Abstract
Double mutants of Escherichia coli dam (DNA adenine methyltransferase) strains with ruvA, ruvB, or ruvC could not be constructed, whereas dam derivatives with recD, recF, recJ, and recR were viable. The ruv gene products are required for Holliday junction translocation and resolution of recombination intermediates. A dam recG (Holliday junction translocation) mutant strain was isolated but at a very much lower frequency than expected. The inviability of a dam lexA (Ind−) host was abrogated by the simultaneous presence of plasmids encoding both recA and ruvAB. This result indicates that of more than 20 SOS genes, only recA and ruvAB need to be derepressed to allow for dam mutant survival. The presence of mutS or mutL mutations allowed the construction of dam lexA (Ind−) derivatives. The requirement for recA, recB, recC, ruvA, ruvB, ruvC, and possibly recG gene expression indicates that recombination is essential for viability of dam bacteria probably to repair DNA double-strand breaks. The effect of mutS and mutL mutations indicates that DNA mismatch repair is the ultimate source of most of these DNA breaks. The requirement for recombination also suggests an explanation for the sensitivity of dam cells to certain DNA-damaging agents.
The dam gene of Escherichia coli encodes a DNA methyltransferase that methylates adenine in -GATC- sequences in double-stranded DNA (17). Mutant strains lacking this enzyme display a pleiotropic phenotype including increased mutability, hyperrecombination, and increased sensitivity to DNA-damaging agents. In addition, dam bacteria have an increased number of single-strand breaks in DNA compared to wild type. The phenotypes displayed by dam mutants are consistent with multiple roles of unmethylated, methylated, and hemimethylated -GATC- sequences in cellular physiology. These include regulation of gene expression and strand discrimination during replication-associated DNA mismatch repair (17).
An additional feature of dam strains is inviability when combined with mutant alleles of recA, recB, recC, or noninducible (Ind−) lexA (19). The lexA inviability suggests a requirement for derepression of one or more SOS genes. The SOS response is induced following treatments that damage DNA or inhibit DNA replication (6). About 20 genes (including recA, lexA, and ruvAB) that are negatively regulated by LexA are derepressed following cleavage of the LexA repressor. Treatments that induce the SOS regulon do so by activating the coprotease activity of RecA (“activated RecA”), resulting in LexA cleavage. RecA protein also catalyzes 3′-single-strand invasion of homologous DNA and is, therefore, essential in the recombination process (15).
Peterson et al. (24) showed that dam bacteria with a temperature-sensitive lexA allele were viable at 42°C but not at 30°C, indicating the requirement for derepressed expression of one or more LexA-regulated SOS genes. In addition, Peterson et al. (24) found higher basal-level expression (two- to sixfold) of several SOS genes (including recA, lexA, sulA, uvrA, uvrB, uvrD, dinD, and recF) in dam mutants than in wild type. However, since other genes are also induced by LexA cleavage, it was not possible to determine which are required for dam viability.
In the present communication, the SOS genes required for viability of dam strains have been identified. They are recA and ruvAB, the latter encoding enzymes that translocate Holliday junctions (15, 29). Two other non-SOS genes have also been identified. The recG gene product can also catalyze translocation (15), and dam recG mutants are probably inviable. It is also shown that expression of the ruvC gene product, a Holliday junction resolvase (15, 29), is also required for dam mutant viability. The requirement for recA, recB, recC, recG, ruvA, ruvB, and ruvC gene expression indicates that recombination is essential for dam mutant viability.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli recipient strains used are derived from AB1157 and are described in Table 1. Annotated descriptions of strains beginning with GM can be found at http://www.ummed.edu/pub/d/dam/dstrains.html. Hfr donors derived from KL14 begin transfer from min 69 and, therefore, transfer the closely linked dam and aroK genes (min 75) as early markers. Plasmids pGB2 (3) and pGB2ruvAB (26), which are derived from pSC101, are compatible with plasmids precA (10) and precAP67W (10), which are derivatives of pBR322.
TABLE 1.
Escherichia coli K-12 strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source and/or reference |
|---|---|---|
| E. coli K-12 strains | ||
| AB1157 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rfbD1 mgl-51 rpoS396(Am) rpsL31(Strr) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1 | E. A. Adelberg |
| AB1874 | F-42 (F-lac)/lac-19 | E. A. Adelberg |
| AM207 | As AB1157 but recR252::mTn10 | R. G. Lloyd |
| AM547 | As AB1157 but ΔruvAC65 | R. G. Lloyd |
| DE407 | lexA3 mal::Tn9 sulA211 rpsL31 Δ(lac-gpt)5 mtl-1? thi-1? | D. Ennis (4) |
| CS81 | As AB1157 but ruvB52 eda-51::Tn10 | R. G. Lloyd |
| GM698 | As KL14 but dam-13::Tn9 | Lab stock |
| GM2807 | As KL14 but dam-16::Kan | Lab stock |
| GM2835 | As KL14 but aroK17::Cam | Lab stock |
| GM7362 | As DE407 but mutL218::Tn10 | This work |
| GM7363 | As DE407 but mutS215::Tn10 | This work |
| GS1481 | As AB1157 but ΔruvC64::Kan | R. G. Lloyd |
| JC10990 | AS AB1157 but recF332::Tn3 | M. Volkert |
| JC13028 | As AB1157 but recJ147(Ts) | S. Lovett |
| KD2250 | As AB1157 but recQ1803::Tn3 | H. Nakayama |
| KL14 | Hfr relA1 spoT1 thi-1 | K. B. Low |
| KM353 | As AB1157 but recD1901::Tn10 | K. M. Murphy |
| N2057 | As AB1157 but ruvA60::Tn10 | R. G. Lloyd |
| N2446 | As AB1157 but recJ284::Tn10 | R. G. Lloyd |
| N3793 | As AB1157 but ΔrecG263::Kan | R. G. Lloyd |
| Plasmids | ||
| precA | A pBR322 derivative with the recA gene under pTac control, Ampr | K. Knight (10) |
| precAP67W | A P67W mutant derivative of precA causing coprotease constitutivity | K. Knight (10) |
| pGB2 | Cloning vector derived from pSC101, Spcr | B. Michel (26) |
| pGB2ruvAB | pGB2 with cloned ruvAB genes | B. Michel (26) |
Conjugation.
Donors and recipients were grown to logarithmic phase (1 × 108 to 2 × 108/ml) in Difco brain heart (BH; 20 g/liter) broth and mixed at a ratio of 1:10, respectively. After 60 min at 37°C, mating was terminated by vigorous blending and the cells were diluted and plated on BH solidified with 1.6% Difco agar and containing 100 μg of streptomycin per ml and, where necessary, 40 μg of kanamycin per ml or 10 μg of chloramphenicol per ml. When required, spectinomycin was added to 50 μg/ml, and the presence of this agent during mating did not significantly affect the yield of recombinants where donors were sensitive to it. Ampicillin was added to media at 100 μg/ml when required but was not present in the mating mixtures. Plates were incubated for 1 to 2 days at 37°C before scoring. Recombination frequencies are given as number of recombinants per 100 donors. F-lac transfer was measured by mating logarithmic-phase cultures at a ratio of one donor to five Lac− recipients for 60 min at 37°C and determining the percentage of Lac+ recipients by plating on MacConkey (Difco) agar containing 100 μg of streptomycin/ml.
Plasmid stability.
Cells were diluted to 100 to 200/ml in BH broth and grown to saturation (1 × 109 to 2 × 109/ml) at 37°C in the absence of antibiotics. The cultures were diluted and plated on BH medium with and without ampicillin. A dilution containing 100 to 200 cells was used for the next cycle. This procedure was repeated several times.
RESULTS
Crosses with recD, recF, recJ, and recR strains.
In order to identify which SOS (and other) genes are required for survival of dam mutants, mutations in genes that are inviable in a dam strain have been sought. As dam recA and dam recBC double mutants are inviable (21), other genes affecting recombination and repair have been tested. Previous work has shown that recF, umuC, dinA, dinB, dinC, recN, recO, and recQ derivatives of dam mutants could be constructed and are, therefore, not required for viability (23, 24). To extend this spectrum, the results shown in Table 2 indicate that the dam-13::Cam allele could be efficiently introduced into recD, recF, recR, and recJ mutant recipients by conjugation. The recF allele used here is a null mutation in contrast to the recF143 point mutation used previously (24). The recD mutation has been shown to result in a reduced level of RecBCD exonuclease V activity but is fully RecBC recombination proficient (1). The viability of the recombination-proficient dam recD double mutant is in contrast to the inviability of dam recBC bacteria that are expected to be recombination deficient.
TABLE 2.
Effect of various rec mutations on recombination frequency in Hfr dam-13::Cam × F− Strr crosses and F-lac transfera
| Recipient strain | Recombination frequency | F-lac transfer (%) |
|---|---|---|
| AB1157 (wild) | 0.45 | 18 |
| KM353 (recD1901::Tn10) | 1.4 | |
| JC10990 (recF312::Tn3) | 1.0 | |
| N3793 (ΔrecG263::Kan) | 0.005 | |
| N2446 (recJ284::Tn10) | 0.62 | |
| KD2250 (recQ1803::Tn3) | 1.2 | |
| AM207 (recR252::mTn10 | 1.0 | |
| N2057 (ruvA60::Tn10) | 0 | 15 |
| CS81 (ruvB52) | 0 | 12 |
| GS1481 (ΔruvC64::Kan) | 0 | 9 |
| AM547 (ΔruvAC65) | 0 | 12 |
Conjugation and selection of Camr Strr recombinants were carried out as described in Materials and Methods. Recombination frequency is the number of recombinants per 100 Hfr donors. This represents 1.6 × 103 Camr Strr recombinants in 50 μl for the wild-type (AB1157) cross. Zero indicates no recombinants in 50 μl of undiluted mating mixture. The percentage of the recipient population receiving F-lac is indicated.
The viability of a recJ284 dam double mutant was unexpected, as it was previously shown that a dam recJ77 double mutant could not be constructed (24). An additional strain, JC13028 [recJ147(Ts)], containing a temperature sensitivity mutation, was mated at 32°C with a dam-16::Kan donor, and Kanr Strr recombinants were obtained at a wild-type frequency. None of 100 recombinants tested were temperature sensitive for growth. On balance, it appears that the recJ77 allele may be anomalous and that dam recJ strains are viable. For Salmonella enterica serovar Typhimurium, it has also been found that dam recJ mutants are viable (27).
Crosses with a recG strain.
The recombination frequency (expressed as the number of recombinants per 100 Hfr donors) of the dam-13::Cam donor with the ΔrecG263::Kan recipient was very low (Table 2). This was unexpected, because a control cross, with the same Hfr donor background but bearing an aroK17::Cam mutation (which is closely linked to dam [16]), yielded recombinants at a frequency at least 50-fold higher (Table 3). The latter result confirms the observation by Lloyd (14), who noted only about a threefold decrease in recombination frequency in crosses of a recG recipient with an HfrH donor. Furthermore, a high frequency of dam-13::Cam transfer to the ΔrecG263::Kan recipient was expected, as the recG+ allele (located at min 82 on the genetic map) should be transferred early by the dam (min 75) donor. Indeed, 49 of 50 of the Camr recombinants had the expected dam recG+ (Camr Kans) genotype.
TABLE 3.
Effect of various ruv mutations on recombination frequency in Hfr aroK17::Cam × F− Strr crossesa
| Recipient | Recombination frequency |
|---|---|
| AB1157 | 1.03 |
| N2057 (ruvA60::Tn10) | 0.12 |
| CS81 (ruvB52) | 0.31 |
| GS1481 (ΔruvC64::Kan) | 0.10 |
| AM547 (ΔruvAC65) | 0.19 |
| N3793 (ΔrecG263::Kan) | 0.28 |
Conjugation and selection of Camr Strr recombinants were carried out as described in Materials and Methods. Recombination frequency is the number of recombinants per 100 Hfr donors.
To test the possibility that a chromosomal duplication encompassing either dam or recG to form a heteroallelic partial diploid occurred, two of the rare Camr Kanr recombinants were grown for about 100 generations in the absence of antibiotics. These isolates exhibited phenotypes identical to those of the same cultures grown in the presence of antibiotics, i.e., sensitivity to 2-aminopurine (a diagnostic test for dam) and UV light (a recG phenotype) and an equal plating efficiency on media with and without chloramphenicol or kanamycin. This result makes the presence of a chromosomal duplication unlikely. The result also rules out the acquisition of a mutS or mutL suppressor mutation, since dam strains with such suppressors are resistant to 2-aminopurine (21).
Crosses with ruv strains.
No recombinants were obtained by using a dam donor with any ruv mutant recipient when undiluted mating mixtures were placed on selective media (Table 2). This extends our previous observation that a dam derivative of a strain with an uncharacterized ruv mutation could not be constructed (24). Recipient strains with mutations in ruvA, ruvB, or ruvC or a deletion removing all three genes failed to yield dam recombinants (Table 2). The recombination deficiency in these crosses is not due to lack of genetic transfer, because F-lac can be transferred to the ruv recipients at near-wild-type frequency (Table 2). The ruv strains are recombination proficient when mated with an aroK17::Cam donor and show only a 3- to 10-fold reduction in recombination frequency compared to wild type (Table 3). This confirms previous data obtained by Lloyd (14), who noted only a three- to fourfold reduction in recombination frequency with ruv mutants with an HfrH donor. The large reduction in recombination frequency with the dam donor and ruv recipients suggests that these combinations are lethal. The lack of conditional dam or ruv alleles, however, prevents a direct test of the inviability of these combinations. The probable lethality is of interest because ruvA and ruvB are LexA-regulated SOS genes and could be candidates for the unknown SOS genes needed for dam viability.
The ruvA60::Tn10 mutation in strain N2057 has a polar effect on the contiguous ruvB gene, and strains bearing it are RuvAB−. Plasmid pGB2ruvAB was introduced into N2057 and then mated with GM2807, a Kanr dam donor. No Kanr Strr recombinants were obtained with N2057 (ruvA60::Tn10), but the recombination frequency was increased by over 1,000-fold when pGB2ruvAB was present (Table 4). The plasmid, therefore, efficiently complements the ruvA mutation for inviability with dam and for sensitivity to UV light (data not shown). The level of recombination obtained with the pGB2ruvAB/ ruvA60::Tn10 strain was used as a control for the remaining crosses in Table 4.
TABLE 4.
Effect of ruvAB and recA plasmids on recombination frequency in Hfr dam16::Kan × F− ruvA or lexA crosses and F-lac transfera
| Recipient strain | Recombination frequency | F-lac transfer (%) |
|---|---|---|
| N2057 (ruvA60::Tn10) | 0 | |
| pGB2/N2057 | 0 | |
| pGB2ruvAB/N2057 | 0.5 | |
| DE407 (lexA3 sfiA211) | 0.005b | 34 |
| pGB2ruvAB/DE407 | 0.005b | 41 |
| precA/DE407 | 0.007b | |
| precA/pGB2ruvAB/DE407 | 0.4 | 42 |
| precAP67W/pGB2ruvAB/DE407 | 0.19 | 30 |
| GM7362 (DE407 mutS453) | 0.1 | |
| GM7363 (DE407 mutL451) | 0.15 | |
| precA/pGB2ruvAB/GM7362 | 0.5 | |
| precA/pGB2ruvAB/GM7363 | 0.5 |
Conjugation and selection of Kanr Strr recombinants were carried out as described in Materials and Methods. Recombination frequency is the number of recombinants per 100 donors. This represents 1.04 × 103 Kanr Strr recombinants in 50 μl for the wild-type (pGB2ruvAB/N2057) cross. Zero indicates no recombinants in 50 μl of undiluted mating mixture. The percentage of the recipient population receiving F-lac is indicated.
The few recombinants recovered were all determined to be lexA+ (Cams) Kanr Strr.
Crosses with a lexA3 (Ind−) recipient.
Strain DE407, a distant derivative of AB1157, has the noninducible lexA3 allele, thereby preventing derepression of the SOS regulon (including the recA, ruvAB, and uvr genes) (4). This strain is, therefore, very sensitive to UV irradiation. A mal::Tn9 (Camr) insertion is closely linked to the lexA3 allele. Conjugation between DE407 (Strr) and the dam (Kanr) donor produced Kanr Strr recombinants at a low level (Table 4). Further examination of 100 rare recombinants indicated that they were Cams and not sensitive to UV irradiation. That is, they were lexA+ due to the transfer of this gene from donor to recipient (the lexA gene is located at min 91). This result serves as an internal control showing that some gene transfer and recombination must have occurred. The results above show that no bona fide dam lexA3 (Kanr Strr Camr) recombinants were recovered.
The inviability of dam ruvAB and dam recA bacteria led to the hypothesis that overexpression of these SOS genes might be sufficient to allow dam lexA3 cells to be viable. To test this idea, plasmids encoding wild-type RecA, RuvA, and RuvB were introduced into strain DE407 (lexA3) prior to mating with the dam donor. The presence of either precA or pGB2ruvAB did not significantly alter the Kanr Strr recombination frequency compared to that with the plasmidless lexA3 recipient (Table 4). An almost-1,000-fold increase in Kanr Strr recombinants was detected, however, when both ruvAB and recA plasmids were harbored in the lexA3 recipient (Table 4). One hundred of these recombinants were shown to be Camr, indicating the presence of the lexA3 allele. A similar recombination frequency was obtained when the coprotease constitutive (P67W) RecA-encoding plasmid was substituted for the wild-type recA, indicating that recombination ability is not substantially impaired by the mutation.
The differences in recombination frequency of the various plasmid-containing DE407 strains are not due to differential abilities to receive genetic material, because all strains show similar frequencies of transconjugants when mated with an F-lac donor (Table 4).
Mutations affecting mismatch repair suppress dam lexA3 lethality.
Inactivation of mismatch repair by mutation in either mutS or mutL allows dam derivatives of recA, recB, or recC cells to be constructed (21, 28). To test if a similar situation applies with lexA3, mutS453 (GM7362) and mutL451 (GM7363) derivatives of DE407 were constructed and used as recipients in matings with a dam donor. The results in Table 4 show that dam lexA3 (Kanr Strr) recombinants were recovered at frequencies only three- to fourfold less than those of the control. This reduction was consistent from experiment to experiment, and wild-type levels of recombinants were formed only when the recA and ruvAB plasmids were present in the mismatch repair-deficient lexA3 recipients (Table 4). One hundred Kanr Strr recombinants from each cross above were shown to be Camr, indicating the presence of the lexA3 allele. These data indicate that abrogation of mismatch repair removes almost all the cause(s) for dam lexA3 lethality.
Plasmid stability.
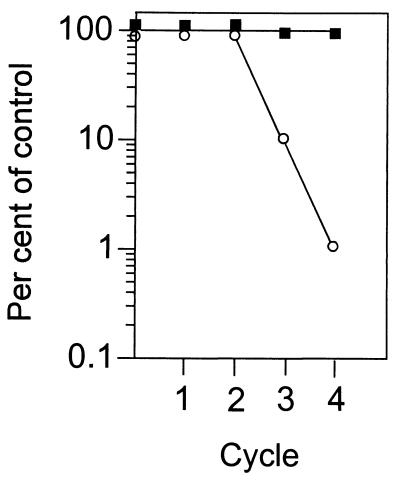
If the recA and ruvAB plasmids are essential for viability in a dam lexA3 strain, then these plasmids should appear to be stable in this strain in the absence of antibiotic selection. The results in Fig. 1 indicate that indeed the recA plasmid is stable in the dam lexA3 strain but much less so in the control lexA3 parent. There was no significant loss of the ruvAB plasmid from either strain during these cycles of growth, presumably due to the presence of the stabilizing par function in pGB2 (3) that ensures efficient plasmid segregation into daughter cells.
FIG. 1.
Loss of the precA plasmid from dam+ and dam mutant lexA3 strains. Cells were grown from 100 to 200 per ml to saturation (1 × 109 to 2 × 109/ml) for the indicated number of cycles in the absence of ampicillin. The number of cells growing on BH medium with and without ampicillin is shown. Unfilled circles and filled squares represent the dam+ lexA3 and dam mutant lexA3 strains, respectively.
DISCUSSION
For a dam mutant to be viable, expression of the recA, recB, recC, ruvA, ruvB, ruvC, and most likely recG genes is essential. The level of expression from the chromosomal copies of recB, recC, recG, and ruvC is sufficient for survival of a dam or dam lexA3 cell, but higher levels of the SOS-regulated RecA and RuvAB proteins are necessary. The precise amount of these proteins required for survival is not yet known, but the level of expression from the plasmids can be estimated. The copy number of the pSC101-based pGB2 vector is about five per cell, and the ruvAB genes bear their own promoter. A fivefold increase over the chromosomal level of RuvAB is realistic because attempts to clone the genes in pBR322-based vectors (copy number of 15 to 20) have not been successful (26). The recA gene is present in a pBR322 derivative transcribed from the tac promoter. The uninduced level of RecA is about 10-fold above that of the wild type (K. Knight, personal communication). The 5- and 10-fold overproduction of RuvAB and RecA, respectively, is similar to the derepressed level from the chromosomal genes in fully SOS-induced wild-type cells (6). Indeed, the induced level of RuvAB and RecA in an SOS-constitutive recA441 mutant is sufficient to allow a dam derivative to be constructed (24).
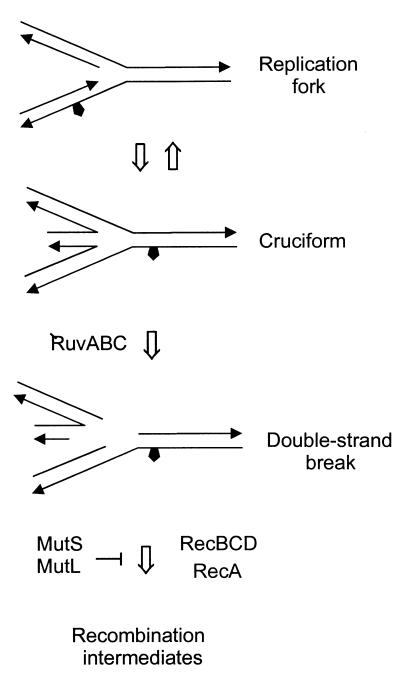
ruvABC recG double mutants are deficient in conjugational recombination, whereas the single mutants are not (14). This suggests an overlap in the function of RuvABC and RecG activity. Both RecG and RuvAB bind specifically to Holliday junctions but appear to have opposing helicase directionality (15). In contrast to conjugal recombination, dam ruvABC mutants are inviable (Table 2), indicating that RecG cannot substitute for RuvABC. The processing of recombination intermediates for viability in dam cells is, therefore, different from that during conjugation and may require the divergent properties of both RecG and RuvABC. For example, in dam mutants perhaps both the 5′- and the 3′-single-strand overhangs that are generated by replication fork collapse (Fig. 2) can be paired with the homologous strand by RecA. To promote strand assimilation by branch migration, the Holliday junction would need to be translocated 5′ to 3′ in one case (the RuvAB polarity) and 3′ to 5′ (the RecG polarity) in the other.
FIG. 2.
Model for the generation of double-strand breaks in dam bacteria (8, 11). A replication fork approaches a nick releasing a chromosomal arm the end of which becomes a substrate for RecBCD action. The 3′ single strand thus produced is made to synapse by RecA to produce a Holliday junction that can be processed by RuvABC. Resolution of the Holliday junction restores the replication fork. See the work of Kuzminov (11) for further details of this model.
An explanation for the low-level recovery of dam recG double mutants is that in the viable recombinants a mutated form of RuvAB can substitute for RecG. This is plausible because the recombination frequency in these experiments approaches the spontaneous mutation frequency. Another possibility is that RuvAB, but not RecG, helps to displace RecA from recombination intermediates. Finally, an unknown suppressor mutation may be present in the viable dam recG mutants. As noted in Results, unstable duplications seem a less likely possibility.
Mutations in the mutS or mutL genes allowed the recovery of dam recombinants with strain DE407 (Table 4), suggesting that mismatch repair is instrumental in causing the breaks in DNA of dam cells. However, the level of recombinants was lower than that of wild type and was increased to that level by the presence of the ruvAB and recA plasmids (Table 4). The inability of the mut mutations to completely restore the wild-type recombination frequency suggests that, in addition to mismatch repair, some other cellular function might be affecting the level of derepression of the LexA regulon. These data confirm the observation that, although dam cells lacking mismatch repair have fewer nicks in DNA (28), the SOS regulon is still induced (23), suggesting some persistent inducing signal.
Why is recombination of vital importance in a dam bacterium? Inactivation of mismatch repair by mutation in either mutL or mutS allows the construction of dam recA (7, 21) and dam recBC (28) bacteria and decreases the level of DNA breaks (28). A complex of MutS, MutL, and MutH is required for efficient repair, and a mutation inactivating any one of these proteins results in mismatch repair deficiency (22). The requirement for recombination must be related to the presence of single-strand DNA breaks (19), which arise due to MutH endonuclease activity at unmethylated GATC sites during mismatch repair (2). A simple explanation for the recombination requirement is that occasional double-strand breaks arise due to MutH cleavage at the same unmethylated GATC sequence that contains a nick in the complementary strand (2, 7). The number of such double-strand breaks is expected to be low because their persistence in cells should be lethal. Indeed, they are detected in dam bacteria only in the absence of RecBCD and even then at a low frequency (28). The conclusions from the present work indicate that such double-strand break repair requires the recA, recBC, and ruvABC gene products. The model predicts that increasing the level of MutH should increase the frequency of double-strand breaks. The presence of a multicopy plasmid encoding mutH, however, does not appear to sensitize dam cells for viability (data not shown).
Another or an additional explanation for the role of recombination in dam cells involves the frequent single-strand interruptions in DNA and the model proposed by Kuzminov and Stahl (11, 12) and Horiuchi and Fujimura (8) for the collapse and repair of replication forks. Indeed, Kuzminov (11) used the phenotypic properties of dam incompatibility with recA and lexA as a foundation for his model (Fig. 2). When a replication fork encounters a single-strand nick in the DNA of dam cells, one of the chromosomal arms dislocates from the chromosome due to the formation of a double-strand break. To reestablish a functional replication fork, the wayward chromosomal arm needs to be recombined back into the chromosome. This requires processing by RecBCD to produce a 3′-invasive strand after encountering a Chi site and the action of RecA to make this strand synapse with its homologue. After formation and translocation of a Holliday junction, resolution by RuvC restores the replication fork (Fig. 2). As multiple replication forks are present in growing bacteria, replication fork collapse is expected to occur frequently, requiring increased recombination capacity. This explains the high subinduced level of SOS genes in dam bacteria and the requirement for elevated RecA and RuvAB levels. This model (11) also explains the inviability of dam mutants with mutations in either the polA or lig genes (20) due to the production of excess double-strand breaks as well as the hyperrecombination phenotype of dam mutants (18). A further prediction based on this model is that dam priA (primosome) mutants should be inviable because replication restart at sites of collapsed replication forks requires reassembly of the primosome complex (13). This prediction is currently being tested.
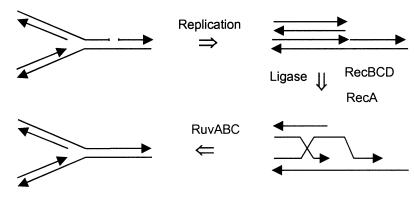
Seigneur et al. (26) have proposed a model to explain how replication fork arrest in E. coli rep mutants, which lack a replicative helicase, leads to formation of double-strand breaks. (Replication fork arrest should be distinguished from replication fork collapse in the model discussed above.) Briefly, the RuvABC proteins are responsible for the formation of double-strand breaks, and the substrate is thought to be a cruciform (Holliday junction) formed by the annealing of the two new DNA daughter strands (Fig. 3). If RecBCD acts on the double-stranded end of the annealed strands before RuvAB, then the breaks are prevented either through the initiation of RecA-RuvABC-dependent recombination or by a recombination-independent resection of the annealed duplex (26). This model is probably not applicable to dam mutants because the rep and dam mutants have different phenotypes (17, 26), a key one being that rep recA double mutants are viable while dam recA mutants are not. The repA recA mutant is viable because of the recombination-independent action of RecBCD referred to above. Although this model may not be applicable during normal growth of dam mutants, it may be important when the replication fork is arrested at drug-induced mismatches (see below).
FIG. 3.
Origin of double-strand breaks in DNA with drug-induced lesions. DNA containing a lesion (pentagon) such as O6meG is replicated, but the polymerase has stalled while mismatch repair is attempted. Stalling can also occur at any polymerase-blocking lesion. The new DNA strands anneal to form a cruciform structure that can be acted upon by RuvABC, producing a double-strand break (only one of the two possible double-strand break configurations is shown). Alternatively, RecBCD and RecA can act on the tail of the newly synthesized annealed strands of the complete or broken cruciform to form a recombination intermediate. See the work of Seigneur et al. (26) for further details on the formation and processing of cruciform structures.
The recombination requirement for dam mutant survival may also explain the increased sensitivity of this strain to DNA damage provoked by alkylating agents (9) and cisplatin (5). DNA damage inflicted by these agents would increase the requirement for repair-associated recombination. Recombination proteins would become limiting, and drug-induced gaps or chromosome breaks would not be repaired, eventually leading to cell death. The importance of recombination pathways in the repair of cisplatin damage has recently been demonstrated (31).
DNA mismatch repair sensitizes dam cells to the cytotoxic action of alkylating agents, specifically those that produce O6-methylguanine (O6meG) (9). It was proposed previously (9) that O6meG paired with either C or T is a substrate for mismatch repair recognition. That is, all possible O6meG base pairs are subject to mismatch repair. The specific binding of E. coli MutS to O6meG base pairs has been reported previously (25). Consequently, upon replication of the O6meG-containing strand a futile cycle of mismatch repair ensues. As the replicative polymerase, PolIII, synthesizes mismatch repair tracts (22), this event would cause polymerase stalling. The requirement for recombination in dam mutants reported here suggests an additional action at O6meG lesions. The blocked fork could lead to annealing of the newly synthesized strands to produce a cruciform structure (Fig. 3) and production of a chromosome double-strand break by RuvABC as proposed by Seigneur et al. (26). Alternatively, recombination initiated by RecBCD at the tail of the annealed new strands could lead to restoration of the replication fork as described by Seigneur et al. (26). The binding of MutS and MutL to O6meG base pairs in the cruciform structure or in subsequent recombination intermediates (Fig. 3) might effectively abort recombination in a manner similar to that described for base mismatches in phage fd-M13 heteroduplexes (30). This antirecombinogenic action of MutS and MutL would return the DNA to a cruciform configuration where RuvABC could cleave it. In the absence of mismatch repair, the replication fork would not be arrested at O6meG mismatches and the cells would have a greater chance for survival. Experiments are in progress to test the predictions of this model.
ACKNOWLEDGMENTS
I thank all those investigators who contributed strains and plasmids to ensure the success of these experiments. Bevin Engelward, Pat Foster, Anders Løbner-Olesen, Benedicte Michel, and Te Wu provided suggestions that improved the manuscript.
This work was supported by a Howard Hughes Medical Institute Research Resources Program for Medical Schools Award to the University of Massachusetts Medical School.
REFERENCES
- 1.Amundsen S K, Taylor A F, Chaudhury A M, Smith G R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 3.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 4.Ennis D G, Fisher B, Edmiston S, Mount D W. Dual role for Escherichia coli RecA protein in SOS mutagenesis. Proc Natl Acad Sci USA. 1985;82:3325–3329. doi: 10.1073/pnas.82.10.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fram R J, Cusick P S, Wilson J M, Marinus M G. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol Pharmacol. 1985;28:51–55. [PubMed] [Google Scholar]
- 6.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 7.Glickman B W, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci USA. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horiuchi T, Fujimura Y. Recombination rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karran P, Marinus M G. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 10.Konola J T, Nastri H G, Logan K M, Knight K L. Mutations at pro67 in the RecA protein P-loop motif differentially modify coprotease function and separate coprotease from recombination activities. J Biol Chem. 1995;270:8411–8419. doi: 10.1074/jbc.270.15.8411. [DOI] [PubMed] [Google Scholar]
- 11.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuzminov A, Stahl F W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Xu L, Sandler S, Marians K J. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd R G. Conjugal recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 16.Lobner-Olesen A, Marinus M G. Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J Bacteriol. 1992;174:525–529. doi: 10.1128/jb.174.2.525-529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marinus M G. Methylation of DNA. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 782–791. [Google Scholar]
- 18.Marinus M G, Konrad E B. Hyper-recombination in dam mutants of Escherichia coli K-12. Mol Gen Genet. 1976;149:273–277. doi: 10.1007/BF00268528. [DOI] [PubMed] [Google Scholar]
- 19.Marinus M G, Morris N R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974;85:309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- 20.Marinus M G, Morris N R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975;28:15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- 21.McGraw B R, Marinus M G. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 22.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 23.Peterson K R, Mount D W. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J Bacteriol. 1993;175:7505–7508. doi: 10.1128/jb.175.22.7505-7508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson K R, Wertman K F, Mount D W, Marinus M G. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201:14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen L, Samson L. The Escherichia coli MutS DNA mismatch binding protein specifically binds O6-methylguanine DNA lesions. Carcinogenesis. 1996;17:2085–2088. doi: 10.1093/carcin/17.9.2085. [DOI] [PubMed] [Google Scholar]
- 26.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 27.Torreblanca J, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T C, Smith K C. Inviability of dam recA and dam recB cells of Escherichia coli is correlated with their inability to repair double-strand breaks produced by mismatch repair. J Bacteriol. 1986;165:1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–214. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 30.Worth L, Clark S R M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zdraveski, Z. Z., J. A. Mello, M. G. Marinus, and J. M. Essigman. Multiple pathways of recombination define cellular responses to cisplatin. Chem. Biol., in press. [DOI] [PubMed]