Highlights
-
•
New cell model to investigate the impact of loss of integrin α3 in podocytes.
-
•
In this novel model, genes of the extracellular matrix and adhesome are mostly downregulated.
-
•
Loss of integrin α3 results in changes of cell adhesion and spreading.
Abbreviations: A3−, integrin alpha3 deficient cells AB8/13 is a conditionally immortalized podocyte cell line carrying a temperature-sensitive T antigen as transgene, in text and figures the abbreviation Podo was used for simplicity; HK2, human kidney-2; ILNEB, interstitial lung disease, nephrotic syndrome and epidermolysis bullosa; PodoA3−, integrin α3 negative podocytes
Keywords: ILNEB, Integrin α3, Skin blistering, Nephrotic syndrome, Kidney, CRISPR/Cas9, Podocyte
Abstract
Integrin α3β1 is a cell adhesion receptor widely expressed in epithelial cells. Pathogenic variants in the gene encoding the integrin α3 subunit ITGA3 lead to a syndrome including interstitial lung disease, nephrotic syndrome, and epidermolysis bullosa (ILNEB). Renal involvement mainly consists of glomerular disease caused by loss of adhesion between podocytes and the glomerular basement membrane.
The aim of this study was to characterize the impact of loss of integrin α3 on human podocytes.
ITGA3 was stably knocked-out in the human podocyte cell line AB8/13, designated as PodoA3−, and in human proximal tubule epithelial cell line HK2 using the targeted genome editing technique CRISPR/Cas9. Cell clones were characterized by Sanger sequencing, quantitative PCR, Western Blot and immunofluorescence staining. RNASeq of integrin α3 negative cells and controls was performed to identify differential gene expression patterns.
Differentiated PodoA3− did not substantially change morphology and adhesion under standard culture conditions, but displayed significantly reduced spreading and adhesion when seed on laminin 511 in serum free medium. Gene expression studies demonstrated a distinct dysregulation of the adhesion network with downregulation of most integrin α3 interaction partners. In agreement with this, biological processes such as “extracellular matrix organization” and “cell differentiation” as well as KEGG pathways such as “ECM-receptor interaction”, “focal adhesion” and the “PI3K-Akt signaling pathway” were significantly downregulated in human podocytes lacking the integrin α3 subunit.
Introduction
As a main receptor linking epithelial cells to basement membranes (BM) [1], integrin α3β1 is involved in multiple physiological and pathological processes [2], [3]. Integrin α3β1 plays a crucial role in skin integrity and kidney organogenesis, and in podocyte function as demonstrated in mouse models [4], [5], [6]. In humans, biallelic loss-of-function mutations in the gene encoding the integrin α3 subunit, ITGA3, lead to a syndromic junctional epidermolysis bullosa subtype, with nephrotic syndrome and interstitial lung disease (ILNEB, MIM 614748) [7]. Skin fragility is mild in patients with ILNEB, and becomes apparent in the first months of life or remains unnoticed. In accordance with this, patients’ keratinocytes constitutionally lacking the α3 subunit demonstrated an activated phenotype, a compensatory change in the repertoire integrin α subunits and a shift from a laminin-, to a suitable fibronectin-rich micromilieu [8]. Intriguingly, in ILNEB patients, the renal manifestations are variable, involving the glomerulus and the urinary tubular tract in the case of null alleles [8], [9], [10], [11], [12], or lacking in the case of certain amino acid substitutions located in the N-terminus [13], [14]. Besides, α3 is a player in acquired renal disorders. It was shown that integrin α3 is upregulated in podocytes of early-stage diabetic nephropathy and downregulated in advanced stages of diabetic nephropathy [15], [16]. Loss of foot processes by loss of adhesion of podocytes to the glomerular basement membrane or reduced cell interaction leads to a dysfunctional filtration barrier, and, eventually to glomerulosclerosis and renal failure [5], [17], [18]. Thus ultimately, actin dynamics at focal adhesions is a common endpoint and putative therapeutic target for proteinuric kidney diseases [19].
Here, we generated ITGA3 negative human podocytes and proximal tubule epithelial cell lines by CRISPR/Cas9 and performed unbiased gene expression studies by RNASeq to address the global impact of loss of this integrin subunit and question whether compensatory mechanisms are present, like previously reported in keratinocytes.
Results
Generation and characterization of integrin α3 negative renal epithelial cell lines
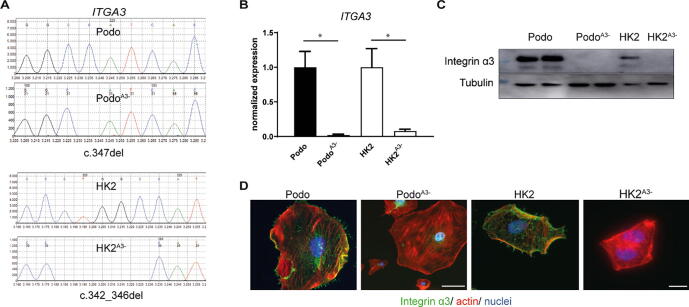
To analyze the function of the integrin α3 subunit in kidney epithelial cells, we generated α3 knock-out podocytes (AB8/13) [20] and HK2 cells [21] using CRISPR/Cas9 lentivirus with a guide RNA directed to the exon 3 of ITGA3 (see Material & Methods). By disrupting the ITGA3 locus, we established clones of podocytes (PodoA3−) and HK2 cells (HK2A3−) with homozygous ITGA3 frameshift deletions (Fig. 1A). Sequencing of the genomic DNA of PodoA3− revealed the homozygous deletion of one C nucleotide in exon 3, c.347del, leading to a premature termination codon, p.His116Ilefs*11. The DNA sequence analysis of HK2A3− disclosed the homozygous deletion of four nucleotides, c.342_346del, and premature termination codon formation, p.Gly115Serfs*4 (Fig. 1A). Both mutations lead to ITGA3 mRNA decay and absence of the protein, as determined by qPCR, Western Blot and immunofluorescence staining (Fig. 1B-D). We mainly focused on podocytes in respect to analysis of cellular functions and biological relevance and used HK2 cells due to their easy manipulation in culture.
Fig. 1.
Characterization of integrin α3 negative podocytes and HK2-cells. (A) DNA Sanger sequencing revealed the deletion of one nucleotide in the podocyte clone (PodoA3−), respectively-five nucleotides in the HK2-clone (HKA3−). (B), (C) Further confirmation of the knock-out of integrin α3 was obtained at gene expression level and protein level (in duplicate for PodoA3−). (D) Immunofluorescence staining for integrin α3 demonstrates its absence in PodoA3− and HK2A3−. Scale bar: 20 μm.
PodoA3− can terminally differentiate
Podocytes are terminally differentiated cells that express specific proteins including podocin, Wilms tumor 1 (WT1) and integrin α3. AB8/13 is a conditionally immortalized podocyte cell line carrying a temperature-sensitive T antigen as transgene. The tsA68 T antigen requires a culture temperature of 33 °C [22]. Under growth-permissive conditions, AB8/13 podocytes can proliferate, increase the cell number and display a cobblestone morphology [23]. Thus, it was important to analyze whether the newly generated podocyte cell lines, without or with ITGA3 deletion can differentiate and express specific markers. By placing the cells in growth-restrictive conditions (37 °C) allowing them to differentiate, they changed their morphology to a cuboidal and arborized appearance with an ordered array of actin fibers and increased in size as shown by immunofluorescence staining of fibrillary actin (Suppl. Fig. 1A). Expression analysis of genes coding for specialized proteins associated with slit-pores (NPHS2), filaments (SYNPO) and a podocyte-specific transcriptional factor (WT1) revealed upregulation of ITGA3, NPHS2 and WT1, as well as a downregulation for SYNPO validating our models as differentiated podocytes (Suppl. Fig. 1B). For the further experiments, differentiated podocytes were used.
Shape and adhesion of PodoA3−
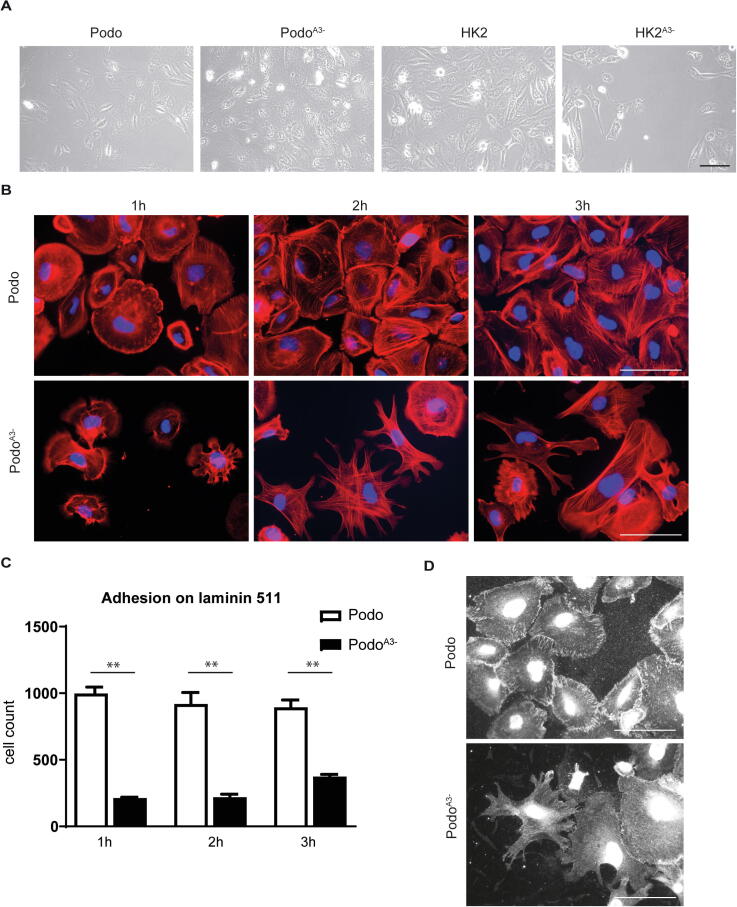
Under standard culture conditions, there were no gross differences, neither in cell survival, nor in shape of the integrin α3 negative podocytes, as compared to the control normal counterparts (Fig. 2A). However, in serum free medium on laminin 511 as a substrate, PodoA3− adhered significantly less than Podo, and demonstrated multiple protrusions and retractions and delayed spreading (Fig. 2B and C). In line with this, when α3 was absent, staining of phosphorylated paxilin (Y118), a marker of focal adhesion signaling, displayed small signals with an less regular distribution at cell periphery as compared to control cells (Fig. 2D).
Fig. 2.
Cell shape and adhesion of PodoA3−. (A) Observation of the cells in culture showed no significant visual differences between the groups. Scale bar: 100 μm. (B), (C) Adhesion and spreading assay on laminin 511 demonstrating significant decreased adhesion and spreading of PodoA3−. (D) Immunofluorescence staining of phosphorylated paxilin (Y118). Scale bar = 100 µm.
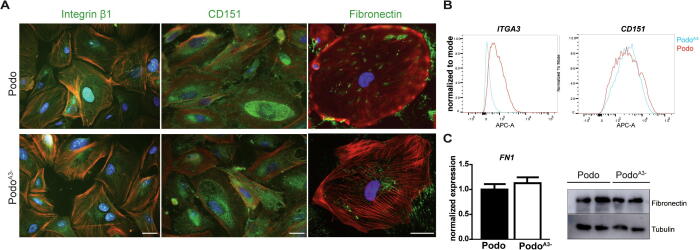
In adherent cells, integrin β1 which forms heterodimers with the α3 subunit, and tetraspanin CD151 which is one important molecule in linking podocytes to the GBM and thus facilitating firm adhesion [3] did not show significant changes in their distribution in PodoA3− as compared to controls. Fibronectin demonstrated a patchy distribution in control cells and was more scattered in the absence of α3, but the total protein amounts were comparable (Fig. 3).
Fig. 3.
Integrin α3 interaction partners. (A) Immunofluorescence staining for integrin α3 interaction partners and fibronectin. Scale bar: 20 μm. (B) Upper right panel, FACS analysis for ITGA3 and CD151 in Podo and PodoA3−. (C) Lower right panel, fibronectin mRNA (FN1) and protein levels are similar Podo and PodoA3−.
Dysregulation of gene expression in integrin α3 renal epithelial cell lines
We employed RNASeq to obtain a global unbiased characterization of the dysregulated genes and signaling pathways of integrin α3 negative renal epithelial cells. RNASeq yield a mean number of raw reads of 52,645,083.33 and a mean of 51,992,049.5 of clean reads (Suppl. Table 1).
The number of mapped reads was between 89.2 and 91.5% among the samples (Suppl. Table 2).
Pearson correlation indicated a nearly linear correlation for each cell line (Supplementary Fig. 2A) validating the quality of the biological replicates. For further analysis we used the readcount value from the gene expression level analysis and DESeq was used for samples with biological replicates [24]. There were 12,545 and 12,256 genes expressed in both podocyte and HK2 cell lines, while 713 and 395 genes were only expressed in integrin α3 negative, and 333 and 1044 only in control cells, respectively (Suppl. Fig. 2B). Volcano plots of the differentially expressed genes show that lack of integrin α3, significantly impacts gene expression in these cell models: 1899 genes were up and 1786 were down regulated between PodoA3− and Podo, and 3046 genes were up and 3577 were downregulated between HK2A3− and HK2 (Suppl. Fig. 2C). The significantly (p-value <0.05) dysregulated genes are included in Supplementary Tables 3 and 4 (see separate Excel files). Among the most upregulated genes in PodoA3− were IFI27 (log2FC = 5.7), BST2 (log2FC = 5.4), CCL5 (log2FC = 5.0), EDAR (log2FC = 4.9), FGB (log2FC = 4.8), IL2RA (log2FC = 4.7) and EDARADD (log2FC = 4.6) that belong to biological processes related to “cellular response to cytokine stimulus”. In PodoA3− ITGA3 had a log2FC of −3.8, while the most downregulated genes (log2FC < −6.0) did not functionally cluster. They included genes like NRXN3 (log2FC = −8.5) encoding a neuronal cell surface protein that may be involved in cell recognition and cell adhesion, TLR4 (log2FC = −8.3), SALL1 (log2FC = −8) and COL5A1 (log2FC = −6.6) (Suppl. Fig. 2D).
Dysregulated biological processes and pathways in podocytes lacking integrin α3
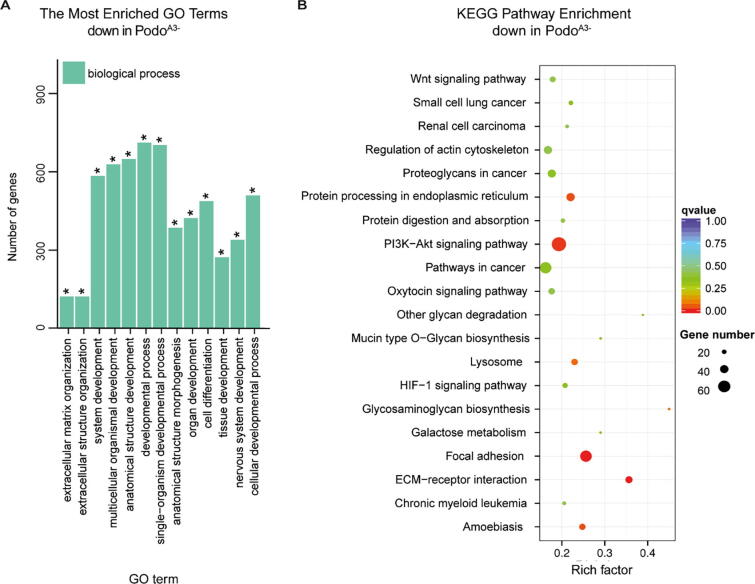
To further specify the impact of loss of integrin α3 on podocytes, all significantly up- and downregulated genes in PodoA3− were classified according to the biological processes (gene ontology, GO, category) (Suppl. Table 5). Most significantly deregulated were “single-organism process” (p = 9.67E−19), “system development” (p = 9.82E−19) and “cell differentiation” (p = 8.59E−16) (Fig. 4A). Upregulated genes were related to “mitotic cell cycle” (p = 1.53E−20) and “cell cycle process” (p = 4.39E−16), while downregulated genes were assigned to the biological processes “extracellular matrix organization (ECM)” (p = 7.03E−26), “system development” (p = 5.72E−20), “cell differentiation” (p = 1.84E−11), “biological adhesion” (p = 1.55E−8) and “cell migration” (p = 8.5E−10) (Suppl. Table 5).
Fig. 4.
Dysregulated biological processes and pathways in PodoA3−. (A) GO enrichment analysis for downregulated biological processes included extracellular matrix (ECM) organization and cell differentiation (B) ECM-receptor interaction, focal adhesion and PI3K-Akt signaling pathway were significantly downregulated in PodoA3− (KEGG database).
Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes database (KEGG) (kappa score ≥0.6, min. genes 3). This indicated that no pathway was significantly upregulated, while “ECM-receptor interaction” (corrected p = 0.0001), “focal adhesion” (p = 0.0001) and the “PI3K-Akt signaling pathway” (p = 0.015) were significantly downregulated in PodoA3− (Fig. 4B, Supplementary Table 6).
Dysregulation of the adhesion network in integrin α3 negative podocytes
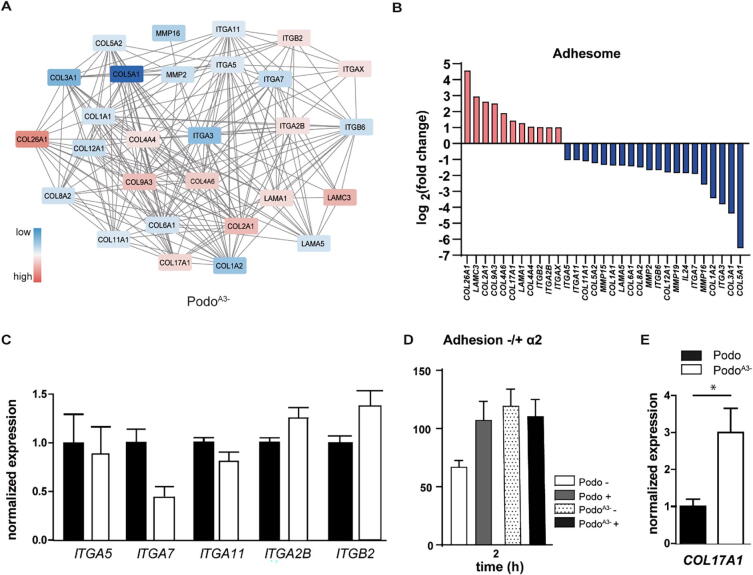
Since ECM organization and ECM-receptor interaction were among the most downregulated terms, we addressed the question how the integrin adhesome is changed and which mechanisms might compensate for adhesion of PodoA3− on substrates like plastic or collagen IV (not shown). Functional protein association networks predicted using STRING (https://string-db.org/) and Cytoscape version 3.7.0 (https://www.cytoscape.org/) revealed that most regulated genes in integrin α3 negative versus control cells encode proteins that clustered around the integrin α3 subunit (Fig. 5A).
Fig. 5.
Lack of α3 integrin in PodoA3− impacts the adhesome. (A) Analysis of interaction network of adhesion proteins identified in PodoA3− compared to the control (String & Cytoscape). Continuous mapping was created with blue and red according to fold change of gene expresson. (B) Gene expression for the adhesome showing log2(fold change) values in integrin α3 deficient podocytes. (C) Statistically not significant results by quantitative PCR (qPCR). (D) Adhesion analysis without (−) and with (+) integrin α2 blocking antibody on collagen IV (statistically insignificant). (E) Upregulated gene expression for COL17A1 in PodoA3− confirmed by qPCR. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The mRNA levels of the integrin subunits α7, α5, α11 which form heterodimers with β1 were downregulated, whereas α2B, αx, and β2 were upregulated (Fig. 5A–C). To further investigate compensatory mechanisms in PodoA3−, cells were treated with well-established blocking antibodies to the integrin α2 subunit for two hours, and adhesion on collagen IV was assessed. This treatment decreased adhesion of PodoA3−, but not of Podo, suggesting that other subunits contribute to adhesion of podocytes in the absence of α3 (Fig. 5D). In addition, another transmembrane adhesion molecule, type XVII collagen was upregulated in PodoA3− as compared to Podo, contributing to cell-matrix adhesion in the absence of integrin α3 (Fig. 5E).
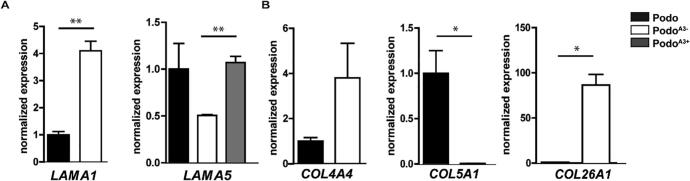
Among the extracellular ligands of integrins, laminin α1 normally expressed during development was upregulated, whereas the α5 chain of the major laminin 521 in the mature glomerular basement membrane [25] was downregulated in PodoA3− as compared to Podo, on mRNA level (Fig. 6A). Re-expression of integrin α3 in PodoA3+ lead to a rescue of LAMA5 expression implying a direct interdependence between receptor and its extracellular ligand (Fig. 6A). In the absence of α3β1, the mRNA levels of integral basement membrane collagens, COL4A4 and COL4A6 were upregulated. COL26A1 mRNA was strongly upregulated and COL5A1 mRNA was downregulated (Fig. 6B). Genes encoding other basement membrane associated proteins, such as nidogen 2 (NID2), fibrillin 1 (FBN1), aggrin (AGRN), tenascin C (TNC) and hemicentrin 1 (HMCN1) were downregulated in PodoA3−, suggesting a broad impact of loss of this integrin subunit on the overall composition of the GBM (Suppl. Table 3, separate Excel file).
Fig. 6.
Dysregulation of genes for extracellular ligands and proteins in PodoA3−. LAMA1 was significantly upregulated in PodoA3− as confirmed by qPCR, while LAMA5 was downregulated in qPCR. Using the retroviral system, full length ITGA3 was cloned in PodoA3−. LAMA5 was highly expressed after treatment. (B) Dysregulation of different collagens confirmed by qPCR.
Discussion
In this study, we generated a novel model of integrin α3 negative podocytes using CRISPR/Cas9 technology [26]. These cells can terminally differentiate, and preserve their morphology and adhesion in standard culture conditions. By employing RNASeq, we identify the main changes of the podocyte-matrix adhesion complex and the regulations of cell-matrix signaling which occur when these podocytes that lose the α3 integrin subunit. This cell autonomous model reflects the primary impact of loss of this integrin receptor subunit on the cells, and does not aim to consider the crosstalk between glomerular cells like other studies [27].
The gene expression pattern of the AB8/13 podocyte cell line used in this study is largely consistent with previous transcriptomic and proteomic studies in mouse and human systems [27], [28]. In PodoA3−, we found that 19% of the genes encoding extracellular proteins were significantly regulated, most of them downregulated. Notably, we observed that in the absence of integrin α3β1, expression of LAMA1, a component of immature glomerular basement membrane was upregulated, while LAMA5 found in mature glomerular basement membrane was downregulated as compared to control podocytes [29], [30]. Our results suggest that podocytes shift their integrin repertoire in the absence of the α3 subunit. β1 binding α subunits – α5, α7 and α11 - were downregulated. Decreased adhesion after α2 blocking of PodoA3−, but not of control suggest this as the main β1 binding integrin in the absence of α3. In addition, αxβ2 integrins were upregulated. Accordingly, collagen IV, the main integrin α2β1 and αxβ2 ligand was also upregulated in PodoA3−. Although these integrins are not known to assure cell-matrix adhesion in podocytes, they might contribute to adhesion in vitro in this model. In agreement with this gene expression pattern, shape and adhesion of podocytes devoid of the integrin α3 subunit appeared unchanged on plastic and on collagen IV when the medium included 10% FBS (not shown). FBS provides fibronectin, which might lead to cell-adhesion through non-laminin dependent integrins, e.g. α5β1 [31]. Since ITGA5 was downregulated in PodoA3−, we assume that this integrin does not play a distinct role in cell adhesion in α3 negative podocytes, but rather α2β1. When the medium lacked FBS, adhesion and spreading of PodoA3− on laminin 511 was significantly reduced, and cells displayed multiple protrusions and retractions. Together these observations suggest that in this cell model, in the absence of α3, collagen mediated adhesion is preferred.
Our results on PodoA3− are in line with a recent study that dissected how basement membrane ligands determine cell shape and the adhesome. Specifically, epithelial cells spread faster on type IV collagen than on laminin 511 or 521, and adapted a circular shape with a large lamellipodium [6]. A combination of α1β1, α2 β1 and α5β1 was engaged when podocytes spread on collagen IV while integrin α3β1 was utilized to a greater degree by podocytes when spreading on laminin [6]. These mechanisms seem to apply to different epithelial cells [6].
While most genes encoding components of the integrin adhesome were downregulated in PodoA3−, previous gene expression studies of integrin α3 deficient keratinocytes demonstrated an upregulation of most of the integrin adhesome suggesting compensatory mechanisms in this cell type [32]. For instance, COL5A1 was distinctly downregulated in kidney cells whereas an upregulation of this gene was found in integrin α3 deficient keratinocytes. So far, little is known about the function of type V collagen in kidneys. In one previous study type V collagen was decreased in type I diabetic patients suffering from a progressive early function decline [33]. COL17A1, recently identified in the glomerular basement membrane was one of the few genes for transmembrane adhesion proteins probably compensatory upregulated in PodoA3−. Deficiency of type XVII collagen was previously shown to cause effacement of podocyte foot processes [34]. An explanation for the differential impact of loss of integrin α3 in kidney and skin epithelial cells might be that adhesion and wound healing in skin are compensated by e.g. integrin α6β4 in the absence of integrin α3β1 [35]. In contrast, in podocytes the regeneration capacity is limited and the mechanism of impaired foot process stability leads to fibrosis [5], [36], [37]. One key player in the observed compensatory mechanisms, might be the PI3K/Akt signaling pathway which is found downregulated in PodoA3−. In contrast, we demonstrated previously that in keratinocytes Akt-activation is increased in integrin α3 negative keratinocytes compared to normal keratinocytes [32]. We suggest that this mechanism in skin is necessary for e.g. wound healing, which might be one factor for a mild skin but severe kidney phenotype in ILNEB [38].
In summary, in this study, we knocked-out ITGA3 in a human podocyte cell line. Loss of the integrin α3 subunit in podocytes lead to significant changes in gene expression in particular of the integrin adhesome, the extracellular matrix and the PI3K-Akt signaling pathway.
Material and methods
Cells
The normal podocyte cell line (AB8/13) immortalized by SV40-T gene and the proximal tubule epithelial cell line (HK2) immortalized by E6 and E7 genes from HPV from human kidney were kindly provided by Dr. Tobias Huber [39]. Podocytes were cultured either in 33 °C 5% CO2 or 37 °C 5% CO2, in Roswell Park Memorial Institute (RPMI) 1640 (Life Technologies GmbH) with 1% l-glutamin (Life Technologies GmbH), 10% fetal bovine serum (FBS, Life Technologies GmbH) and RPMI supplements (5 mg/500 ml insulin–transferrinsodium selenite supplement, Roche; 0,5 ml/500 ml sodium pyruvate MEM 100 ml, Life Technologies; 0,5 ml/500 ml MEM NON Essential, Life Technologies; 2,5 ml/500 ml Herpes Buffer 1 M, Life Technologies). Human kidney-2 (HK2) cells derive from an immortalized proximal tubule epithelial cell line from normal adult human kidney widely.[21] HK2 cells were cultured in 37 °C 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) with 10% FBS, 2% l-glutamine and 1% sodium pyruvate.
Generation of the ITGA3 knock-out podocytes and HK2 cells using CRISPR/Cas9 lentivirus
About 0.3 to 0.4*106 podocytes or HK2 cells were seeded in 6-well plates and incubated overnight (for podocytes 33 °C 5% CO2, for HK2 cells 37 °C 5% CO2). Subsequently, 2 µl polybrene, 10 µl Cas9 and 10 µl CRISPR were added (LentiArray Cas9 Lentivirus, Life Technologies, Supplementary Table 7). After another two days, cells were selected by adding blasticidin (5 µg/ml) and puromycin (2 µg/ml) [26].
Subcloning of cell lines
Confluent CRISPR/Cas9 treated cells were trypsinized, spun down, and counted on a Neubauer counting chamber. Cells were plated at a density of 1–2 cells per 100 µl on a 96-well plate, splitted at confluency by trypsin digestion (Pan Biotech) and resuspended on the next larger flask.
Induction of podocyte differentiation
Podocytes grew either undifferentiated at the permissive temperature of 33 °C (in 5% CO2) for proliferation or at the nonpermissive temperature of 37 °C (in 5% CO2) for 7–14 days to inactivate the SV40 T antigen and allow the cells to differentiate [20].
Cell adhesion assay
Differentiated podocytes described above were used for this assay. Coverslips on 24-wells plates were coated with human laminin 511 (1 µg/cm2, CC160, ECMatrix-511 E8 Laminin Substrate, Sigma-Aldrich over night) at 37 °C. Plates were washed with PBS and blocked for 30 min with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Podocytes were resuspended in medium without FBS and equal numbers (4*103) were seeded onto the plates. Cells were incubated at 37 °C for different time points (60 min, 120 min, 180 min). Non-adherent cells were washed away with PBS and adherent cells on the coverslips were fixed using 2% paraformaldehyde (PFA) in PBS for 15 min. PFA solution was washed away using PBS and cells were stained by 4′,6-Diamidin-2-phenylindol (DAPI) (1:2000, Sigma-Aldrich Chemie GmbH) and phalloidin (1:2000, Invitrogen) for 1 h at room temperature. The antibodies were washed away using PBS three times and coverslips were transferred onto specimen slides using one drop of DAKO fluorescence mounting medium. Adherent cells were quantified using ImageJ.
Fluorescence activated cell sorting (FACS)
Podo and PodoA3− were stained after 5 min of trypsin-detachment and 3 times washing in FACS-buffer (DMEM + 10%FCS) with anti-integrin α3 antibody (Millipore P1B5; 1:50) or anti-CD151 antibody (Abcam 11G5a, 1:100) for 15 min at room temperature. Staining was visualized by counterstaining with anti-mouse secondary antibody Alexa594 (Invitrogen) (1:2000). Isotype controls consisted of mIgG1 antibodies counterstained with Alexa594 as before (not shown). Dead cells were excluded during analysis according to FSC/SSC settings and DAPI staining; 10,000 living cells were acquired. Analysis was performed on a FACS CantoII (BD) with data analysis using FlowJo V10.7.2.
DNA isolation
DNA from cells were isolated after trypsin digestion and adding of 20 µl proteinase K (Qiagen) to the cell pellet. DNA isolation was performed by using QIAmp DNA FFPE Mini Kit (Qiagen, Hilden, Germany).
RNA isolation and qPCR
RNA from differentiated cells that were cultured as described above (in RPMI and 10% FBS) was isolated after trypsin digestion and adding of RLT buffer (Qiagen) and mercaptoethanol to the cell pellet. RNA isolation was performed by using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) as described previously. RNA was transcribed into cDNA (First Strand cDNA Synthesis Kit 100, Life Technologies GmbH) and subjected to quantitative real-time PCR using iQ™ SYBR® Green Supermix and Biorad CFX96 Real-Time PCR Detection System (Bio-Rad, Munich, Germany). The data were analyzed using the Bio-Rad CFX Manager Software (version 1.5). Primers are listed in Supplementary Table 8.
Immunofluorescence staining
Cells were seeded on uncoated coverslips in 24-wells plates and allowed to grow for two days, fixed using 2% PFA in PBS as described above. Cells were then permeabilized using 0.1% Triton X-100 in PBS for 5 min. After washing with PBS, 100 µl of primary antibody diluted in 1% BSA in PBS was added to the coverslips and left to incubate overnight at 4 °C. On the next day, cells were washed three times using PBS. Nuclei were visualized with DAPI (1:2000 in PBS, Sigma-Aldrich Chemie GmbH) and cytoskeleton was visualized by phalloidin staining (1:2000 in PBS, Invitrogen). Images were captured by using immunofluorescence microscopy (Zeiss Axio Imager, Zeiss, Germany). Primary and secondary antibodies are included in Supplementary Tables 9 and 10.
Protein extraction and immunoblotting
The two cell types were lysed with a buffer containing 25 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, 1 mM PEFA-Bloc, 2 mM EDTA and protease inhibitor cocktail set (Merck Chemicals) and phosphatase inhibitor cocktail (Sigma-Aldrich). For immunoblotting, equal amounts of proteins were separated on 10% SDS-PAGE under reducing conditions and immunoblotted and then transferred onto nitrocellulose. For detection of laminin α5, equal amounts of proteins were separated on 4% SDS-PAGE under reducing conditions and immunoblotted, and then transferred onto PVDF. The membranes were incubated with primary antibodies overnight at 4 °C, followed by incubation with horseradish peroxidase coupled secondary anti-mouse, anti-goat and anti-rabbit IgG antibodies (Biorad) for one hour at room temperature. Visualization followed with the ECL detection reagent (Amersham, Billerica, USA) and the Fusion system (PeQlab, Germany). The intensities of the bands were quantified with ImageJ [10]. Primary and secondary antibodies are included in Supplementary Tables 9 and 10.
RNA sequencing and quality control
RNA isolated from cells as previously described was measured for its integrity by running agarose gel (Thermo Fischer). Samples were submitted to Novogene and were sequenced by NovaSeq 6000 (Illumina).
Data analysis and statistical analysis
Interactive network of the proteins was visualized by using the software STRINGDB (https://string-db.org/) and Cytoscape (http://www.cytoscape.org/). Genes regulated at least 2-fold [±1 log2] in RNA-sequencing were selected for analysis.
For correlation analysis between samples, Pearson correlation was used, as an important evaluating indicator to test the reliability of the experiment. The more the correlation coefficient is close to 1, the higher the similarity of the samples. Co-expression Venn diagram among groups of significantly expressed genes were screened (default threshold of FPKM values was set to 1) and then summarized to draw the venn diagrams presenting the number of different expressed genes in each group and the overlaps between groups. Volcano plots were used to infer the overall distribution of different expression genes. For the experiment with biological replicates, as the DESeq has already eliminated the biological variations, our threshold was set as: padj < 0.05.
For comparative analysis between keratinocytes and podocytes lacking integrin α3, we used our generated data from RNASeq (PodoA3−) and compared these data to previously obtained data by microarrays (KeraA3−) [32]. Genes that were regulated 2-fold were included in the analysis.
Data is presented as mean ± standard error of the mean (SEM), when applicable. We assessed differences between groups with Welch's t-test, assuming non-parametric distribution (GraphPad Prism, version 5.03). Differences were considered statistically significant when p ≤ 0.05.
GO enrichment and KEGG pathways
Gene Ontology(GO, https://www.geneontology.org/) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. GO types include cellular component, biological process and molecular function while we focused on biological processes. All differentially expressed genes genes with GO annotation were analyzed according to GO enrichement analysis. Hypergenometric P-value was calculated. For KEGG pathways enrichment analysis, differential expressed genes with pathway annotation were analyzed using hypergenometric t-test. Significance level was set at corrected p-value <0.05 for both GO and KEGG analysis.
Molecular cloning, site mutagenesis
Integrin α3 full length cDNA was amplified via PCR (forward primer with BglII restriction site 5′-actaaagatctatgctgtcatccactgacttta-3′ and reverse primer with HpaI restriction site 5′-caagttaactcaatcctgaccgccggt-3′) with Phusion high fidelity DNA polymerase (Fermentas, St. Leon-Rot, Germany). PCR products were ligated into the retroviral vector pMIG (Bioss centre for biological signalling studies, University Freiburg), using the restriction sites for BglII and HpaI and T4 DNA ligase (New England Biolabs, Frankfurt, Germany). The plasmids were transformed into competent Escherichia coli (DH5α, Invitrogen), DNA was purified and positive clones were confirmed by sequencing.
Transfection and retroviral transduction
Three µg of the retroviral construct and the helper plasmids pHit60 and pVSV-G were co-transfected into 30% confluent Hek293 cells with Superfect transfection reagent (Qiagen), according to the manufacturers’ instructions. Production of amphotropic retroviral particles was stimulated by adding 5 mM sodium-butyrate (Sigma-Aldrich) for 8 h. 24 and 48 h after stimulation, the supernatant containing retroviral particles was collected, sterilized trough a 0.22 µm syringe filter (Nalgene, Thermo Scientific), polybrene was added at a final concentration of 5 µg/ml (Millipore, Schwalbach, Germany) and cleared supernatants were stored at −80 °C. Podo and PodoA3− were transduced with 100 µl retroviral supernatant in 1 ml podocyte growth medium twice. Positively transduced cells were selected by mono-flow cytometry sorting employing the IRES GFP cassette in pMIG vector. Sorted new cell lines were designated according to the expression of the recombinant proteins: as Podo (empty vector), PodoA3− and PodoA3+.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
L.F., L.T. and Y.H. were supported by the Else-Kröner-Fresenius foundation. C.H. was supported by the Deutsche Forschungsgemeinschaft (DFG) CRC/SFB 1140. The authors thank Dr. Tobias Huber for providing the cell lines AB8/13 and HK2.
Author contributions
CH designed the study and analyzed the results; LF performed most of the experiments, analyzed the results and prepared the figures; SBS, YW, YH, JL, LT, PE performed experiments and analyzed data. All authors interpreted the data. LF and CH drafted the manuscript. All authors revised the manuscript and approved the submitted version.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2022.100119.
Contributor Information
L.H. Frommherz, Email: leonie.frommherz@med.uni-muenchen.de.
C. Has, Email: cristina.has@uniklinik-freiburg.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sachs N., Sonnenberg A. Cell-matrix adhesion of podocytes in physiology and disease. Nat. Rev. Nephrol. 2013;9(4):200–210. doi: 10.1038/nrneph.2012.291. [DOI] [PubMed] [Google Scholar]
- 2.Longmate W., DiPersio C.M. Beyond adhesion: emerging roles for integrins in control of the tumor microenvironment. F1000Res. 2017;6:1612. doi: 10.12688/f1000research.11877.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pozzi A., Zent R. Integrins in kidney disease. J. Am. Soc. Nephrol. 2013;24(7):1034–1039. doi: 10.1681/ASN.2013010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreidberg J.A., Donovan M.J., Goldstein S.L., Rennke H., Shepherd K., Jones R.C., Jaenisch R. Alpha 3 beta 1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122(11):3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- 5.Sachs N., Kreft M., van den Bergh Weerman M.A., Beynon A.J., Peters T.A., Weening J.J., Sonnenberg A. Kidney failure in mice lacking the tetraspanin CD151. J. Cell Biol. 2006;175(1):33–39. doi: 10.1083/jcb.200603073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randles M.J., Lausecker F., Humphries J.D., Byron A., Clark S.J., Miner J.H., Zent R., Humphries M.J., Lennon R. Basement membrane ligands initiate distinct signalling networks to direct cell shape. Matrix Biol. 2020;90:61–78. doi: 10.1016/j.matbio.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Has C., et al. Integrin alpha3 mutations with kidney, lung, and skin disease. N. Engl. J. Med. 2012;366(16):1508–1514. doi: 10.1056/NEJMoa1110813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y., et al. Constitutional absence of epithelial integrin alpha3 impacts the composition of the cellular microenvironment of ILNEB keratinocytes. Matrix Biol. 2018;74:62–76. doi: 10.1016/j.matbio.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Shukrun R., et al. A human integrin-alpha3 mutation confers major renal developmental defects. PLoS ONE. 2014;9(3):e90879. doi: 10.1371/journal.pone.0090879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yalcin E.G., et al. Crucial role of posttranslational modifications of integrin alpha3 in interstitial lung disease and nephrotic syndrome. Hum. Mol. Genet. 2015;24(13):3679–3688. doi: 10.1093/hmg/ddv111. [DOI] [PubMed] [Google Scholar]
- 11.Nicolaou N., et al. Gain of glycosylation in integrin alpha3 causes lung disease and nephrotic syndrome. J. Clin. Invest. 2012;122(12):4375–4387. doi: 10.1172/JCI64100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y., Balasubramanian M., Humphreys N., Waruiru C., Brauner M., Kohlhase J., O'Reilly R., Has C. Intronic ITGA3 mutation impacts splicing regulation and causes interstitial lung disease, nephrotic syndrome, and Epidermolysis Bullosa. J. Invest. Dermatol. 2016;136(5):1056–1059. doi: 10.1016/j.jid.2015.11.031. [DOI] [PubMed] [Google Scholar]
- 13.Colombo E.A., Spaccini L., Volpi L., Negri G., Cittaro D., Lazarevic D., Zirpoli S., Farolfi A., Gervasini C., Cubellis M.V., Larizza L. Viable phenotype of ILNEB syndrome without nephrotic impairment in siblings heterozygous for unreported integrin alpha3 mutations. Orphanet. J. Rare Dis. 2016;11(1) doi: 10.1186/s13023-016-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen-Barak E., Danial-Farran N., Khayat M., Chervinsky E., Nevet J.M., Ziv M., Shalev S.A. A nonjunctional. JAMA Dermatol. 2019;155(4):498. doi: 10.1001/jamadermatol.2018.5368. [DOI] [PubMed] [Google Scholar]
- 15.Sawada K., et al. Upregulation of alpha3beta1-integrin in podocytes in early-stage diabetic nephropathy. J. Diabetes Res. 2016;2016:9265074. doi: 10.1155/2016/9265074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H.C., et al. Altering expression of alpha3beta1 integrin on podocytes of human and rats with diabetes. Life Sci. 2000;67(19):2345–2353. doi: 10.1016/s0024-3205(00)00815-8. [DOI] [PubMed] [Google Scholar]
- 17.Hamano Y., Grunkemeyer J.A., Sudhakar A., Zeisberg M., Cosgrove D., Morello R., Lee B., Sugimoto H., Kalluri R. Determinants of vascular permeability in the kidney glomerulus. J. Biol. Chem. 2002;277(34):31154–31162. doi: 10.1074/jbc.M204806200. [DOI] [PubMed] [Google Scholar]
- 18.Pavenstadt H., Kriz W., Kretzler M. Cell biology of the glomerular podocyte. Physiol. Rev. 2003;83(1):253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 19.Sever S., Schiffer M. Actin dynamics at focal adhesions: a common endpoint and putative therapeutic target for proteinuric kidney diseases. Kidney Int. 2018;93(6):1298–1307. doi: 10.1016/j.kint.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saleem M.A., O’Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 21.Ryan M.J., Johnson G., Kirk J., Fuerstenberg S.M., Zager R.A., Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45(1):48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 22.Shankland S.J., Pippin J.W., Reiser J., Mundel P. Podocytes in culture: past, present, and future. Kidney Int. 2007;72(1):26–36. doi: 10.1038/sj.ki.5002291. [DOI] [PubMed] [Google Scholar]
- 23.Sakairi T., Abe Y., Kajiyama H., Bartlett L.D., Howard L.V., Jat P.S., Kopp J.B. Conditionally immortalized human podocyte cell lines established from urine. Am. J. Physiol. Renal Physiol. 2010;298(3):F557–F567. doi: 10.1152/ajprenal.00509.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen B.P., Gil S.G., Carter W.G. Deposition of laminin 5 by keratinocytes regulates integrin adhesion and signaling. J. Biol. Chem. 2000;275(41):31896–31907. doi: 10.1074/jbc.M006379200. [DOI] [PubMed] [Google Scholar]
- 26.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byron A., Randles M.J., Humphries J.D., Mironov A., Hamidi H., Harris S., Mathieson P.W., Saleem M.A., Satchell S.C., Zent R., Humphries M.J., Lennon R. Glomerular cell cross-talk influences composition and assembly of extracellular matrix. J. Am. Soc. Nephrol. 2014;25(5):953–966. doi: 10.1681/ASN.2013070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boerries M., Grahammer F., Eiselein S., Buck M., Meyer C., Goedel M., Bechtel W., Zschiedrich S., Pfeifer D., Laloë D., Arrondel C., Gonçalves S., Krüger M., Harvey S.J., Busch H., Dengjel J., Huber T.B. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83(6):1052–1064. doi: 10.1038/ki.2012.487. [DOI] [PubMed] [Google Scholar]
- 29.Miner J.H., et al. The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8–11, and cloning of a novel alpha3 isoform. J. Cell Biol. 1997;137(3):685–701. doi: 10.1083/jcb.137.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naylor R.W., Morais M., Lennon R. Complexities of the glomerular basement membrane. Nat. Rev. Nephrol. 2021;17(2):112–127. doi: 10.1038/s41581-020-0329-y. [DOI] [PubMed] [Google Scholar]
- 31.Akiyama S.K. Integrins in cell adhesion and signaling. Hum. Cell. 1996;9(3):181–186. [PubMed] [Google Scholar]
- 32.Pazzagli C., et al. Absence of the integrin alpha3 subunit induces an activated phenotype in human keratinocytes. J. Invest. Dermatol. 2017;137(6):1387–1391. doi: 10.1016/j.jid.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Merchant M.L., Perkins B.A., Boratyn G.M., Ficociello L.H., Wilkey D.W., Barati M.T., Bertram C.C., Page G.P., Rovin B.H., Warram J.H., Krolewski A.S., Klein J.B. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J. Am. Soc. Nephrol. 2009;20(9):2065–2074. doi: 10.1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurskainen T., Moilanen J., Sormunen R., Franzke C.-W., Soininen R., Loeffek S., Huilaja L., Nuutinen M., Bruckner-Tuderman L., Autio-Harmainen H., Tasanen K. Transmembrane collagen XVII is a novel component of the glomerular filtration barrier. Cell Tissue Res. 2012;348(3):579–588. doi: 10.1007/s00441-012-1368-x. [DOI] [PubMed] [Google Scholar]
- 35.Kligys K.R., et al. alpha6beta4 integrin, a master regulator of expression of integrins in human keratinocytes. J. Biol. Chem. 2012;287(22):17975–17984. doi: 10.1074/jbc.M111.310458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longmate W.M., Dipersio C.M. Integrin regulation of epidermal functions in wounds. Adv. Wound Care (New Rochelle) 2014;3(3):229–246. doi: 10.1089/wound.2013.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiPersio C.M., Zheng R., Kenney J., Van De Water L. Integrin-mediated regulation of epidermal wound functions. Cell Tissue Res. 2016;365(3):467–482. doi: 10.1007/s00441-016-2446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitsialos G., Chassot A.-A., Turchi L., Dayem M.A., LeBrigand K., Moreilhon C., Meneguzzi G., Buscà R., Mari B., Barbry P., Ponzio G. Transcriptional signature of epidermal keratinocytes subjected to in vitro scratch wounding reveals selective roles for ERK1/2, p38, and phosphatidylinositol 3-kinase signaling pathways. J. Biol. Chem. 2007;282(20):15090–15102. doi: 10.1074/jbc.M606094200. [DOI] [PubMed] [Google Scholar]
- 39.Huber T.B., Edelstein C.L., Hartleben B., Inoki K., Jiang M., Koya D., Kume S., Lieberthal W., Pallet N., Quiroga A., Ravichandran K., Susztak K., Yoshida S., Dong Z. Emerging role of autophagy in kidney function, diseases and aging. Autophagy. 2012;8(7):1009–1031. doi: 10.4161/auto.19821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.