Abstract
One of the primary difficulties in treating patients with genetic syndromes is diagnosing their condition. Many syndromes are associated with characteristic facial features that can be imaged and utilized by computer-assisted diagnosis systems. In this work, we develop a novel 3D facial surface modeling approach with the objective of maximizing diagnostic model interpretability within a flexible deep learning framework. Therefore, an invertible normalizing flow architecture is introduced to enable both inferential and generative tasks in a unified and efficient manner. The proposed model can be used (1) to infer syndrome diagnosis and other demographic variables given a 3D facial surface scan and (2) to explain model inferences to non-technical users via multiple interpretability mechanisms. The model was trained and evaluated on more than 4700 facial surface scans from subjects with 47 different syndromes. For the challenging task of predicting syndrome diagnosis given a new 3D facial surface scan, age, and sex of a subject, the model achieves a competitive overall top-1 accuracy of 71%, and a mean sensitivity of 43% across all syndrome classes. We believe that invertible models such as the one presented in this work can achieve competitive inferential performance while greatly increasing model interpretability in the domain of medical diagnosis.
Index Terms—: Genetic Syndrome, Normalizing Flow, Interpretable Machine Learning, 3D Shape Model
I. Introduction
Diagnosing human genetic syndromes is a complex and difficult process due to their diversity, subtle differences between subjects with different syndromes, and their rarity. Genetic testing is the ideal way to diagnose afflicted subjects, but these tests are expensive, genetic experts are often scarce, and many syndromes exist for which the genetic profile is not yet known. As a supplement to genetic testing, computer-assisted phenotyping based on facial images or scans has been proposed as a low-cost, easy to utilize, and entirely non-invasive strategy for genetic syndrome screening [1].
The facial morphology associated with a syndrome can be quite distinctive and it has been shown that facial shape features are useful diagnostic indicators for many syndromes [1]–[4]. Experienced clinical geneticists will often use facial morphology as a preliminary diagnostic indicator prior to genetic testing. The development of robust and fully automatic computational pipelines to analyze facial morphology would, therefore, allow non-expert clinicians all across the globe to utilize a unified quantitative understanding of syndromic facial morphology to support clinical decision making processes at a very low cost.
Face-based syndrome detection models have been developed for both 2D facial images and 3D facial surface scans. State-of-the-art 2D approaches such as [5], [6] commonly use deep learning-based discrimintative models. However, single uncalibrated 2D images cannot capture facial morphology with the fidelity of 3D surface imaging [4]. Furthermore, deep discriminative models often lack interpretability, which makes it difficult for clinicians to understand what information a model uses to make inferences. Model interpretability is particularly important in medical applications as many clinicians hesitate to introduce black box models into their decision making process. On the other hand, state-of-the-art approaches relying on more informative 3D surface scans like [4] commonly utilize point-based generative shape models [7] that are equipped with an inference mechanism (e.g., a regularized discriminant analysis variant as proposed in [4]). While these models are more interpretable than solely discriminative ones, they usually rely on simplified facial representations (e.g., a sparse set of landmarks) and simplified probabilistic assumptions (e.g., Gaussian distributions). Both of those properties restrict model flexibility in ways that may impact model performance. 3D facial morphology may not follow a Gaussian distribution, and sparse landmarks may be incapable of capturing important, subtle shape details.
In this work, we propose a novel deep learning-based, invertible 3D facial surface modeling approach. The main novelty of our method is two-fold: (1) In contrast to standard facial shape analysis methods used for syndrome data modeling (often limited to Gaussian distributions), our NF model can learn complex non-Gaussian conditional face distributions. (2) Our model is fully invertible and, as a result of this, is highly multi-functional. Specifically, the proposed model is the first non-Gaussian 3D facial shape model with the ability to (1) infer syndrome diagnosis and other demographic variables given a high-resolution 3D facial surface scan, (2) generate modal, randomly sampled, and counterfactual 3D faces using demographic information, and (3) analyze the magnitude of facial variation between and within demographic groups (e.g., males vs. females) in a fully probabilistic way. The proposed normalizing flow approach efficiently handles all tasks within a single unified probabilistic model. The results of the evaluation show how this multi-functionality can help non-technical clinicians to intuitively understand and gain confidence in the model and its inference process through, for example, counterfactual visualizations. To the best of our knowledge, a deep invertible model of 3D syndromic facial morphology has not been proposed before.
(A). Related Work
1). Face-based Syndrome Classification:
In contrast to the volumes of work on general facial shape modeling and recognition [8], approaches specifically designed to diagnose genetic syndromes are scarce. Many available machine learning-based syndrome classification methods [5], [6], [9]–[12] rely on 2D frontal facial images of the subject as they are widely available in a clinical setting. However, this usually restricts the set of input features to projected geometric information and texture data, which may limit the achievable classification accuracy [9]. This problem can be alleviated by using 3D geometric information from 3D surface scans [3], [4] directly acquired via 3D scanning techniques [1]. As a low-cost alternative to real 3D data acquisition, some authors propose to infer 3D shape information for diagnostic purposes from 2D images [13], [14] by fitting a 3D face model [15]. Most approaches were developed and evaluated using a small number of syndrome classes.
2). Generative 3D Facial Shape Modelling:
Facial shape modelling generally aims at estimating a low-dimensional manifold of typical shape variations together with a probability density based on available high-dimensional training data (e.g., contours or meshes). Historically, this has been mostly achieved by linear approaches [7], [16] that define linear subspaces and use simple (often Gaussian) densities (see [8] for an overview). Over the years, those efforts have led to a multitude of so-called 3D morphable face models (3DMMs; e.g., [15], [17], [18]). While many 3DMMs successfully disentangle certain semantically meaningful factors of variation like identity and expressions, conditioning them on additional demographic variables is not common or straightforward (e.g., in [18] several 3DMMs are built to independently capture age and sex variations) and available solutions for conditional shape modeling [19] typically rely on Gaussian distributions.
3). Deep Learning-based 3D Facial Shape Modelling:
More recently, the first deep learning-based 3DMMs have been proposed [20]–[25] that make use of specifically adapted versions of variational auto-encoders (VAE) and generative adversarial networks (GAN). In contrast to traditional 3DMMs, their inherent non-linearity allows them to represent more complex manifolds and probability densities, which may lead to models that better capture the data [21]. A unique challenge associated with processing 3D surface meshes is their special graph-like structure. Hence, popular operations widely used in imaged-based deep learning solutions either need to be specifically adapted (e.g., spectral graph convolutions [21]–[23]) or the data needs to be reparameterized accordingly [25]. Although VAEs and GANs are excellent for generating visually convincing synthetic data samples, they are not well suited for tasks that involve evaluating the likelihood of samples. Both GANs and VAEs require computationally expensive Monte-Carlo integration, which can be intractable for high dimensional data, or lower-bound approximations to estimate likelihood values [26], [27]. In contrast, invertible normalizing flow models are designed to support efficient and exact likelihood evaluation.
4). Normalizing Flows:
Normalizing flows (NF) are a recently proposed class of deep learning model. A NF model represents a learnable bijective function (see reviews in [28], [29]). NFs are most commonly applied to generative manifold and density estimation tasks much like VAEs and GANs [30]. However, in contrast to VAEs where two separate models (encoder and decoder) are trained to map to and from a latent variable space, NFs are able to perform encoding and decoding using the forward and inverse directions of a single unified model. This avoids consistency issues often seen when modeling both directions independently. Furthermore, unlike GANs and VAEs, the likelihood of a NF model can be evaluated efficiently and exactly. This allows for direct maximum likelihood-based training, Bayesian inference of condition variables, and estimation of information theoretic measures like KL-divergence and differential entropy.
Recently, the first NF models operating on point clouds or mesh data were described (e.g., [31], [32]), but we are not aware of any work specifically using NFs to build syndromic 3D face models.
II. Methods
In this section, we will first introduce our notation and describe necessary data pre-processing steps before our NF architecture is described in Sec. II-C. We then describe in Sec. II-D how the model can be used to perform Bayesian inference of syndrome classes and demographic variables before different probabilistic interpretability mechanisms are described in Secs. II-E and II-F.
The overarching goal of our modelling approach is to efficiently approximate the distribution of human facial surface morphology conditioned on genetic syndrome diagnosis, age, and sex. For model training, we assume a population of npop subjects to be given. Each tuple (Si, yi) consists of a subject’s 3D facial surface mesh Si and a set of associated factors yi = {agei, sexi, syndi} with , sexi ∈ {male, female}, and syndrome diagnosis syndi ∈ Γ, where Γ is a set of clinical genetic syndrome classes including a class for unaffected (non-syndromic) people.
A. Reference Surface and Registration
Depending on the actual 3D scanning and reconstruction techniques employed to capture the facial scans Si of the training population, the number of vertices to represent each discrete surface and their topology may vary considerably between subjects. We, therefore, normalize all scans to a reference topology with a fixed number of vertices located at corresponding locations for each subject. This is done by first registering a fixed template mesh with |V | = nvert vertices to all npop scans Si (see Sec. III-B for details). This results in non-linear transformations that are used to propagate the template’s vertices to the subject scans. Finally, positional and rotational information is removed and each surface is vectorized by stacking the 3D point coordinates of all nvert vertices to obtain vectors .
B. Manifold Estimation
Facial surface meshes produced by modern scanners commonly contain tens of thousands of vertices resulting in very high-dimensional surface vectors . Estimating a probability density on this very high-dimensional space is both computationally challenging and unnecessary since the positions of neighboring points on densely sampled facial surfaces will naturally have high mutual information. Therefore, we construct our model as a probability density on a sub-manifold of the ambient data space (see [30] for a description of how manifold and density estimation relate within a NF framework).
As in standard linear shape modeling approaches [7], we assume that the surface vectors si can be accurately represented by a nsub-dimensional Euclidean manifold of maximum data variation F estimated using principal components analysis (PCA) of the training samples. F is then spanned by the first nsub principal components with descending eigenvalue magnitude and centered at the training sample mean . More specifically, all elements of sub-manifold F are identified by the set . Surface vectors si are projected to F in a least squares sense [7], which results in low-dimensional vectors with . nsub is selected to be as low as possible without negatively impacting syndrome classification performance so that the manifold projection removes only diagnostically unimportant information. The manifold projection can be easily inverted as in standard shape models [7] via .
C. Model Architecture
Let denote the training tuples consisting of the dimensionality-reduced face representations and the associated conditioning factors. We now aim to estimate the conditional probability distribution pF (f |y) of facial surface morphology on manifold F. Mathematically, our NF model represents a bijective function with trainable parameters θ that maps elements z of a nsub-dimensional latent space Z to a nsub-dimensional space F of facial morphology f while imposing conditions y. We chose a simple Gaussian base distribution pZ (z) = pZ (z|y) = N (0, I) for the latent variable space Z. This allows us to express the the potentially non-Gaussian conditional distribution of interest pF (f |y) using the change of variables theorem [33]:
Here, denotes the Jacobian determinant of g−1 (f; y, θ). To convert Eq. (1) into a tractable NF model, function g(·; y, θ) must be specified such that it is efficiently invertible and possesses a tractable jacobian determinant. We first split into a chain of nlay simpler sub-functions (called layers in NF models). The different types of layers used in our model will be described first, followed by a summary of how the layers are composed to create the full NF model (see also Fig. 1).
Fig. 1.
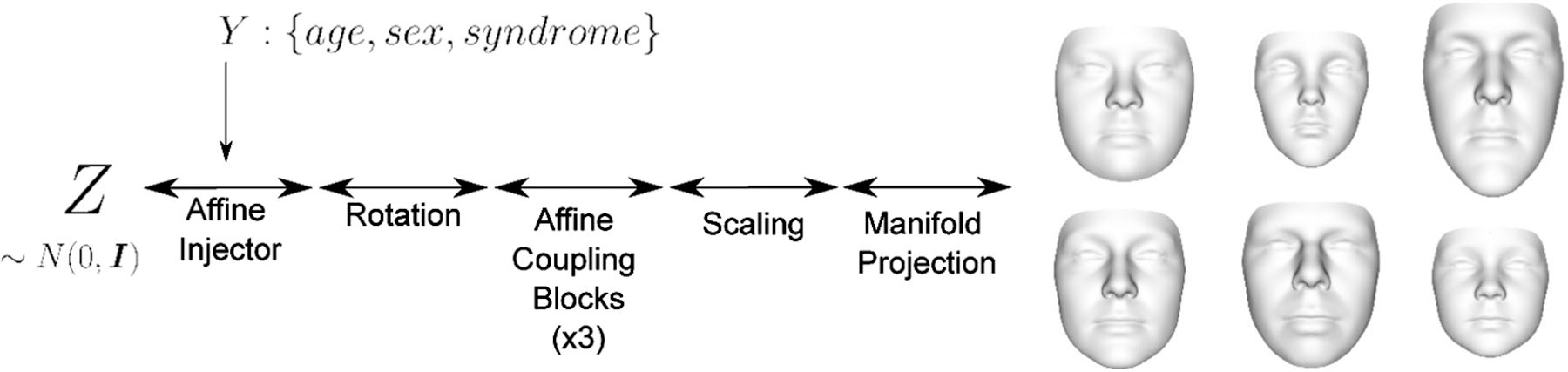
The proposed normalizing flow-based 3D facial shape model is a non-linear, invertible bijection (indicated by bidirectional arrows) between a normally distributed latent space Z and a linear manifold embedded in a space of 3D facial surfaces. The bijection is modeled as a composition of bijective layers (affine injector, rotation, …) that successively transform the normal latent density pZ (z) to match the complex density on the manifold of 3D faces. Furthermore, the bijection is conditional on demographic variables age, sex, and genetic syndrome diagnosis. The synthetic faces on the right represent maximally probable faces (modes) on the linear manifold that are produced by the model for different combinations of demographic variables. The characteristic Down syndrome facial phenotype is clearly recognizable in the first column, first row. See sections II-B and II-C for a detailed explanation of the model architecture and mathematical notation.
1). Affine Injector:
Within our NF model, the first layer is the only conditional layer. It applies an affine transformation w =gi(u; y, θi) determined by the conditions y to each dimension of the layer’s input to generate the output :
The scaling s(·; θi) and translation t(·; θi) functions can be complex neural networks as the inverse of the layer can be computed without having to invert s(·; θi) or t(·; θi) via
and its Jacobian has a simple triangular structure [34]. We will refer to this type of layer as an affine injector following [35]. We choose s(·; θi) and t(·; θi) to be fully-connected neural networks (two hidden layers, 100 neurons per layer and ELU activations) with partially shared weights θi. In consideration of the available training data, we also encode the assumption that the magnitude of facial shape variation along each dimension of the latent space does not depend on syndrome class by excluding synd as an input to the scaling network s(·; θi).
2). Rotation:
The second layer in our model is a trainable rotation layer. Rotation is an appealing transformation in this context due to its close relation to PCA and related methods for learning linear bases of data spaces. We use the Cayley transform to produce a smooth parameterization of the special orthogonal group SO(nsub) as discussed in [36]. Later in the model, we also use fixed random rotations to mix information between dimensions. Rotations are easily invertible and always have a Jacobian determinant of unity.
3). Affine Coupling Blocks:
The next layers of our NF model are affine coupling layers that were first introduced in [34]. Affine coupling layers also define an invertible affine transformation w = gi(u; θi) between their inputs and their outputs . Here, u1 and w1 denote the first nsub/2 dimensions and u2 and w2 represent the second half of the input and output vectors, respectively. The element-wise affine transformation is then defined as:
In our model, we use a volume preserving (and differential entropy preserving) variant of affine coupling layers where the Jacobian determinant of each layer is constrained to unity (see [37] for details). In practice, this constraint has a strong regularizing effect and can be enforced by subtracting the mean from the vector produced by the scaling function s(·, θi) such that it sums to zero. We choose s(·; θi) and t(·; θi) to be fully-connected neural networks (two hidden layers, 32 neurons per layer and ELU activations) with shared weights θi.
Permuting or mixing the inputs after each affine coupling layer is necessary because otherwise interactions between dimensions would be restricted. Therefore, we create affine coupling blocks consisting of two affine coupling layers separated by a permutation that reverses dimension order. At the end of each affine coupling block, we also place a random, fixed rotation that mixes the data as proposed in [33].
4). Scaling:
The final layer of our model is a fixed scaling layer. The fixed parameters of the layer are set once at the start of training according to the standard deviation of each dimension of the dimensionality reduced training data. The purpose of the final layer is to ensure that data representations are normalized as they pass through the other layers of the model while maintaining the information associated with data magnitude in the loss function via the Jacobian determinant of the scaling layer.
5). Layer Composition:
The composition order of the various flow layers used to create g(·; y, θ) is shown in Fig 1. The trainable layer parameters can be optimized using maximum likelihood training. We chose a multivariate Gaussian distribution with identity covariance as a prior for pZ (z) resulting in the negative log-likelihood loss
for training data . Although the optimization is carried out on the low-dimensional shape representations fi, it is equivalent to an optimization on manifold F in the high-dimensional ambient data space. This is due to the definition of F as a Euclidean manifold; the manifold projection described in Sec. II-B has a constant Jacobian determinant independent of the input data. [30], [38].
D. Bayesian Inference
To infer the syndrome class syndi of a subject i from facial surface morphology fi and demographic variables agei and sexi, the flow model can be used along with Bayes’ theorem to compute a posterior distribution:
Here, we can see that the inclusion of age and sex as conditioning variables enables our model to account for the effects of age and sex on facial morphology when making inferences. Furthermore, an identical approach can be used to infer subject age or sex. Although this is less relevant for clinical applications, it nicely demonstrates the multi-functionality of the proposed model. In addition to a trained flow model, we require a joint distribution for the condition variables p(age, sex, synd) in order to perform inference. In this work, we use a naïve prior that assumes condition variable independence:
In a clinical application the joint distribution of condition variables could be set and manipulated by the clinician using prior knowledge about the application context.
E. Face Generation
Data generation capabilities improve interpretability by enabling a model to visually answer questions about what information it has learned and what information it uses to make inferences. Two of the three interpretablity mechanisms of the proposed NF model demonstrated in this work involve data generation.
1). Demographic Specific Face Generation:
The first type of interpretability mechanism is intended to answer questions about what facial representations the model has learned for a particular demographic (e.g., “What do 8 year old males with Down syndrome look like?”). To address this, the model can be used to generate randomly sampled and maximally probable faces (modes) to exemplify general trends and typical variability. Clinicians can then visually assess the facial characteristics the model has learned.
For this task, our NF model can be used in the same way as a conditional VAE or GAN without requiring any additional training. First, a latent sample is drawn from the latent prior. The sample is then mapped using the NF and the specified condition variables from the latent space to the data space and rendered as a 3D surface mesh. Our model can also generate modes (maximally probable faces) for specific demographics by mapping the origin of the latent space (the mode of the latent prior) to the data space in the same way. This property of mode preservation is a consequence of our model architecture enabled through use of the affine injector layer and volume preserving coupling layers.
2). Counterfactual Face Generation:
The second interpretability mechanism is intended to answer questions about what facial information the model uses to justify particular inferences (e.g., “Why did the model infer the syndrome class of this subject as unaffected instead of Down syndrome?”). To address this, the model can be used to generate a counterfactual face, which represents what the model would expect a given subject to look like if they belonged, hypothetically, to a counterfactual demographic group. This counterfactual face visually shows, by contrast with the subjects true face, what facial information was used by the model to justify that particular inference. Counterfactual representations have been shown to be highly effective when explaining a models decision making process to non-technical users [39].
For this task, the original subject face is first mapped to the latent space using the NF and the predicted (or true) condition variables. Next, the subject’s latent representation is mapped back to the data space using the inverse direction of the flow and a different set of counterfactual condition variables. The original and counterfactual faces can then be visually compared.
F. Variation Analysis
The third interpretability mechanism is intended to answer questions about the magnitude of inter- and intra-demographic facial variation as captured by the model (e.g., “How much overlap is there between the facial morphology of males and females?”). This information can be presented to clinicians to help them assess and evaluate the model’s internal understanding of facial variation within and between demographic groups. Variance, co-variance, and variance-based statistics are commonly used within Gaussian modelling approaches to compute standardized effect sizes and magnitudes of variation. To address these questions using a non-Gaussian model such as the one presented in this work, we propose a more general information-based approach.
For this task, we compute Monte Carlo estimates of differential entropy
and KL divergence
for and between different demographic groups. Integration over F weighted by a probability distribution pF (f |yi) can be numerically approximated by sampling from the model, and the likelihood of samples under different demographic conditions can be efficiently evaluated using Eq. (1).
III. Experiments
The first aim of the evaluation is to show that the developed NF model can accurately infer syndrome diagnosis, age, and sex from 3D faces. We then use the same model to generate interpretability results that can be used by clinicians to evaluate whether the model bases its inferences on reasonable facial shape information, or noise and other imaging artefacts. Since the exact same NF model is used for a multitude of tasks, the results of various evaluation procedures are mutually supportive.
A. Data Description
The 4727 3D facial scans used to train and evaluate our model were acquired using 3DMD facial imaging systems1 and are available through the FaceBase Consortium2. Patients with cranio-facial syndromes were recruited through clinical geneticists at different sites across North America and have a clinical or molecular diagnosis. Each of the 47 syndromes in this analysis is represented by 20 or more subjects. 2600 of the 4727 subjects are presumed to be unaffected by a genetic syndrome. Ethics approval for this study was granted by the Conjoint Health Research Ethics Board (Id #: REB14-0340 REN4) at the University of Calgary
B. Data Pre-Processing
Each subject scan was landmarked with a set of eight guide points using a combination of manual landmarking and an image-based automatic algorithm [40]. An averaged template mesh (see Fig. 2) was then registered to each scan. This template mesh was initially non-linearly mapped to each subject scan using a thin plate spline transformation anchored by the corresponding guide points. Next, the non-rigid iterative closest point algorithm [41] was used to relax the template mesh completely onto the surface of the scan (see Fig. 2 for an example).
Fig. 2.

Top Left: the average template mesh annotated with eight guide points. Bottom Left: an example subject scan annotated with the same eight guide points. Top Right: the subject mesh (with color) overlaid with transformed template (white mesh). Bottom Right: the transformed template.
The topology of the template was designed so that there is a bijective mapping between bilaterally (across the median plane of the template) corresponding vertices. This bijection was used to produce a mirrored and symmetric version of each subject face as a form of data augmentation. Finally, information associated with facial position and rotation was removed from the template registration transformations and a data manifold was estimated using the approach described in Sec. II-B. For this analysis, we chose nsub = 100, which produced a manifold capturing 99.8% of the total variance in the training data.
C. Training and Evaluation
All NF models evaluated were trained for 1500 epochs using the NAdam optimizer with a learning rate of 10−3 and a batch size of 2056 using a Python-based implementation (Tensorflow 2.2.0 and Tensorflow-Probability 0.10.1) and a 2070 Super NVIDIA GPU with 8 GB memory. The full NF model has 68428 trainable parameters in total. Training a single NF model takes less than 1 hour. Mapping individual faces to and from the latent space is very fast (less than a second) and on par with a VAE comparison model. Importantly, the time to evaluate the conditional likelihood pF (fi|yi) of a 3D face fi using the NF model is comparable to performing a forward or inverse transformation. This is essential for efficient Bayesian inference and analysis of variation. Comparatively, approximating the conditional likelihood of a single sample using a cVAE model requires expensive Monte Carlo integration. The time to perform inference using the NF model is slightly longer compared to discriminative comparison models (MLP, PointNet) due to the need to sample multiple conditional likelihood values when computing a posterior distribution. The time to perform an analysis of variation varies with the number of random samples used to produce Monte Carlo estimates of the information-based statistics. The full analysis performed in this work completes in approximately one hour.
1). Inference:
For all inference experiments, model training was performed as described above using Monte Carlo cross validation with ten random train/test splits of the 4727 scans. Maximum a posteriori (MAP) estimates from posterior distributions as defined in Eq. (3) are used for all inference tasks and integer age values are sampled at an interval of one year during inference.
For the sake of comparison with baseline non-linear discriminative models, we also trained and evaluated a multi-layer perceptron (MLP) model (two hidden layers, 100 neurons per layer, and ELU activations) and a PointNet (PN) model [42] following the implementation of keras.io/examples/vision/pointnet/ on the same data splits as used for the proposed NF model. The MLP model uses the same dimensionality reduced data as the NF model while the PN model is applied to a randomly sampled subset of 3038 3D points from the dense surface meshes.
In order to compare the proposed model to previously proposed Gaussian modelling approaches, and to test if non-Gaussian models are valuable in this application, NF models were trained and evaluated with the affine coupling blocks removed from the architecture shown in Fig 1. This ablated architecture (LinearNF) has a bijection g(·; y, θ) that is linear with respect to the input (though not with respect to the condition variable y) so that the induced distribution pF (f |y) is always Gaussian.
2). Face Generation:
The NF model used to generate qualitative face generation results (Figures 3, 4) as well as for analysis of variation results (Sec. IV-C) was trained as described above but using the full set of scans described in Sec.III-A.
Fig. 3.
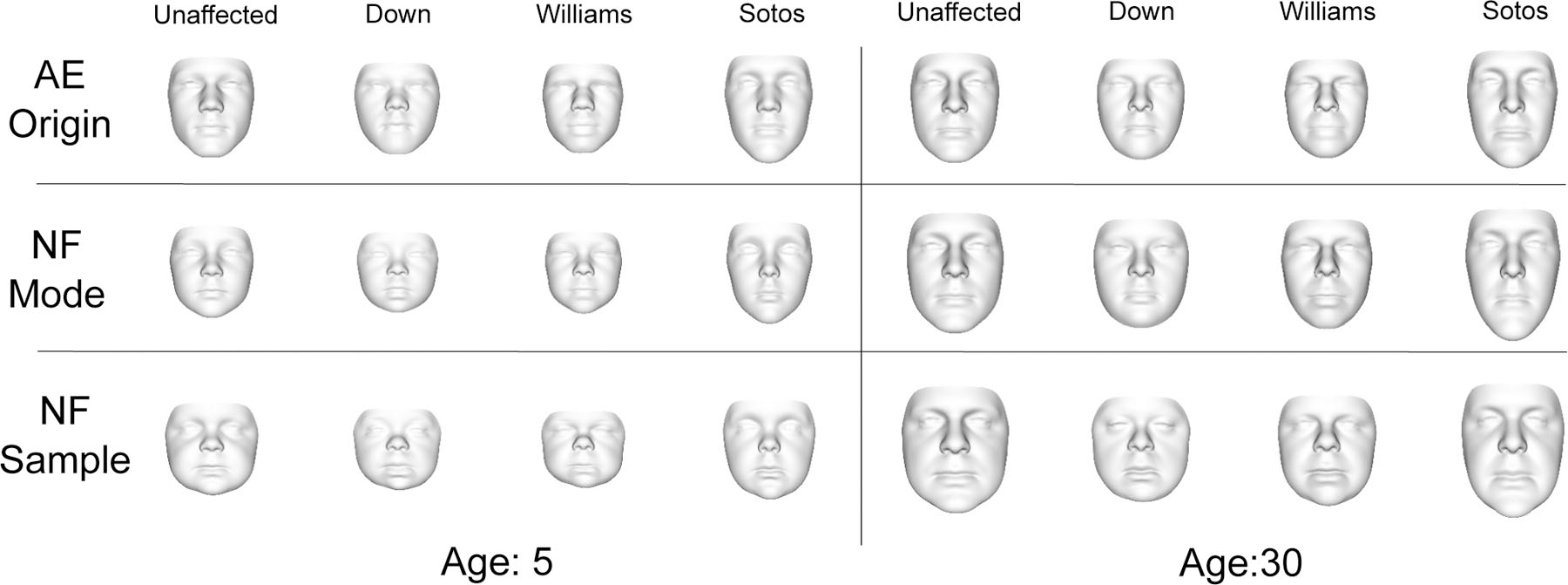
Top Row: Faces corresponding to the origin of the latent space of a cVAE which, unlike the proposed NF model, may not represent the mode of the conditional distribution. Middle Row: modal faces for different syndrome classes at different ages produced by the NF model. Bottom Row: a random sample from the latent space zrand mapped forward through the NF using different syndrome and age conditions. The sex condition was fixed to male.
Fig. 4.
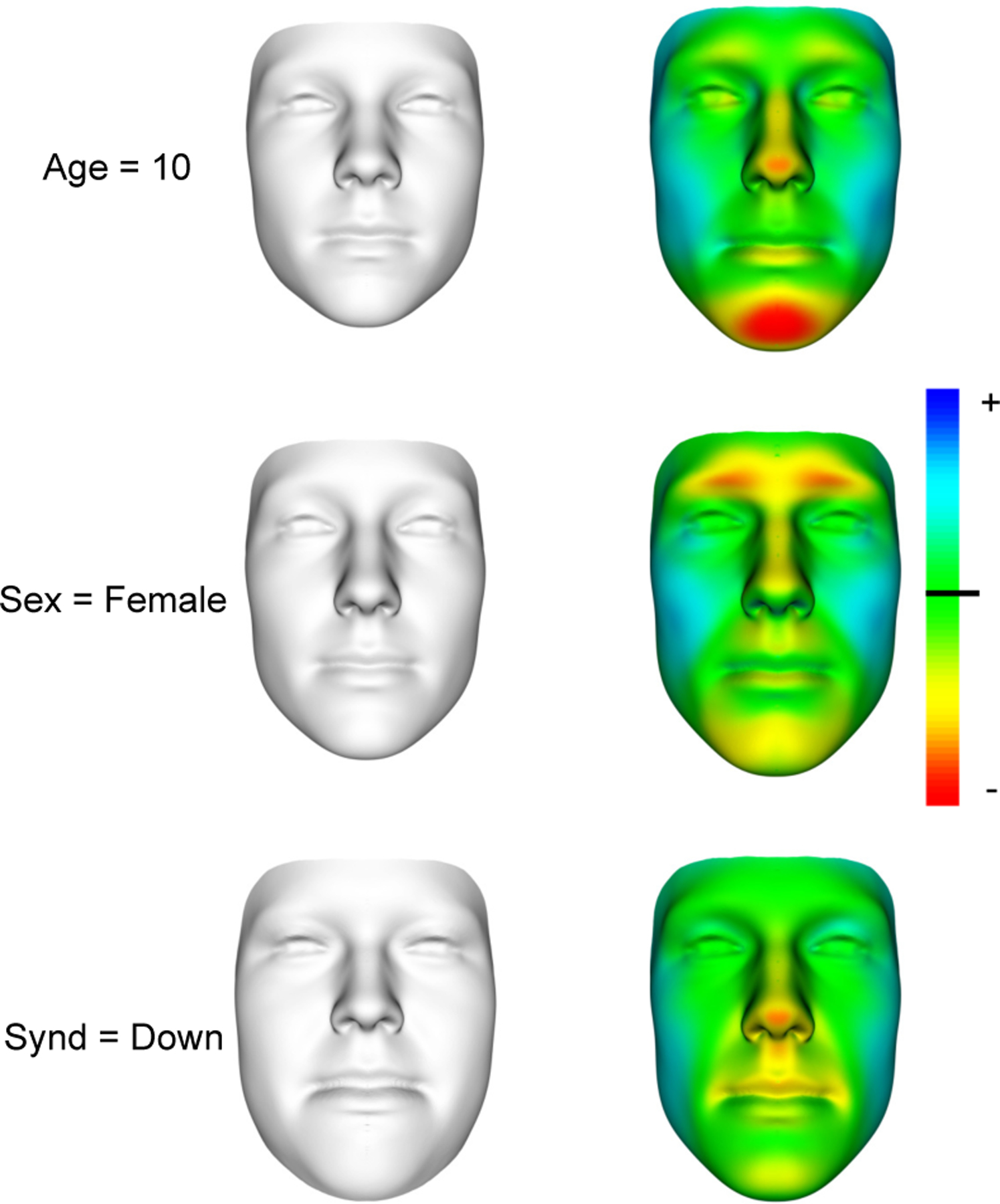
Left column: counterfactual faces for the example subject shown in Fig. 2. Right column: A color map of the shape differences (excluding size information) between original and counterfactual faces overlaid on the original face. Blue indicates an area where the counterfactual shape protrudes outwards compared to the original shape, and red indicates the opposite. The true demographics of the subject are ytrue ={27, Male, Unaffected}. The counterfactual demographics shown in each row are ytop ={10, Male, Unaffected}, ymiddle ={27, Female, Unaffected}, and ybottom ={27, Male, Down}.
For a quantitative comparison with another non-linear generative model, we trained a conditional variational auto-encoder (cVAE) on the same, dimensionality reduced, face data as the proposed NF model to represent the same conditional distribution of facial morphology pF (f |y). The auto-encoder has a latent space dimensionality of nsub and an isotropic Gaussian latent prior (the same as the NF model). The encoder and decoder are densely connected neural networks (two hidden layers, 100 neurons per layer, and ELU activations).
We quantitatively compared the generative capabilities in terms of cross-validated likeness scores. Likeness scores are calculated by comparing the distributions of intra-class Euclidean distances, and between-class distances (where class indicates whether the data is real or model generated) using the Komolgorov-Smirnov (KS) distance. The larger of the two KS distances between intra-class Euclidean distances and between class Euclidean distances is subtracted from 1 to compute the likeness score. Therefore, a score closer to 1 is better. Likeness scores have been shown to capture important performance aspects of generative models such as creativity, diversity, and inheritance [43]. We compute likeness scores for different demographics using the first of the ten random train/test data splits. All scores were computed using 10,000 random samples.
3). Variation Analysis:
All estimates of KL divergence and differential entropy (Eqs. (5) and (6)) were produced using 50,000 random samples from the latent prior pZ (z), which we found sufficient to produce stable results. KL divergence and differential entropy are always expressed in units of nats/dim. Units of information (nats or bits) per dimension are commonly used to evaluate non-Gaussian generative models [34], [44].
Unlike the case of discrete entropy where there is a natural canonical reference measure (the counting measure), there is no canonical reference measure for differential entropy. Here, we use the Lebesque measure over and express all facial measurements f using units of millimeters. Despite this added complexity, differential entropy can still be interpreted as a measure of relative uncertainty or magnitude of variation. A uniform distribution over a unit cube in with volume 1 will have a differential entropy of 0 nats/dim. More localized distributions will have a smaller (negative) differential entropy and less localized distributions will have a larger (positive) differential entropy. KL divergence has the same interpretation (relative entropy or information gain) for both continuous and discrete probability distributions.
IV. Results
A. Inference
1). Syndrome Inference:
Table I summarizes the results of the inference experiments. The overall accuracy of the NF model is 71%. For 92% of unaffected subjects, the correct unaffected class was selected by the model. Results vary widely for the syndrome classes, which is an effect regularly seen in studies that include a large number of syndrome classes [4]. Averaged across all 47 syndrome classes in the analysis (excluding unaffected subjects), the mean sensitivity was 43%. Compared to the Gaussian model (LinearNF), the non-Gaussian model (NF) showed improved overall accuracy (71% vs. 69%) and mean syndrome sensitivity (43% vs. 38%).
TABLE I.
Top-1 overall accuracy and per-syndrome sensitivities for the syndrome inference task (%).
| NF | LinearNF | MLP | PointNet | |
|---|---|---|---|---|
| Overall accuracy | 71 | 69 | 72 | 47 |
| Mean syndrome sensitivity | 43 | 38 | 41 | 9 |
|
| ||||
| 1p36 Del | 20 | 14 | 23 | 2 |
| 22q 11 2 Del | 36 | 43 | 35 | 7 |
| 4p Del Wolff-Hirschhorn | 53 | 32 | 56 | 11 |
| 5p Del Cri du Chat | 51 | 47 | 57 | 9 |
| Achondroplasia | 68 | 61 | 62 | 14 |
| Angelman | 17 | 19 | 26 | 2 |
| CHARGE | 44 | 41 | 56 | 5 |
| Cardiofaciocutaneous | 19 | 25 | 34 | 7 |
| Cleft Lip Palate | 36 | 27 | 27 | 10 |
| Cockayne | 75 | 67 | 71 | 44 |
| Coffin Siris | 0 | 25 | 0 | 0 |
| Cohen | 57 | 67 | 43 | 4 |
| Cornelia de Lange | 52 | 59 | 56 | 8 |
| Costello | 43 | 41 | 54 | 6 |
| Crouzon | 34 | 21 | 33 | 13 |
| Down | 81 | 66 | 79 | 20 |
| Ehlers Danlos | 40 | 34 | 35 | 9 |
| Fragile X | 25 | 3 | 32 | 0 |
| Goldenhar | 23 | 35 | 27 | 11 |
| Jacobsen | 28 | 21 | 24 | 1 |
| Joubert | 35 | 17 | 22 | 1 |
| Kabuki | 23 | 33 | 30 | 5 |
| Klinefelter | 58 | 67 | 50 | 13 |
| Loeys Dietz | 22 | 22 | 23 | 0 |
| Marfan | 40 | 41 | 35 | 11 |
| Moebius | 9 | 0 | 7 | 0 |
| Mucopolysaccharidosis | 32 | 27 | 39 | 4 |
| Neurofibromatosis | 35 | 31 | 26 | 2 |
| Noonan | 38 | 39 | 34 | 4 |
| Osteogenesis Imperfecta | 26 | 17 | 24 | 7 |
| Pallister Killian | 40 | 41 | 35 | 10 |
| Phelan McDermid | 48 | 34 | 42 | 2 |
| Pierre Robin Sequence | 4 | 9 | 7 | 0 |
| Pitt Hopkins | 38 | 35 | 40 | 1 |
| Prader-Willi | 30 | 12 | 24 | 5 |
| Rett | 40 | 45 | 32 | 5 |
| Rhizomelic Chondro Punctata | 72 | 86 | 59 | 26 |
| Rubinstein Taybi | 43 | 22 | 43 | 17 |
| Russell Silver | 68 | 51 | 62 | 12 |
| Smith Lemli Opitz | 13 | 24 | 28 | 2 |
| Sotos | 60 | 38 | 58 | 9 |
| Stickler | 42 | 21 | 21 | 6 |
| Treacher Collins | 63 | 65 | 64 | 12 |
| Trisomy 18 | 74 | 26 | 68 | 13 |
| Turner | 71 | 69 | 58 | 8 |
| Unaffected | 93 | 94 | 96 | 79 |
| Williams | 78 | 71 | 64 | 12 |
| X Linked Hypohidrotic Ectodermal | 47 | 51 | 31 | 1 |
The MLP results were generally similar to those of the NF model, but come without the additional interpretability and multi-functionality of the proposed NF model. Overall accuracy and mean syndrome sensitivity were both within two percentage points. The MLP model struggled with the same syndromes as the NF model (Coffin Siris sensitivity 0% and Pierre Robin Sequence sensitivity 7%), and performed well on similar syndromes (Down sensitivity 79%, Cockayne sensitivity 71%, Williams sensitivity 64%). The sensitivity of the MLP model identifying unaffected individuals was slightly better (96%).
The PointNet model performed the worst overall as well as for each individual syndrome. We believe this is primarily an issue of the high input dimensionality and low sample size (as low as 20 subjects for some syndromes). The comparison between the PointNet model and the MLP model (which uses dimensionality reduced data) suggests that the manifold projection step in our approach provides useful regularization for this setup. Furthermore, the PointNet model and other similar models [45] were designed for more challenging tasks in which mesh vertex ordering is unknown and variable. Mesh topology and vertex order are fixed in our dataset using registration to a reference mesh (see section II-A). This property of vertex permutation invariance may also contribute to the inferior performance of PointNet. Like the MLP and most other discriminative models, PointNet has no ability to generate 3D facial surfaces, and no ability to analyze inter- or intra-demographic facial variation.
2). Age Inference:
The mean absolute error between predicted and true age was 4.4 years for unaffected subjects and 11.9 years for patients with syndromes. For unaffected subjects, the mean standard deviation of the posterior age distribution was 3.5 years, indicating that the model tends to be slightly overconfident in its age estimates. In general, age estimation was more accurate for younger subjects. This trend was also mirrored in the variation analysis results (see Fig. 6).
Fig. 6.
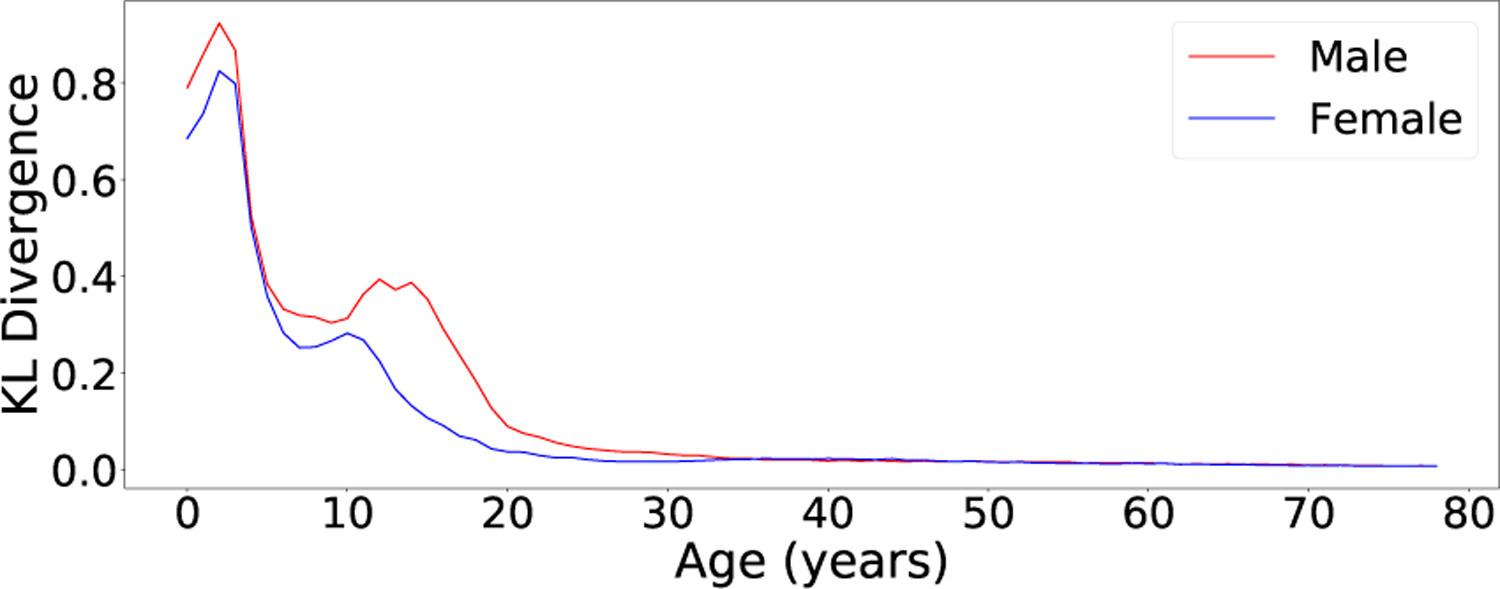
The KL divergences DKL(pF (f|yi)||pF (f|yj)) for different values of agei with agej = agei +1. The syndrome condition was fixed (syndi = syndj = Unaffected). In general, facial morphology changes fastest at very young ages and during puberty.
3). Sex Inference:
91% of unaffected subjects and 66% of patients with syndromes were classified as the correct sex using MAP estimation. Sex estimation was more accurate for older subjects most likely because sex-specific facial features develop later in life. This trend was also mirrored in the variation analysis results (see Fig. 7).
Fig. 7.
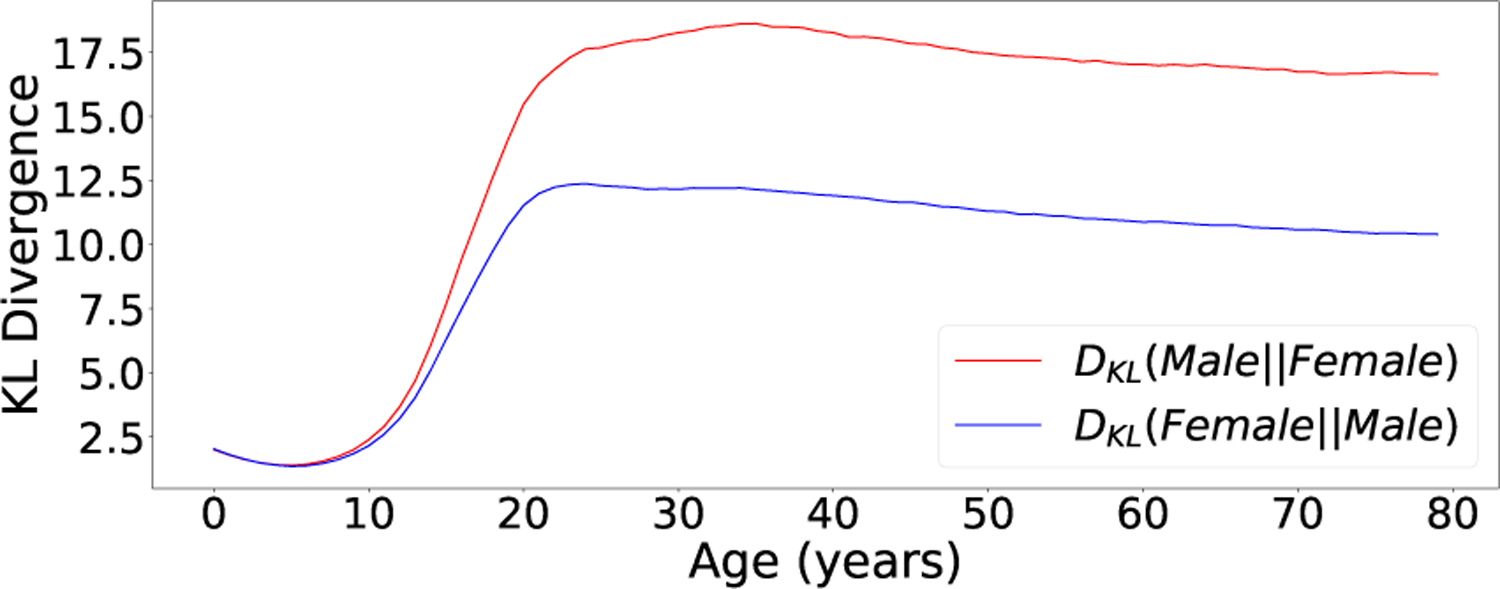
The KL divergences DKL(pF (f|yi)||pF (f|yj)) between sex specific distributions at different ages (agei = agej ). The syndrome condition was fixed (syndi = syndj = Unaffected). In general, sex divergences increase with age until adulthood.
B. Face Generation
1). Demographic Specific Face Generation:
Fig. 3 shows maximally probable faces (modes) and random samples produced by the NF model for a selection of different syndrome and age conditions. The modes and samples exhibit facial features characteristic of the different syndromes and age groups (e.g., small faces for young ages, wide faces for Down syndrome, long faces for Sotos syndrome). Additional visualizations of samples and modes from the NF model are provided as supplementary files (see Appendix II).
Fig. 3 also shows faces generated using a cVAE model. These faces correspond to the origin of the latent space which, unlike the proposed NF model, may not represent the mode of the conditional distribution. Although these faces lack a probabilistic interpretation, they also exhibit features characteristic of the different syndrome classes. The effect of age appears to be less prominent compared to the NF modes.
Table II shows cross-validated likeness scores for selected demographics that are well represented within our data. Both NF models outperform the cVAE model for all demographics included in our evaluation. The Gaussian (LinearNF) and non-Gaussian (NF) flow models performed similarly overall. NF showed small improvements over LinearNF in some demographics and was slightly outperformed in other demographics. In general, likeness scores are better for demographics with more training and evaluation subjects.
TABLE II.
Likeness scores for different generative models and different demographics.
| Demographic (# of evaluation subjects) | NF | LinearNF | cVAE |
|---|---|---|---|
| Unaffected, 5–10 yrs (48) | 0.66 | 0.61 | 0.10 |
| Unaffected, 10–20 yrs (117) | 0.89 | 0.82 | 0.65 |
| Unaffected, 20–30 yrs (321) | 0.94 | 0.94 | 0.74 |
| Unaffected, 30–40 yrs (134) | 0.87 | 0.89 | 0.70 |
| Marfan, 5–40 yrs (21) | 0.81 | 0.83 | 0.51 |
| Down, 5–40 yrs (9) | 0.40 | 0.39 | 0.44 |
2). Counterfactual Face Generation:
Fig. 4 shows counterfactual faces produced using an example subject previously unseen by the model. By contrasting the counterfactual faces with the original face, we can see what information the model uses to make inferences. With respect to this example subject (ytrue ={27 yrs, Male, Unaffected}), the model expects that: a younger subject would have a smaller face with a less pronounced nose and chin, a female subject would have a smaller face and less pronounced brow, nose, and chin, and a Down syndrome patient would have a wider, flatter face.
C. Variation Analysis
1). Differential Entropy Analysis:
We first calculated the differential entropy of the marginal distribution of facial morphology h(pF (f )) = 483 nats/dim, marginalizing the condition variables by integration with respect to the naïve joint distribution of condition variables (see Eq. (4)). Next, we calculated the conditional differential entropy of facial morphology given all condition variables h(pF (f |y)) = 424 nats/dim. Thus, differential entropy decreased by 12% on average when age, sex, and syndrome diagnosis are specified. This result indicates that intra-demographic facial variation is larger than inter-demographic variation with respect to age, sex, and syndrome class. Furthermore, we computed partially conditional entropies h(pF (f |synd)) = 460 nats/dim, h(pF (f |age)) = 463 nats/dim, and h(pF (f |sex)) = 478 nats/dim. These results indicate that the sex condition has the least effect on facial morphology compared to the other condition variables.
Fig. 5 shows the differential entropies of age- and sex-specific distributions. In general, total facial morphological variation was greater for males and older demographics. Our model architecture assumes that differential entropy is invariant with respect to syndrome class.
Fig. 5.
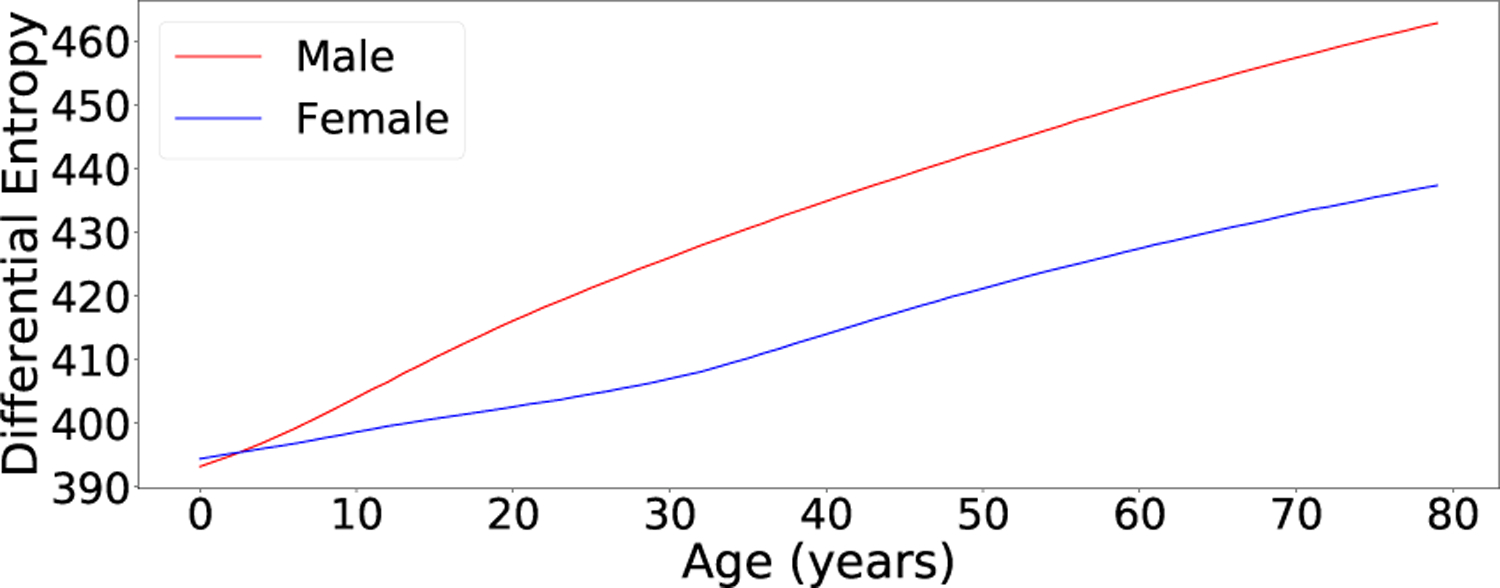
The differential entropies h(pF (f|yi)) for different values of agei and sexi with syndi fixed (our model assumes that differential entropy is invariant with respect to syndrome class). In general, total facial morphological variation is greater for males and older demographics.
2). KL Divergence Analysis:
Fig. 6 shows the KL-divergences induced by increasing the age condition by 1 year for unaffected subjects of both sexes at different ages. These values reflect the rate at which the distribution of facial morphology changes at different ages across both sexes. In general, facial morphology changes faster at younger ages. This pattern is also mirrored in the age inference experiments where the MAP age estimates are more accurate for younger subjects. Some other interesting patterns are the spikes in KL divergence that occur around 10–15 years. These are likely attributable to the onset of puberty; the female spike happens slightly earlier and the male spike continues more into the late teens and early twenties.
Fig. 7 shows KL-divergences between sex-specific distributions at different ages. In general, sex divergences increase with age up until 25 to 30 years after which they gradually decline. This pattern is also mirrored in the sex inference experiments where the MAP sex estimates are more accurate for older subjects.
Overall, the variation analysis results quantitatively show that the model has learned patterns of facial variation that are in agreement with what is generally known about syndromic facial morphology, facial development, and sexual dimorphism. The results provide an additional perspective from which clinicians can evaluate patterns (e.g., the development of sexually dimorphic facial features with age) that are also observed in the inference and generation results.
V. Discussion
For the task of predicting genetic syndrome diagnosis from a 3D facial surface scan in a challenging setup with 48 classes, the model performed very well (overall top-1 accuracy of 71%, and a mean sensitivity of 43% across all syndrome classes). It is important to note that the face-based computer-assisted diagnosis of genetic syndromes is an extremely difficult problem as there is a large number of syndrome classes and there often exists a considerable overlap between the facial morphological distributions associated with different genetic syndromes. A particularly useful property of our probabilistic model is its inherent ability to directly quantify and visualize those overlaps (see Figs. 6 and 7).
Aside from the domain-specific challenges mentioned above, we believe that the classification results are also affected by the sample sizes within our training data. For some classes, only twenty patients were available (e.g., Coffin Siris), which is a very small sample for deep learning applications. Performance for those minority classes could likely be improved by collecting additional data. Due to the rarity of genetic syndromes and the large number of different syndromes, data collection is challenging within this domain.
As a result of patient anonymization processes, we do not have the ability to perform a direct comparison with 2D image-based approaches using this data. We believe that a future study comparing 2D and 3D facial representations for syndrome diagnosis would be highly valuable.
Despite these limitations, we believe that our results are highly clinically relevant and the models very useful. Compared to discriminative baseline models, the syndrome classification performance of the proposed NF model is similar (MLP) or better (PointNet) while being far more interpretable and multi-functional. In addition to inferential tasks, the NF model is able to perform multiple generative tasks (sample, mode, and counterfactual generation) as well as extensive analyses of inter- and intra-demographic facial variation that the MLP and PointNet models cannot perform. Compared to a Gaussian generative model (LinearNF), our non-Gaussian architecture achieves a higher overall accuracy and mean syndrome class sensitivity. We also expect the more flexible, non-Gaussian model to benefit more from a larger training sample size. Compared to the limited number of previous studies also specifically aiming at differentiating a large number of syndrome classes using 3D facial data such as [4], the syndrome inference results are competitive. However, it is very challenging to compare scores that are generated using different training and validation data. Furthermore, the generative results and information-based statistics produced by our models provide additional insight into demographic-specific facial morphological variation that may be useful to clinicians to study characteristics of different syndromes.
VI. Conclusion
In this work, we proposed a novel 3D facial surface model, which can be used to infer syndrome diagnosis and other demographic variables given a high-resolution 3D facial scan. With the goal of maximizing model interpretability within a single, flexible deep learning framework, an invertible normalizing flow architecture was designed that discards the commonly employed Gaussian assumption and can seamlessly handle high dimensional 3D data as well as a large number of syndrome classes. The proposed model is the first non-Gaussian 3D facial shape model with the ability to (1) infer syndrome diagnosis and other demographic variables given a high-resolution 3D facial surface scan, (2) generate modal, randomly sampled and counterfactual 3D faces using demographic information, and (3) analyze the magnitude of facial variation between and within demographic groups (e.g., males vs. females) in a fully probabilistic way. Our evaluation demonstrates that a unified invertible architecture achieves competitive inferential performance while enabling much greater interpretability through multiple mechanisms that do not require any additional model training or modification. To the best of our knowledge, a deep invertible model of 3D facial morphology has never been proposed before, neither for general purposes in computer vision nor specifically for genetic syndromes. Furthermore, this work, for the first time, describes the use of invertible flow models to analyze the magnitude of inter- and intra-demographic morphological variation using entropy-based statistics. We believe that invertible models such as the one presented in this work have the potential to greatly increase model interpretability in the domain of medical diagnosis.
Supplementary Material
Acknowledgments
This research was funded by the National Institutes of Health (U01-DE024440), the Canada Research Chairs program, as well as by the River Fund at Calgary Foundation.
Footnotes
See www.facebase.org for more information and how to access the data.
Contributor Information
Jordan J. Bannister, Biomedical Engineering Graduate Program, University of Calgary, Canada
Matthias Wilms, Department of Radiology, the Alberta Children’s Hospital Research Institute, and the Hotchkiss Brain Institute, University of Calgary, Canada.
J. David Aponte, Department of Cell Biology and Anatomy, the Alberta Children’s Hospital Research Institute and the McCaig Bone and Joint Institute, University of Calgary, Canada.
David C. Katz, Department of Cell Biology and Anatomy, the Alberta Children’s Hospital Research Institute and the McCaig Bone and Joint Institute, University of Calgary, Canada
Ophir D. Klein, Program in Craniofacial Biology and the Department of Orofacial Sciences, University of California, San Francisco, USA
Francois P.J. Bernier, Department of Medical Genetics and the Alberta Children’s Hospital Research Institute, University of Calgary, Canada
Richard A. Spritz, Human Medical Genetics and Genomics Program and the Department of Pediatrics, University of Colorado School of Medicine, USA
Benedikt Hallgrímsson, Department of Cell Biology and Anatomy, the Alberta Children’s Hospital Research Institute and the McCaig Bone and Joint Institute, University of Calgary, Canada.
Nils D. Forkert, Department of Radiology, the Alberta Children’s Hospital Research Institute, and the Hotchkiss Brain Institute, University of Calgary, Canada
References
- [1].Thevenot J, López MB, and Hadid A, “A survey on computer vision for assistive medical diagnosis from faces,” IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 5, pp. 1497–1511, 2017. [DOI] [PubMed] [Google Scholar]
- [2].Hart T and Hart P, “Genetic studies of craniofacial anomalies: clinical implications and applications,” Orthodontics & craniofacial research, vol. 12, no. 3, pp. 212–220, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hammond P and Suttie M, “Large-scale objective phenotyping of 3d facial morphology,” Human mutation, vol. 33, no. 5, pp. 817–825, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hallgrímsson B, Aponte JD, Katz DC, Bannister JJ, Riccardi SL, Mahasuwan N, McInnes BL, Ferrara TM, Lipman DM, Neves AB et al. , “Automated syndrome diagnosis by three-dimensional facial imaging,” Genetics in Medicine, pp. 1–12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz PM, Kamphausen SB, Zenker M et al. , “Identifying facial phenotypes of genetic disorders using deep learning,” Nature medicine, vol. 25, no. 1, pp. 60–64, 2019. [DOI] [PubMed] [Google Scholar]
- [6].Jin B, Cruz L, and Gonc N¸alves, “Deep facial diagnosis: Deep transfer learning from face recognition to facial diagnosis,” IEEE Access, 2020. [Google Scholar]
- [7].Cootes TF, Taylor CJ, Cooper DH, and Graham J, “Active shape models-their training and application,” Computer vision and image understanding, vol. 61, no. 1, pp. 38–59, 1995. [Google Scholar]
- [8].Egger B, Smith WAP, Tewari A, Wuhrer S, Zollhoefer M, Beeler T, Bernard F, Bolkart T, Kortylewski A, Romdhani S, Theobalt C, Blanz V, and Vetter T, “3d morphable face models—past, present, and future,” ACM Trans. Graph, vol. 39, no. 5, 2020. [Google Scholar]
- [9].Zhao Q, Okada K, Rosenbaum K, Kehoe L, Zand DJ, Sze R, Summar M, and Linguraru MG, “Digital facial dysmorphology for genetic screening: Hierarchical constrained local model using ica,” Medical Image Analysis, vol. 18, no. 5, pp. 699–710, 2014. [DOI] [PubMed] [Google Scholar]
- [10].Cerrolaza JJ, Porras AR, Mansoor A, Zhao Q, Summar M, and Linguraru MG, “Identification of dysmorphic syndromes using landmark-specific local texture descriptors,” in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), 2016, pp. 1080–1083. [Google Scholar]
- [11].Boehringer S, Guenther M, Sinigerova S, Wurtz RP, Horsthemke B, and Wieczorek D, “Automated syndrome detection in a set of clinical facial photographs,” American Journal of Medical Genetics Part A, vol. 155, no. 9, pp. 2161–2169, 2011. [DOI] [PubMed] [Google Scholar]
- [12].Shukla P, Gupta T, Saini A, Singh P, and Balasubramanian R, “A deep learning frame-work for recognizing developmental disorders,” in 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), 2017, pp. 705–714. [Google Scholar]
- [13].Tu L, Porras AR, Boyle A, and Linguraru MG, “Analysis of 3d facial dysmorphology in genetic syndromes from unconstrained 2d photographs,” in International Conference on Medical Image Computing and Computer-Assisted Intervention 2018. Springer, 2018, pp. 347–355. [Google Scholar]
- [14].Kumov V and Samorodov A, “Recognition of genetic diseases based on combined feature extraction from 2d face images,” in 2020 26th Conference of Open Innovations Association (FRUCT), 2020, pp. 1–7. [Google Scholar]
- [15].Gerig T, Morel-Forster A, Blumer C, Egger B, Luthi M, Schönborn S, and Vetter T, “Morphable face models-an open framework,” in 2018 13th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2018), 2018, pp. 75–82. [Google Scholar]
- [16].Lüthi M, Gerig T, Jud C, and Vetter T, “Gaussian process morphable models,” IEEE transactions on pattern analysis and machine intelligence, vol. 40, no. 8, pp. 1860–1873, 2017. [DOI] [PubMed] [Google Scholar]
- [17].Blanz V and Vetter T, “A morphable model for the synthesis of 3d faces,” in Proceedings of the 26th annual conference on Computer graphics and interactive techniques, 1999, pp. 187–194. [Google Scholar]
- [18].Booth J, Roussos A, Ponniah A, Dunaway D, and Zafeiriou S, “Large scale 3d morphable models,” International Journal of Computer Vision, vol. 126, no. 2–4, pp. 233–254, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blanc R, Reyes M, Seiler C, and Székely G, “Conditional variability of statistical shape models based on surrogate variables,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2009, pp. 84–91. [DOI] [PubMed] [Google Scholar]
- [20].Ranjan A, Bolkart T, Sanyal S, and Black MJ, “Generating 3d faces using convolutional mesh autoencoders,” in Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 704–720. [Google Scholar]
- [21].Bouritsas G, Bokhnyak S, Ploumpis S, Bronstein M, and Zafeiriou S, “Neural 3d morphable models: Spiral convolutional networks for 3d shape representation learning and generation,” in Proceedings of the IEEE International Conference on Computer Vision, 2019, pp. 7213–7222. [Google Scholar]
- [22].Jiang Z-H, Wu Q, Chen K, and Zhang J, “Disentangled representation learning for 3d face shape,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 11 957–11 966. [Google Scholar]
- [23].Cheng S, Bronstein M, Zhou Y, Kotsia I, Pantic M, and Zafeiriou S, “Meshgan: Non-linear 3d morphable models of faces,” arXiv preprint arXiv:1903.10384, 2019. [Google Scholar]
- [24].Dai H and Shao L, “Pointae: Point auto-encoder for 3d statistical shape and texture modelling,” in Proceedings of the IEEE International Conference on Computer Vision, 2019, pp. 5410–5419. [Google Scholar]
- [25].Bagautdinov T, Wu C, Saragih J, Fua P, and Sheikh Y, “Modeling facial geometry using compositional vaes,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 3877–3886. [Google Scholar]
- [26].Kingma DP and Welling M, “Auto-encoding variational bayes,” CoRR, vol. abs/1312.6114, 2014. [Google Scholar]
- [27].Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2. MIT Press, 2014, p. 2672–2680. [Google Scholar]
- [28].Kobyzev I, Prince S, and Brubaker M, “Normalizing flows: An introduction and review of current methods,” IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 1–1, 2020. [DOI] [PubMed] [Google Scholar]
- [29].Papamakarios G, Nalisnick E, Rezende DJ, Mohamed S, and Lakshminarayanan B, “Normalizing flows for probabilistic modeling and inference,” arXiv preprint arXiv:1912.02762, 2019. [Google Scholar]
- [30].Brehmer J and Cranmer K, “Flows for simultaneous manifold learning and density estimation,” in Advances in Neural Information Processing Systems, Larochelle H, Ranzato M, Hadsell R, Balcan MF, and Lin H, Eds., vol. 33. Curran Associates, Inc., 2020, pp. 442–453. [Google Scholar]
- [31].Pumarola A, Popov S, Moreno-Noguer F, and Ferrari V, “C-flow: Conditional generative flow models for images and 3d point clouds,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2020, pp. 7949–7958. [Google Scholar]
- [32].Yang G, Huang X, Hao Z, Liu M-Y, Belongie S, and Hariharan B, “Pointflow: 3d point cloud generation with continuous normalizing flows,” in Proceedings of the IEEE International Conference on Computer Vision, 2019, pp. 4541–4550. [Google Scholar]
- [33].Ardizzone L, Lüth C, Kruse J, Rother C, and Köthe U, “Guided image generation with conditional invertible neural networks,” arXiv preprint arXiv:1907.02392, 2019. [Google Scholar]
- [34].Dinh L, Sohl-Dickstein J, and Bengio S, “Density estimation using real NVP,” in 5th International Conference on Learning Representations – ICLR 2017, 2017. [Google Scholar]
- [35].Lugmayr A, Danelljan M, Gool LV, and Timofte R, “Srflow: Learning the super-resolution space with normalizing flow,” 2020. [Google Scholar]
- [36].Lezcano-Casado M and Martínez-Rubio D, “Cheap orthogonal constraints in neural networks: A simple parametrization of the orthogonal and unitary group,” 2019. [Google Scholar]
- [37].Sorrenson P, Rother C, and Köthe U, “Disentanglement by nonlinear ica with general incompressible-flow networks (gin),” 2020. [Google Scholar]
- [38].Wilms M, Bannister JJ, Mouches P, MacDonald ME, Rajashekar D, Langner S, and Forkert ND, “Bidirectional modeling and analysis of brain aging with normalizing flows,” in 3rd International Workshop on Machine Learning in Clinical Neuroimaging – in conjunction with MICCAI 2020, 2020. [Google Scholar]
- [39].Jeyakumar JV, Noor J, Cheng Y-H, Garcia L, and Srivastava M, “How can i explain this to you? an empirical study of deep neural network explanation methods,” Advances in Neural Information Processing Systems, 2020. [Google Scholar]
- [40].Bannister JJ, Crites SR, Aponte JD, Katz DC, Wilms M, Klein OD, Bernier FPJ, Spritz RA, Hallgrímsson B, and Forkert ND, “Fully automatic landmarking of syndromic 3d facial surface scans using 2d images,” Sensors, vol. 20, no. 11, p. 3171, Jun 2020. [Online]. Available: 10.3390/s20113171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Amberg B, Romdhani S, and Vetter T, “Optimal Step Nonrigid ICP Algorithms for Surface Registration,” 2007 IEEE Conference on Computer Vision and Pattern Recognition, 2007. [Google Scholar]
- [42].Charles RQ, Su H, Kaichun M, and Guibas LJ, “Pointnet: Deep learning on point sets for 3d classification and segmentation,” in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 77–85. [Google Scholar]
- [43].AGuan LM, S., “A novel measure to evaluate generative adversarial networks based on direct analysis of generated images,” Neural Computing and Applications, 2021.
- [44].Kingma DP and Dhariwal P, “Glow: Generative flow with invertible 1x1 convolutions,” in Advances in neural information processing systems, 2018, pp. 10 215–10 224. [Google Scholar]
- [45].Guo Y, Wang H, Hu Q, Liu H, Liu L, and Bennamoun M, “Deep learning for 3d point clouds: A survey,” IEEE Transactions on Pattern Analysis Machine Intelligence, vol. 43, no. 12, pp. 4338–4364, dec 2021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


