 1, PNun′(−10) (strain LM86); lane 2, PNun+ (strain LM87); lane 3, Δ[PNun-PNun′] (strain LM89); lane 4, PNun(−10) (strain RK422); lane 5, nun::Tn10 (strain JO350); lane 6, strain MC1000 (the nonlysogenic parent); lane 7, PLac-nun (strain RW4055). The first six strains were grown to 1 × 109 to 2 × 109 cells per ml and the seventh strain was grown to 4 × 108 to 6 × 108 cells per ml in LB broth at 37°C before extraction. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to the culture of RW4055, which contains a plasmid with a PLac-nun fusion, 50 min before the cells were harvested. The production of Nun by this strain was much reduced if IPTG was omitted (data not shown).
Further analysis of PNun.
We estimated the amount of Nun per cell by comparing immunoblots of lysogens to those of known quantities of purified Nun protein (Fig. 3). There are 120 to 360 molecules of Nun per cell in a culture grown to 1 × 109 to 2 × 109 cells/ml in LB broth. This number is roughly consistent with the activity of β-galactosidase per cell in comparable cultures of an HK022 lysogen containing a nun::lacZ translational fusion (strain RW4072) (the calculation assumes that the fusion protein is as active as native β-galactosidase [26]). The specific activity in this strain and another strain with a PNun-lacZ transcriptional fusion (RK444) increased from three- to sixfold as the cultures proceeded from mid-log phase (0.5 × 108 to 1.5 × 108 cells/ml) into stationary phase (1 × 109 to 2 × 109 cells/ml). The mechanism and biological significance of this increase are unknown.
To determine if any factors are required for PNun activity, we analyzed transcripts generated from PNun and PNun(−10) templates using purified E. coli RNA polymerase holoenzyme (E-ς70). In a multiround transcription reaction that included linear wild-type template and radioactive substrate, a runoff transcript of the predicted size was observed (data not shown). The corresponding transcript from the PNun(−10) mutant template was not detected. We were also unable to see a transcript in similar reactions with a template that contained PNun′. This result is consistent with our primer extension experiments and with results from lacZ fusions that show that PNun′ is less active than PNun. Finally, we determined that purified E-ς70 utilizes the same transcription start site for PNun in vitro as it does in vivo by performing primer extension analysis on RNA generated in multiround transcription reactions (data not shown). These results suggest that no additional factor besides E. coli polymerase containing the ς70 subunit is absolutely required for transcription from PNun.
DISCUSSION
Our experiments show that PNun is the major promoter for expression of the nun gene in an HK022 lysogen. This promoter thus joins a fairly short list of identified promoters that are active in repressed prophages. We estimate that there are several hundred molecules of Nun per cell in early-stationary-phase cells, and perhaps one-sixth to one-third that number in exponentially growing cultures. The mechanism and biological significance of this growth phase regulation are unknown. Since Nun is stable in vivo (21), its low level probably reflects, at least in part, the weakness of PNun. Measurements of the activity of comparable promoter-lacZ fusions suggest that PNun is about 1% as strong as fully derepressed HK022 pL (R. A. King and R. A. Weisberg, unpublished results). This result is qualitatively consistent with the relative strengths of the two promoters in vitro. The ς70 consensus is not well conserved in PNun, and the transcript is unusual in that it starts with a U residue. These factors may contribute to the low activity of this promoter.
Nun acts by binding stoichiometrically to a stem-loop formed by the transcript of the boxB element in the nut sites of the nascent λ pL and pR transcripts (5). It then interacts with and arrests elongating RNA polymerase (12, 13, 33). The arrested transcription elongation complexes are stable in vitro, but the arrested polymerase probably disassociates from the template in vivo (24, 25, 29). The efficiency of Nun action is remarkable: it completely blocks λ lytic growth and cell killing and reduces to about 1% the expression of reporter genes that are fused to λ pL or pR and preceded by either nutL or nutR, respectively. The reduction persists for more than 1 h after derepression (24) (Table 1). λ pL directs the synthesis of 10 to 20 transcripts/min in vivo (15), and the strength of λ pR is likely to be similar. It is unlikely that a few hundred molecules of Nun could arrest such a large number of transcription elongation complexes if Nun were consumed during the reaction. Therefore, we suggest that Nun bound to arrested elongation complexes is released in active form after termination and recycles to newly synthesized transcripts where it acts again.
RNA polymerase molecules that initiate at PNun do not transcribe the putL antitermination site (Fig. 1) and should therefore efficiently terminate at downstream transcription terminators. One of these terminators is within the IS903 insertion element located immediately downstream of nun (8, 21). Termination of nun transcripts is probably advantageous to HK022 and its lysogens because the prophage kil, cIII, xis, and int genes lie downstream of transcription terminators in the pL operon, and the products of these genes are likely to destabilize the prophage or harm the lysogen. As expected, nun can also be expressed from the pL promoter: we found that more than 10 times the amount of β-galactosidase was produced by a pL-PNun-nun::lacZ gene fusion in the absence than in the presence of repressor (unpublished results). However, lytic growth of HK022 does not require Nun (21).
Mutation of PNun depressed but did not completely prevent Nun production. Nun was undetectable by immunoblot (≤25% of wild type) and did not reduce expression of a pL(λ)-nutL-lacZ fusion in a PNun(−10) lysogen. Nevertheless, it did reduce growth of superinfecting λ (Table 2). Lambda growth could be more severely affected by low Nun levels than is expression of a reporter gene fused to nutL because the phage also has a nutR site, and this could amplify its sensitivity. The source of the residual Nun in the PNun mutant is unknown, but several possibilities are open. First, PNun(−10) retains 1 to 2% of PNun+ promoter activity, as judged by expression of a reporter gene fused to the promoter, and this could be enough to impede the growth of superinfecting λ. Second, PNun′ could be a source of nun transcripts. We have no independent evidence that the PNun′(−10) mutation inactivates PNun′, and we were surprised to find that the Δ[PNun-PNun′] mutant retained considerable Nun activity. An inspection of the fused sequence suggests that this deletion creates a new promoter. Third, pL could contribute to residual nun transcription if repression is incomplete. Finally, pRM could contribute to residual nun transcription.
With respect to the last possibility, Cam et al. (3) detected low-intensity transcription initiating upstream of pL and extending into putL in HK022 lysogens. They also found that a high level of HK022 repressor supplied in trans to a prophage by a plasmid reduced both Nun activity and the expression of a nun::lacZ gene fusion. Cam et al. (3) suggested that transcripts originating at pRM are the principal source of nun expression in a lysogen and that excess repressor reduces nun expression by repressing pRM. An alternative explanation of these results is that PNun responds to repressor. Although inspection of the DNA sequence revealed no obvious HK022 operator site in the vicinity of PNun, high repressor concentrations might relax the specificity of binding. Indeed, Carlson and Little (4) demonstrated binding of repressor to sequences adjacent to an isolated operator under such conditions.
Bacteriophage HK022 resembles other phages of the λ family with respect to its genetic organization. However, in striking contrast to λ, the first gene in the HK022 pL operon is expressed in the presence of the prophage repressor. Our identification of a dedicated, constitutive promoter for nun explains in a simple way the expression of the phage exclusion function of a repressed HK022 prophage. The advantage that Nun expression confers on HK022 is less clear. Nun excludes phages with nut sites of λ specificity; the growth of several other phages, including some with related nut sites, is unaffected (15). Perhaps phage strains with nut sites of λ specificity are important natural competitors of HK022. Alternatively, perhaps Nun has functions that are yet undiscovered.
ACKNOWLEDGMENTS
We are grateful to Orna Resnekov for her assistance with the immunoblots and to Orna Resnekov and Max Gottesman for a critical reading of the manuscript. David Friedman provided Nun antiserum and Randy Watnick provided purified Nun.
REFERENCES
- 1.Baron J, Weisberg R A. Mutations of the phage λ nutL region that prevent the action of Nun, a site-specific transcription termination factor. J Bacteriol. 1992;174:1983–1989. doi: 10.1128/jb.174.6.1983-1989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barondess J J, Beckwith J. bor gene of phage λ, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J Bacteriol. 1995;177:1247–1253. doi: 10.1128/jb.177.5.1247-1253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cam K, Oberto J, Weisberg R A. The early promoters of bacteriophage HK022: contrasts and similarities to other lambdoid phages. J Bacteriol. 1991;173:734–740. doi: 10.1128/jb.173.2.734-740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson N G, Little J W. Highly cooperative DNA binding by the coliphage HK022 repressor. J Mol Biol. 1993;230:1108–1130. doi: 10.1006/jmbi.1993.1229. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay S, Hung S C, Stuart A C, Palmer III A G, Garcia-Mena J, Das A, Gottesman M E. Interaction between the phage HK022 Nun protein and the nut RNA of phage lambda. Proc Natl Acad Sci USA. 1995;92:12131–12135. doi: 10.1073/pnas.92.26.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Court D, Oppenheim A B. Phage lambda's accessory genes. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 251–277. [Google Scholar]
- 7.Dente L, Cesareni G, Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983;11:1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grindley N D, Joyce C M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor Symp Quant Biol. 1981;45:125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Haggard-Ljungquist E, Barreiro V, Calendar R, Kurnit D M, Cheng H. The P2 phage old gene: sequence, transcription and translational control. Gene. 1989;85:25–33. doi: 10.1016/0378-1119(89)90460-5. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 11.Henthorn K S, Friedman D I. Identification of functional regions of the Nun transcription termination protein of phage HK022 and the N antitermination protein of phage lambda using hybrid nun-N genes. J Mol Biol. 1996;257:9–20. doi: 10.1006/jmbi.1996.0142. [DOI] [PubMed] [Google Scholar]
- 12.Hung S C, Gottesman M E. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995;247:428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- 13.Hung S C, Gottesman M E. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 1997;11:2670–2678. doi: 10.1101/gad.11.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King R A, Banik-Maiti S, Jin D J, Weisberg R A. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 15.Liang S, Bipatnath M, Xu Y, Chen S, Dennis P, Ehrenberg M, Bremer H. Activities of constitutive promoters in Escherichia coli. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 16.Matz K, Schmandt M, Gussin G N. The rex gene of bacteriophage lambda is really two genes. Genetics. 1982;102:319–327. doi: 10.1093/genetics/102.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Mosig G, Yu S, Myung H, Haggard-Ljungquist E, Davenport L, Carlson K, Calendar R. A novel mechanism of virus-virus interactions: bacteriophage P2 Tin protein inhibits phage T4 DNA synthesis by poisoning the T4 single-stranded DNA binding protein, gp32. Virology. 1997;230:72–81. doi: 10.1006/viro.1997.8464. [DOI] [PubMed] [Google Scholar]
- 19.Mulligan M E, McClure W R. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acids Res. 1986;14:109–126. doi: 10.1093/nar/14.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberto J, Clerget M, Ditto M, Cam K, Weisberg R A. Antitermination of early transcription in phage HK022. Absence of a phage-encoded antitermination factor. J Mol Biol. 1993;229:368–381. doi: 10.1006/jmbi.1993.1040. [DOI] [PubMed] [Google Scholar]
- 21.Oberto J, Weisberg R A, Gottesman M E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989;207:675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- 22.Oppenheim A B, Gottesman S, Gottesman M E. Regulation of bacteriophage lambda int gene expression. J Mol Biol. 1982;158:327–346. doi: 10.1016/0022-2836(82)90201-7. [DOI] [PubMed] [Google Scholar]
- 23.Powell B S, Rivas M P, Court D L, Nakamura Y, Turnbough C L., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. . (Erratum, 23:1278, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert J, Sloan S B, Weisberg R A, Gottesman M E, Robledo R, Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987;51:483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- 25.Robledo R, Gottesman M E, Weisberg R A. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990;212:635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- 26.Rotman B. Partial loss of activity of individual molecules of aged β-galactosidase. In: Beckwith J R, Zipser D, editors. The lactose operon. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1970. pp. 279–289. [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 28.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 29.Sloan S B, Weisberg R A. Use of a gene encoding a suppressor tRNA as a reporter of transcription: analyzing the action of the Nun protein of bacteriophage HK022. Proc Natl Acad Sci USA. 1993;90:9842–9846. doi: 10.1073/pnas.90.21.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susskind M M, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Susskind M M, Youderian P. Bacteriophage P22 antirepressor and its control. In: Hendrix R W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 347–364. [Google Scholar]
- 32.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 33.Watnick R S, Gottesman M E. Escherichia coli NusA is required for efficient RNA binding by phage HK022 nun protein. Proc Natl Acad Sci USA. 1998;95:1546–1551. doi: 10.1073/pnas.95.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisberg R A, Gottesman M E. Processive antitermination. J Bacteriol. 1999;181:359–367. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarmolinsky M B, Sternberg N. Bacteriophage P1. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 291–437. [Google Scholar]
- 36.Zhu L, Gangopadhyay T, Padmanabha K P, Deutscher M P. Escherichia coli rna gene encoding RNase I: cloning, overexpression, subcellular distribution of the enzyme, and use of an rna deletion to identify additional RNases. J Bacteriol. 1990;172:3146–3151. doi: 10.1128/jb.172.6.3146-3151.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]