Abstract
Affinity selection-mass spectrometry, which includes magnetic microbead affinity selection-screening (MagMASS), is ideal for the discovery of ligands in complex mixtures that bind to pharmacological targets. Therapeutic agents are needed to prevent or treat COVID-19, which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infection of human cells by SARS-CoV-2 involves binding of the virus spike protein subunit 1 (S1) to the human cell receptor angiotensin converting enzyme-2 (ACE2). Like antibodies, small molecules have the potential to block the interaction of the viral S1 protein with human ACE2 and prevent SARS-CoV-2 infection. Therefore, a MagMASS assay was developed for the discovery of ligands to the S1 protein. Unlike previous MagMASS approaches, this new assay used robotics for five-fold enhancement of throughput and sensitivity. The assay was validated using the SBP-1 peptide, which is identical to the ACE2 amino acid sequence recognized by the S1 protein, and then applied to the discovery of natural ligands from botanical extracts. Small molecule ligands to the S1 protein were discovered in extracts of the licorice species, Glycyrrhiza inflata. In particular, the licorice ligand licochalcone A was identified through dereplication and comparison with standards using HPLC with high-resolution tandem mass spectrometry.
Graphical Abstract
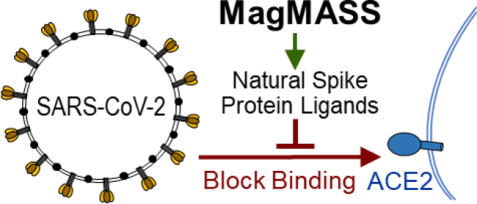
A mass spectrometry-based assay using magnetic microbead affinity-selection screening (MagMASS) was developed to facilitate the discovery of ligands to the spike protein of SARS-CoV-2, which causes COVID-19. By applying MagMASS to the screening of botanical extracts, licochalcone A from licorice was identified as a ligand to the S1 spike protein. Ligands to the virus spike protein have the potential to block virus binding to the human cell receptor ACE2 and inhibit infection.
INTRODUCTION
Natural products have historically been abundant sources of biologically active compounds and new therapeutic agents, although natural products research in the pharmaceutical industry has declined.1,2 To renew interest in natural sources of therapeutic agents and to facilitate the discovery of pharmacologically active compounds in botanical dietary supplements, faster and more efficient approaches are required. Affinity selection-mass spectrometry (AS-MS), which includes size exclusion AS-MS,3 pulsed ultrafiltration AS-MS,4–8 and magnetic microbead affinity selection (MagMASS),9–11 has considerable potential for facilitating the discovery of natural products with affinity for pharmacological targets. Compared with conventional high-throughput screening utilizing fluorescence or absorbance readouts (such as FRET or fluorescence polarization), AS-MS offers advantages such as compatibility with any type of ligand mixture, matrix, or assay buffer and no requirement for fluorescent tags.12 Uniquely, AS-MS does not suffer from interference caused by samples containing fluorophores or chromophores, which are common in natural products. Furthermore, MagMASS is compatible with 96-well screening plate formats and automation, which is not the case for size exclusion AS-MS.13
Magnetic microbeads were originally developed for protein isolation and purification from complex mixtures using approaches such as batch affinity chromatography.14 Our laboratory9 pioneered the development of MagMASS for screening mixtures of small molecules such as botanical extracts and combinatorial libraries to identify small molecules with therapeutic value, and subsequently, other laboratories have adopted this approach.10,11,15–18 Briefly, MagMASS utilizes a pharmacological target such as a receptor or enzyme immobilized on magnetic microbeads. Mixtures of potential ligands are incubated with the immobilized target, unbound compounds are rinsed away with binding buffer while a magnetic field retains the beads, and then ligands are released from the target using a destabilizing solution such as organic solvent for analysis using LC-MS (Figure 1).
Figure 1.
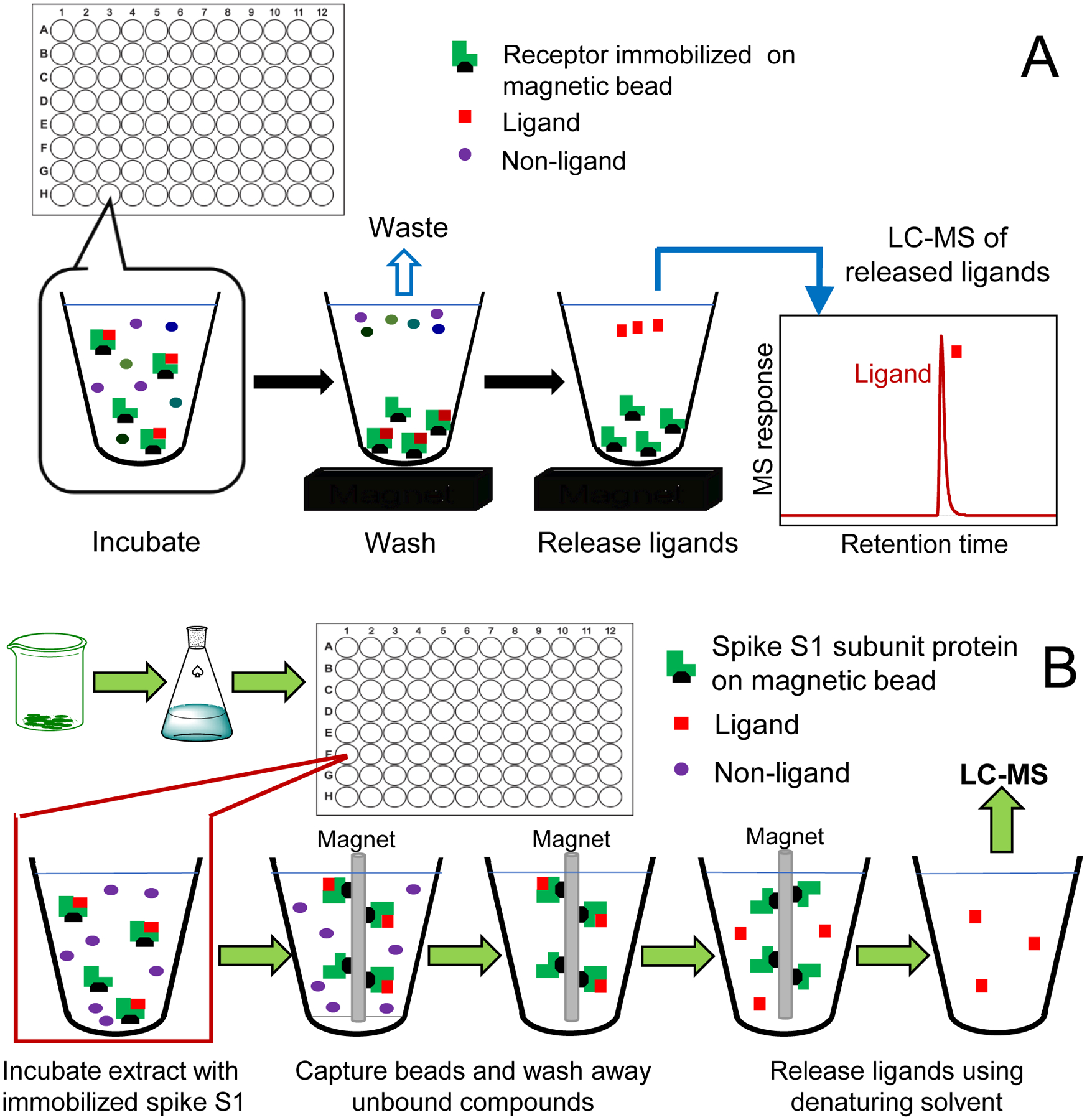
Comparison of (A) our previous 96-well plate MagMASS assay; and (B) the new MagMASS approach utilizing a faster and automated magnetic particle processor.
Responsible for the COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a coronavirus, named after the characteristic crown-like spike proteins on the outer surface of the virus particles (Figure 2). Binding of the spike protein of SARS-CoV-2 to the human cell surface receptor angiotensin converting enzyme-2 (ACE2) is a first step in the infection of human cells.19 Vaccines and antibody therapy against SARS-CoV-2 target the spike protein subunit 1 (S1; residues 331 to 524),20 which has high affinity for the human cell-surface protein ACE2, and prevent the virus from binding to ACE2 and infecting human cells.21 Small molecules with affinity to the SARS-CoV-2 S1 protein also have the potential to function as cell entry inhibitors and prevent infection or shorten the course of SARS-CoV-2 infections.
Figure 2.
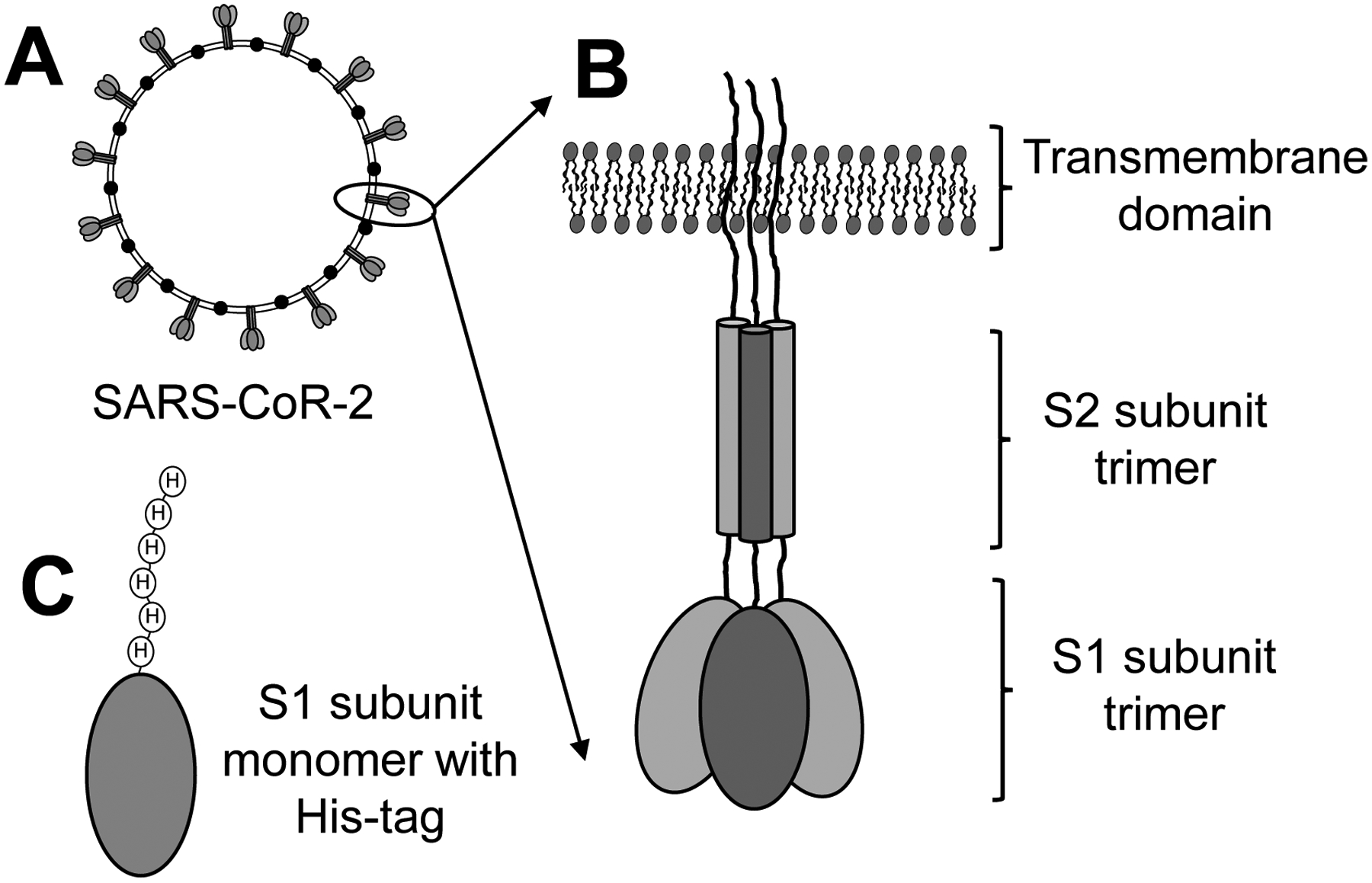
(A) SARS-CoV-2 is a coronavirus named for the transmembrane spike protein on the surface of the infectious virus particle. (B) The spike protein is a trimer consisting of a transmembrane domain, an S2 domain, and an S1 domain which binds to the human receptor ACE2 to initiate the infection of human cells. (C) Recombinant S1 protein monomer containing a histidine tag was immobilized on magnetic beads for use as the target for MagMASS.
A MagMASS assay was developed to discover small molecule ligands to the SARS-CoV-2 S1 protein that might function as cell entry inhibitors and serve as lead molecules for the development of new therapeutic agents that prevent viral infection. The utility of this new assay was demonstrated by screening botanical extracts and discovering ligands to the S1 protein in the licorice species, Glycyrrhiza inflata. In addition, our previous MagMASS approach was enhanced 5-fold with respect to speed and sensitivity through automation using a 96-well plate magnetic particle processor.
EXPERIMENTAL SECTION
Materials and reagents
Recombinant SARS-CoV-2 spike protein S1 (Figure 2) was obtained from RayBiotech (Peachtree Corners, GA, USA). The 23-mer SBP-1 peptide was purchased from LifeTein (Hillsborough, NJ, USA). The custom peptide TVFGLNVWKRYSK was synthesized by Pierce Custom Peptides (ThermoFisher; Rockford, IL). Licochalcone A, imatinib, mycophenolic acid, quinacrine, and HPLC-grade methanol and acetonitrile were purchased from Sigma Aldrich (St. Louis, MO, USA). LC-MS grade formic acid and rapid equilibrium dialysis single use plates were purchased from ThermoFisher (Waltham, MA, USA). Magnetic microbeads derivatized with Ni2+-nitrilotriacetic acid were obtained from EmerTher (Parsippany, NJ, USA). Polypropylene 2-mL conical bottom 96-well plates were purchased from Fisher Scientific (Hanover Park, IL, USA), and ultrapure water was prepared in house using a Milli-Q water purification system (Millipore, MA, USA). Extracts of Dioscorea villosa, Glycyrrhiza glabra, Glycyrrhiza inflata, Humulus lupulus, and Trifolium pratense L. were prepared, botanically authenticated and chemically standardized as previously described.11,22 For example, G. inflata roots were powdered, extracted by percolation with methanol (weight powder/volume of solvent, 1/20), and freeze dried (extraction yield 25% w/w, weight of extract/weight of root powder).22
Affinity selection-mass spectrometry (MagMASS)
Recombinant SARS-CoV-2 spike protein S1 subunit (~72 kDa) containing an N-terminal His-tag (Figure 2) was immobilized on Ni2+-nitrilotriacetic acid derivatized magnetic microbeads as recommended by the manufacturer. Briefly, 100 pmol of S1 protein and 400 μg of Ni2+-nitrilotriacetic acid magnetic microbeads were mixed with 139 μL of 50 mM Tris buffer (adjusted to pH 7.3) containing 137 mM NaCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4, and 2.7 mM KCl. The extract was then added, and the mixture was incubated for 1 h. To confirm the activity of immobilized S1 protein, positive control incubations were carried out using SBP-1, a 23 amino acid peptide with the sequence of IEEQAKTFLDKFNHEAEDLFYQS, which is identical to the ACE2 α1 helix sequence recognized by the SARS-CoV-2 spike protein S1 subunit. As a negative control, denatured S1 protein, which was obtained by incubating intact S1 protein in a water bath at 95 °C for 15 min, was immobilized on identical magnetic microbeads.
Initial MagMASS assays (Figure 1A) were carried out as described previously for the immobilized target, 15-lipoxygenase.11 Briefly, SBP-1 (33 nM) was incubated for 60 min with magnetic microbeads (8 μL, 50 mg/mL) containing 100 pmol of immobilized active or denatured S1 protein in 150 μL binding buffer. After washing twice with 500 μL of 30 mM ammonium acetate to remove unbound ligand while the beads were retained by a magnetic field, ligand was released from the beads using 90% methanol in water (200 μL). The samples were evaporated to dryness and reconstituted in 100 μL of water/methanol (50:50; v/v) containing 189 nM ketoconazole as internal standard for analysis using UHPLC-LC/MS.
Analyses were carried out using a Shimadzu (Kyoto, Japan) Nexera UHPLC system interfaced with a Shimadzu LCMS-8050 triple quadrupole mass spectrometer. SBP-1 and ketoconazole were measured using positive ion electrospray followed by collision-induced dissociation with selected reaction monitoring at unit resolution. The transitions of m/z 701.6 to m/z 136.2 (collision energy −30 V) and m/z 701.6 and m/z 120.2 (collision energy −45 V) were measured for SPB-1 with a dwell time of 25 ms each, and the transitions of m/z 531.1 to m/z 489.2 and m/z 531.1 to m/z 243.9 (collision energy −33 V) were measured for ketoconazole with a dwell time of 20 ms each. Reversed phase UHPLC separation was carried out using a Waters (Milford, MA, USA) Acquity UPLC BEH C18 column (1.7 μm, 130 Å, 2.1 mm × 50 mm) with a 3 min linear gradient from 20% to 80% acetonitrile in 0.1% aqueous formic acid at a flow rate of 0.3 mL/min. The auto-sampler temperature was 4 °C, the injection volume was 10 μL, and the column oven was 40 °C.
To enhance the throughput of MagMASS for botanical extract assays 5-fold, a ThermoFisher KingFisher Flex Magnetic Particle Processor was used for automated processing of one 96-well plate at a time (Figure 1B). Briefly, botanical extracts (10 μg/mL), mixtures of compounds (0.10 μM each), or individual compounds (0.10 μM) were incubated with 100 pmol of immobilized S1 protein per well. Incubations with immobilized denatured S1 protein were used to control for non-specific binding. The magnetic beads containing immobilized S1 protein in each well were captured by the magnetic probe of the KingFisher, and the contents of each well were mixed slowly for 2 out of every 5 min for 1 h. The captured magnetic beads containing immobilized S1 protein and ligands were washed twice for 1 min each in 500 μL of 30 mM ammonium acetate and washed again in 500 μL of high purity water. Ligands were eluted into 200 μL of 90% aqueous methanol (Figure 1B). The samples were evaporated to dryness as described above and reconstituted in 100 μL of water/methanol (50:50) containing 480 nM [d4]-daidzein and 189 nM ketoconazole as internal standards.
The released ligands were analyzed using LC-MS with positive ion or negative ion electrospray on a Shimadzu LCMS-9030 Q-ToF hybrid high-resolution mass spectrometer at a resolving power of 30,000 FWHM. The electrospray interface temperature was 300 °C, and the voltages were 4.5 kV and −3.5 kV for positive and negative ion mode, respectively. The heat block and desolvation line temperatures were 400 °C and 250 °C, respectively. Nitrogen was used as a drying gas at a flow rate of 10 L/min, as a heating gas at 10 L/min, and for nebulization at 3 L/min. Data-dependent product ion tandem mass spectrometry was used such that mass spectra and product ion tandem mass spectra were acquired every 100 ms over the scan range of m/z 100–1200 and m/z 70–1200, respectively. During data dependent acquisition, six dependent events were specified at an intensity threshold of 3000, and the product ion tandem mass spectra were obtained using a collision energy of 35 V with an energy spread of 17 V.
Reversed phase UHPLC separations were carried out using a Waters C18 Cortecs HPLC column (2.7 μm, 2.1 mm × 50 mm) with a 19 min linear gradient from 5% to 95% acetonitrile in water containing 0.1% formic acid at a flow rate of 0.3 mL/min. The column was re-equilibrated at 5% acetonitrile for 2 min between injections. The auto-sampler temperature was 4 °C, the injection volume was 10 μL, and the column oven was 30 °C.
Data processing
The analysis of each pair of chromatograms (intact S1 protein and denatured S1 protein negative control) was carried out using Online XCMS (Scripps Research, La Jolla, CA USA) which is an open source metabolomics software program.23 To expedite data analysis, an in-house Python code was designed that enables comparison of UHPLC-MS data of a botanical extract (aligned with a blank analysis) with the corresponding MagMASS UHPLC-MS data (aligned with denatured receptor control data). This additional step helps eliminate false positives that may result from background noise. High-resolution mass spectra were processed using Shimadzu LabSolutions V5.2 software. Natural product ligands were identified based on their elemental compositions (determined using high-resolution accurate mass measurements), tandem mass spectra, and comparison with authentic standards.
Ligand binding site determination
To determine if binding occurs at the active site of the S1 protein (orthosteric) or at another site (allosteric), natural product ligand binding to the S1 protein was investigated in the presence of a high affinity known orthosteric ligand, SBP-1. If binding of the new ligand was reduced or eliminated through competition with SBP-1, then it would be determined to be an orthosteric ligand.
Determination of equilibrium dissociation constant
The equilibrium dissociation constant for the binding of licochalcone A to the S1 protein was determined by using rapid equilibrium dialysis. The optimum time for equilibration was determined by adding 1 μM S1 protein in phosphate buffered saline, pH 7.2 (300 μL) to the protein chamber and 500 μL blank buffer to the buffer chamber. Licochalcone A was spiked into the protein chamber at a final concentration of 2.5 μM. During incubation at 37 °C on an orbital shaker at 200 rpm, aliquots (30 μL) were sampled from each chamber at 0.5, 1, 2, 3, 4, 5, 6 and 7 h and mixed with equal volumes of buffer. Ice-cold 90% aqueous acetonitrile (300 μL) was added containing 0.1% formic acid and 500 ng/mL ketoconazole (internal standard), vortex mixed, and incubated on ice for 1 h. After centrifugation at 18,000 g for 30 min at 4 °C, the supernatant was removed and ligand concentration was measured using UHPLC-MS/MS. The equilibrium dissociation constant of licochalcone A was determined by incubating the S1 protein with different concentrations of the ligand ranging from 0.05 – 50 μM in triplicate. After 4 h incubation, the concentrations of licochalcone A in the sample and buffer chambers were measured using UHPLC-MS/MS. Data analysis and fitting were carried out using Microsoft Excel (Seattle, WA, USA) and KaleidaGraph v4.1 (Reading, PA, USA).
Computational modeling of hit compounds to the S1 C-terminal domain (CTD)
The computational aided modeling of ligand binding to the SARS-CoV-2 spike S1 protein was carried out using AutoDock Vina24 (Scripps Research). The coordinates of the crystal structure of CTD of the S1 protein were downloaded from Protein Data Bank (PDB, ID number 6LZG). The ChemDraw structures of the ligands were converted to .pdb files using Pymol (Schrödinger, New York, NY). The S1-CTD was loaded into the AutoDock Vina program, the search space was defined around the known orthosteric site, and the file was converted to .pdbqt. Similarly, the ligands were individually loaded and converted to .pdbqt files. The search space x, y and z coordinates, S1-CTD, and ligand were defined on the command prompt in which the Python script developed for the AutoDock Vina was loaded and the run was initiated.
RESULTS AND DISCUSSION
Assay validation
To discover natural ligands to the SARS-CoV-2 spike protein, a MagMASS assay (Figure 1) was developed that utilizes the spike S1 protein (Figure 2) immobilized on magnetic microbeads. To confirm that the immobilized S1 protein retained selectivity for the human cell surface protein ACE2, S1 protein immobilized on magnetic microbeads was incubated with SBP-1, which is a peptide containing the amino acid sequence of human ACE2 to which the SARS-CoV-2 spike protein recognition site (amino acid residues 331 to 524) binds. After processing using MagMASS, SBP-1 was determined to bind to the immobilized S1 protein (positive control) but not to denatured S1 subunit that had been immobilized in an identical manner (Figure 3A).
Figure 3.
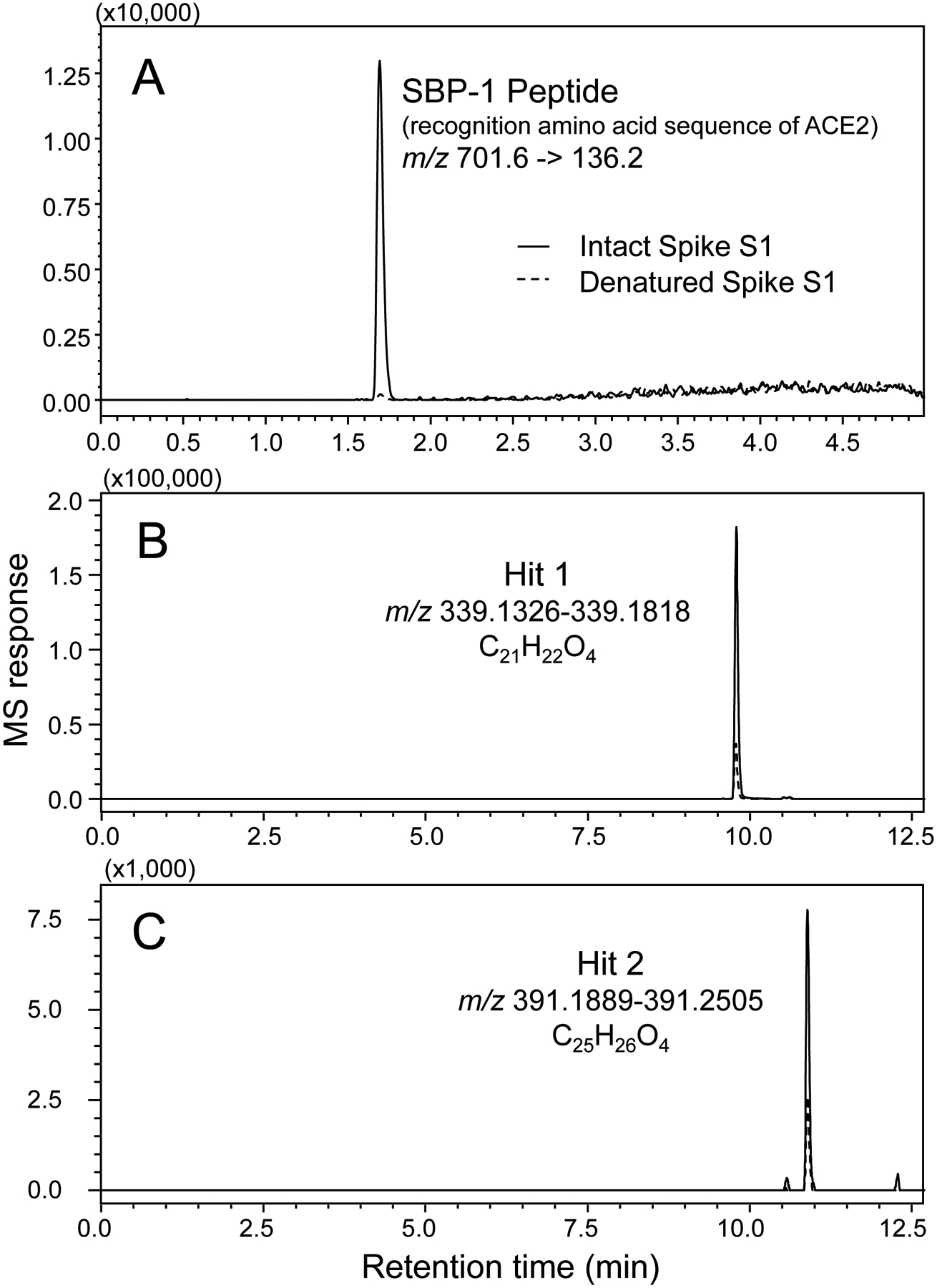
Following affinity selection using MagMASS, positive ion electrospray LC-MS was used to detect ligands that bound to the SARS-CoV-2 spike S1 protein. Selective binding of a ligand to the native S1 protein (solid line) is indicated by peak enhancement relative to denatured S1 protein (dashed line). (A) The positive control SBP-1 peptide bound to the native but not to denatured S1 protein (selected reaction monitoring of the [M+4H]4+ precursor). (B) and (C) High-resolution computer-reconstructed mass chromatograms showing two hits detected during the screening of a licorice root extract (Glycyrrhiza inflata). Hit 1 was identified as licochalcone A by comparison with authentic standard (see Figure 4).
Screening for natural ligands to the SARS-CoV-2 spike S1 protein
The application of MagMASS to the screening of botanical extracts resulted in detection of two compounds from the licorice species Glycyrrhiza inflata that showed specific binding to intact spike S1 protein (Figure 3B). For comparison, no ligands were detected in extracts of wild yam (Dioscorea villosa), hops (Humulus lupulus), red clover (Trifolium pratense), or in the licorice species Glycyrrhiza glabra (data not shown). G. inflata ligands to the spike S1 subunit were detected at retention times of 9.8 (hit 1) and 10.9 min (hit 2) (Figure 3B and 3C) using high-resolution positive ion electrospray mass spectrometry as protonated molecules of m/z 339.1590 and m/z 391.1907, respectively. These accurate mass measurements correspond to elemental compositions of C21H23O4 (theoretical m/z 339.1596; ΔM −1.8 ppm) for hit 1 and C25H27O4 (theoretical m/z 391.1909; ΔM −0.5 ppm) for hit 2.
To identify these natural ligands to the SARS CoV-2 S1 protein, we carried out dereplication by searching databases such as PubChem, METLIN, and GNPS for compounds of these elemental compositions known to occur in G. inflata. Some possible compounds in G. inflata corresponding to an elemental composition of C25H26O4 (hit 2, uncharged molecule) include abyssinone III, licoflavone B, and glyinflanin K. In the absence of authentic standards, confirmation of the identity of this ligand is ongoing. Dereplication of G. inflata compounds with elemental compositions of C21H22O4 (uncharged molecule) indicated several possibilities including licochalcone A, licochalcone C, licochalcone E, lupwigheone, and eurycarpin A. Comparison of the LC-MS retention time and high-resolution tandem mass spectrum of hit 1 eluting at 9.8 min with those of a licochalcone A standard confirmed that this compound was licochalcone A (Figure 4).
Figure 4.
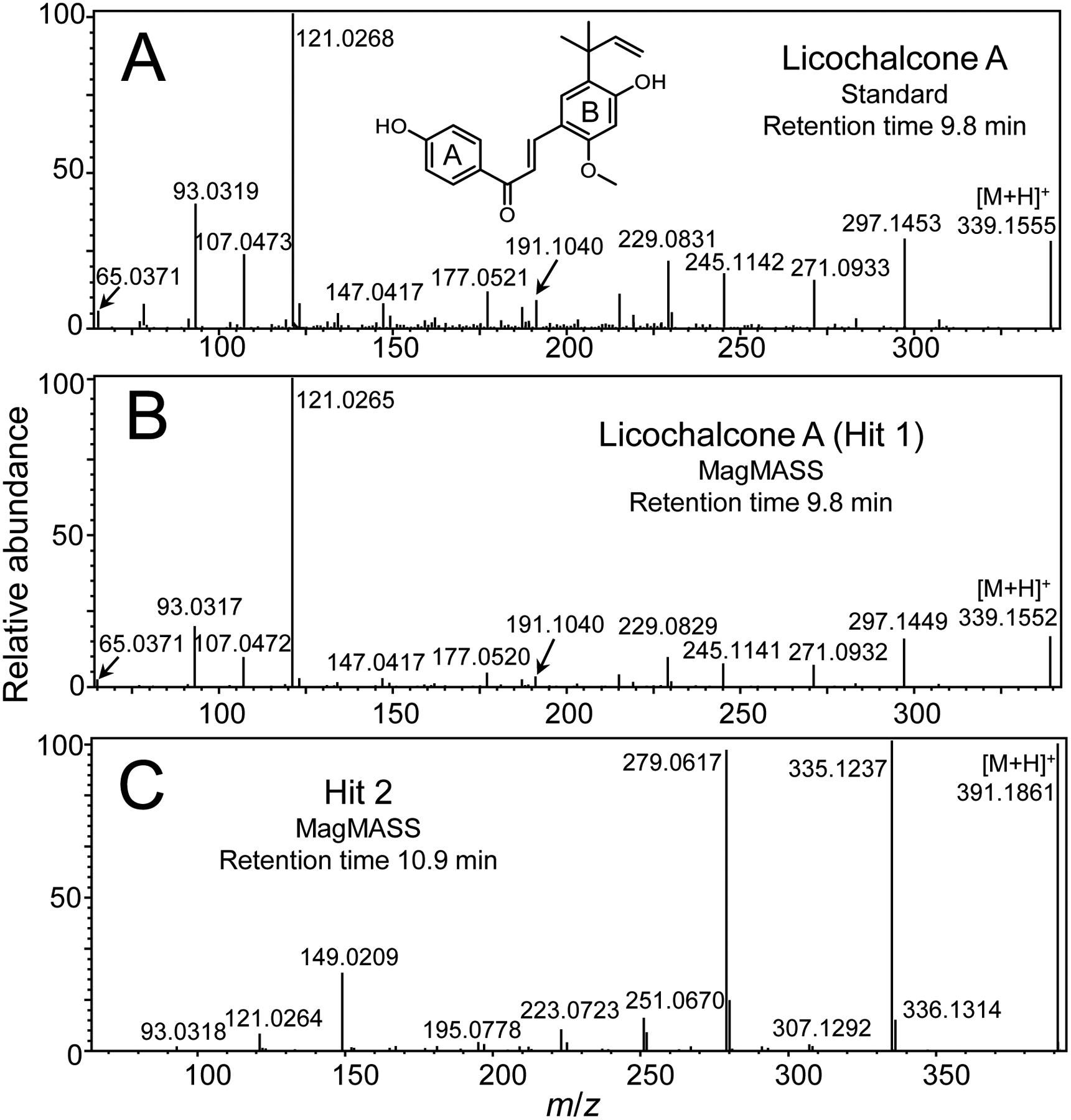
High-resolution positive ion electrospray product ion tandem mass spectra of (A) licochalcone A standard; and SARS-CoV-2 spike S1 protein ligands from G. inflata eluting at (B) 9.8 min (hit 1); and (C) 10.9 min (hit 2) during LC-MS in Figure 3. Hit 1 was identified as licochalcone A through LC-MS/MS comparison with authentic standard.
Binding of licochalcone A to the SARS-CoV-2 S1 protein
The equilibrium dissociation constant (Kd) for the binding of licochalcone A to the SARS-CoV-2 spike protein S1 subunit was determined using equilibrium dialysis. The Kd value was 6.3 ± 1.1 μM, and the time to reach equilibrium was 4 h. Binding of licochalcone A to the active site of the SARS-CoV-2 S1 protein was confirmed for licochalcone A by demonstrating competition for binding with SBP-1. The MagMASS peak area enrichment of licochalcone A was lowered 3-fold when SBP-1 was added to the incubation mixture containing licochalcone A and active S1 protein (Figure 5). This observation strongly suggests that licochalcone A binds at the orthosteric site and competes with the known orthosteric ligand SBP-1.
Figure 5.
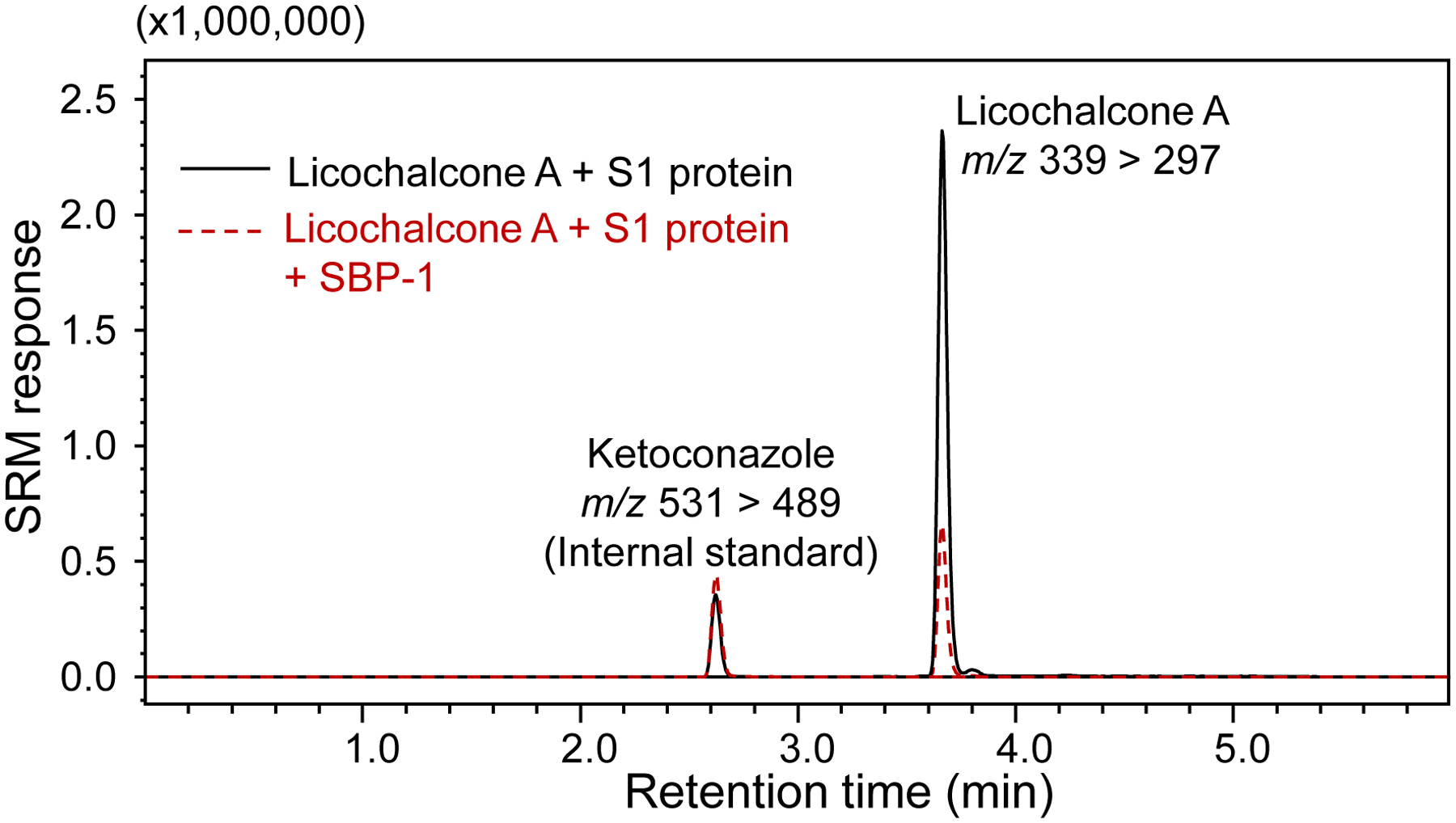
MagMASS assay showing competition between licochalcone A and SBP-1 peptide for binding to the SARS-CoV-2 S1 protein. The binding of 10 μM licochalcone A was reduced in the presence of 100 nM SBP-1 indicating that both ligands compete for the orthosteric binding site on the S1 protein.
Recently, the crystal structure of the C-terminal domain (CTD) of the SARS-CoV-2 S1 protein was solved in complex with the human ACE2 receptor.25 The key interactions involve residues along the S1-CTD interface that contribute to a network of hydrogen-bonds and salt-bridge interactions with ACE2. These S1-CTD residues include A475, N487, E484, Y453, K417, G446, Y449, G496, Q498, T500, G502, Y489, and F486.25 Note that these key interaction residues are part of the S1 protein monomer and not the trimer of SARS-CoV-2 spike protein. Therefore, ligands to the monomer of the S1-CTD alone have the potential to block binding to ACE2. Licochalcone A was modeled into the S1-CTD using AutoDock Vina24 to determine if it binds at the orthosteric site or allosterically. The search scope was based around the SARS-CoV-2 spike CTD orthosteric site (Figure 6A), and the size of the search space in all dimensions was >25 Å to ensure the space was large enough for the ligands to rotate.
Figure 6.
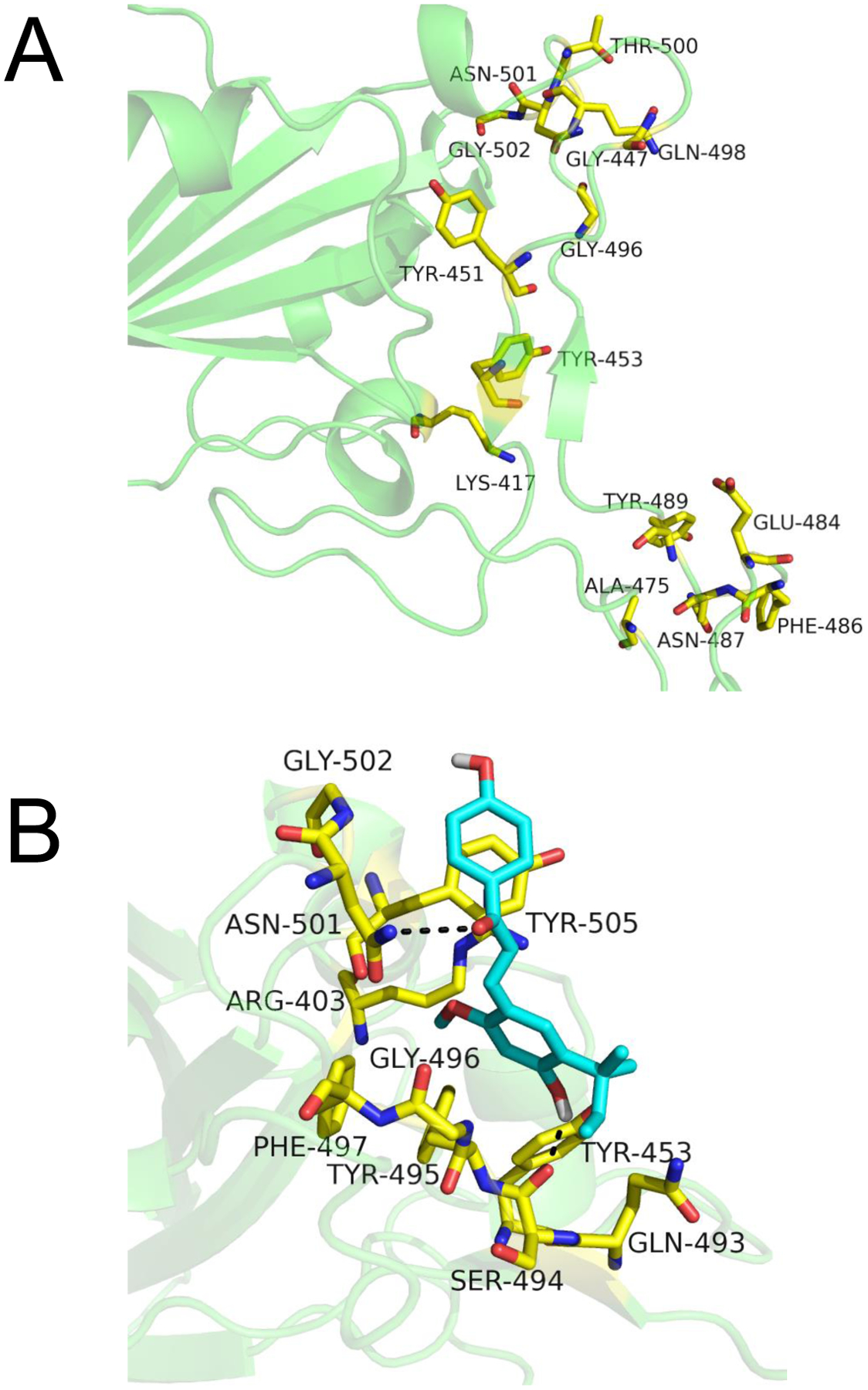
Computational based modeling of the binding of licochalcone A to the SARS-CoV-2 spike S1 protein C-terminal domain (S1-CTD) orthosteric site using AutoDock Vina. A) The active site residues of the S1 protein are shown in yellow. B) licochalcone A (cyan) is predicted to bind at the orthosteric site at −6.5 kcal/mol free energy of binding.
The best predicted binding mode for licochalcone A was at the orthosteric site with a free energy of binding of −6.5 kcal/mol (Figure 6B). The key interactions within 3.5 Å involved hydrogen bonding between the hydroxyl group on the B-ring of licochalcone A and the carbonyl backbone of S494. Additional hydrogen bonding was predicted between the carbonyl group of licochalcone A with the side chain amino group of N501. The A-ring of licochalcone A was in a favorable position to form π - π stacking interaction with the aromatic ring of Y505. The B-ring of licochalcone A was buried in a hydrophobic pocket surrounded by F497, Y495, and G496.
Other SARS-CoV-2 entry inhibitors
Imatinib, mycophenolic acid, and quinacrine have been reported26 to block SARS-CoV-2 replication in cell culture. However, the mechanism of action of these inhibitors remains unknown. To determine if these compounds function by binding to the SARS-CoV-2 spike protein and thereby blocking cell entry, we tested them in the new MagMASS assay. Imatinib, mycophenolic acid, and quinacrine showed no binding to the S1 protein in the MagMASS assay (data not shown), which indicates that these compounds act by a different mechanism.
Yu, et al.,27 reported that glycyrrhizic acid, a toxic constituent of licorice, was a ligand of the S1 protein that could block interaction between the SARS-CoV-2 receptor binding domain and ACE2. The Kd for the binding of glycyrrhizic acid to the S1 protein is 66.8 μM. Although present in the licorice extracts we tested, glycyrrhizic acid was not detected as a ligand to the S1 protein in our assay, probably because its affinity for the S1 protein is more than 10-fold weaker than that of licochalcone A (Kd = 6.3 ± 1.1 μM).
Using an AS-MS assay with immobilized S1 receptor binding domain, which is a 52 kDa subunit of the 108 kDa SARS-CoV-2 S1 protein, Pomplun, et al.,28 screened mixtures of synthetic peptides and found a peptide (TVFGLNVWKAYSK) ligand with a Kd of 250 nM. This peptide did not bind to the active site where the peptide interacts with ACE2, but the alternative binding site was not determined. Surprisingly, the SBP-1 peptide, which should bind to that active site, did not bind anywhere to the S1 receptor binding domain in the assay of Pomplun, et al.28 In our assay, SBP-1 peptide bound to the S1 protein as expected, but the synthetic high affinity peptide reported by Pomplun, et al.,28 did not bind. This suggests that the 108 kDa S1 protein provides a more accurate target for spike protein ligand discovery than the smaller S1 receptor binding domain.
Although vaccines against SARS-CoV-2 are now available, the COVID-19 pandemic continues due to incomplete vaccination of the population and emergence of resistant viral strains. Therefore, the need continues for small molecule therapeutic agents to prevent as well to treat SARS-CoV-2 infections. Due to their chemical diversity, natural products remain important sources of new therapeutic agents and lead compounds for drug development as indicated by the observation that since 1981, two-thirds of all new small molecule drugs approved by the FDA are natural products, mimics, or derivatives of natural products.29 As proof of concept that natural products can prevent SARS-CoV-2 cell entry, the natural product panduratin from the medicinal plant Boesenbergia rotunda was reported recently to be active against SARS-CoV-2 at both pre-entry and post-infection phases.30
In the discovery of panduratin as a potential therapeutic agent against SARS-CoV-2, cell-based assays were carried out using live SARS-CoV-2, which is slower and more labor intensive than AS-MS approaches like MagMASS. Furthermore, we enhanced the throughput of our previous 96-well MagMASS assay 5-fold by incorporating an automated robotic magnetic particle processor (Figure 1). Fast separation of the ligand-receptor complexes from the unbound compounds is essential to minimize the loss of signal due to dissociation of the ligand during sample processing. This is particularly important when the dissociation of the ligand is rapid. In this MagMASS assay for the discovery of ligands to the SARS-CoV-2 S1 protein, 5-fold faster sample processing resulted in 5-fold signal enhancement. An additional feature of our MagMASS assay was the use of data-dependent acquisition of high-resolution product ion tandem mass, which meant that not only elemental composition data but also structural information for ligands were acquired during a single analysis.
CONCLUSIONS
New applications and innovations continue to expand the utility and popularity of AS-MS.12 Among the suite of AS-MS approaches, MagMASS has been the most amenable to automation, and we report here an automated version that is 5-fold faster than has been achieved previously. In addition, we report the application of MagMASS to the discovery of natural ligands to the SARS-CoV-2 spike protein subunit S1, which have the potential to prevent the infection of human cells by SARS-CoV-2 and be developed for use as therapeutic agents. As an application of this new assay, we report the discovery of licorice compounds that bind to the SARS-CoV-2 S1 protein. Like any AS-MS discovery, the activity of these licorice ligands must be determined using follow up functional assays such as cell-based virus neutralization assays using live SARS-CoV-2.
ACKNOWLEDGEMENTS
Funding was provided by NIH grant T32AT010131 from the National Center for Complementary and Integrative Health. We thank Shimadzu Scientific Instruments for providing the Q-ToF mass spectrometer, EmerTher for providing the magnetic microbeads used during this investigation, and Dana Gibbon, Katherine Carter, and Brett Tyler for support from the Center for Genome Research and Biocomputing at Oregon State University.
REFERENCES
- 1.Li JW-H, Vederas JC: Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 325, 161–165 (2009) [DOI] [PubMed] [Google Scholar]
- 2.Shen B: A New Golden Age of Natural Products Drug Discovery. Cell 163, 1297–1300 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell TN, Ramsay J, Rieth SF, Shapiro MJ, Stroh JG: Solution-Based Indirect Affinity Selection Mass Spectrometry—a General Tool for High-Throughput Screening of Pharmaceutical Compound Libraries. Anal. Chem 86, 7413–7420 (2014) [DOI] [PubMed] [Google Scholar]
- 4.van Breemen RB, Huang C-R, Nikolic D, Woodbury CP, Zhao Y-Z, Venton DL: Pulsed Ultrafiltration Mass Spectrometry: A New Method for Screening Combinatorial Libraries. Anal. Chem 69, 2159–2164 (1997) [DOI] [PubMed] [Google Scholar]
- 5.Johnson BM, Nikolic D, van Breemen RB: Applications of Pulsed Ultrafiltration‐Mass Spectrometry. Mass Spectrom. Rev 21, 76–86 (2002) [DOI] [PubMed] [Google Scholar]
- 6.Nikolic D, Habibi-Goudarzi S, Corley DG, Gafner S, Pezzuto JM, van Breemen RB: Evaluation of Cyclooxygenase-2 Inhibitors Using Pulsed Ultrafiltration Mass Spectrometry. Anal. Chem 72, 3853–3859 (2000) [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y-Z, van Breemen RB, Nikolic D, Huang C-R, Woodbury CP, Schilling A, Venton DL: Screening Solution-Phase Combinatorial Libraries Using Pulsed Ultrafiltration/Electrospray Mass Spectrometry. J. Med. Chem 40, 4006–4012 (1997) [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Guo J, Luo Y, Broderick DJ, Schimerlik MI, Pezzuto JM, van Breemen RB: Screening for Ligands of Human Retinoid X Receptor-α Using Ultrafiltration Mass Spectrometry. Anal. Chem 79, 9398–9402 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, van Breemen RB: Development of a Screening Assay for Ligands to the Estrogen Receptor Based on Magnetic Microparticles and LC-MS. Comb. Chem. High Throughput Screen 11, 1–6 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Ma Y, Chen X, Xiong X, Shi S: Screening and Identification of BSA Bound Ligands from Puerariae lobata Flower by BSA Functionalized Fe3O4 Magnetic Nanoparticles Coupled with HPLC–MS/MS. J. Chromatogr. B 887–888, 55–60 (2012) [DOI] [PubMed] [Google Scholar]
- 11.Rush MD, Walker EM, Burton T, van Breemen RB: Magnetic Microbead Affinity Selection Screening (MagMASS) of Botanical Extracts for Inhibitors of 15-Lipoxygenase. J. Nat. Prod 79, 2898–2902 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muchiri RN, van Breemen RB: Affinity Selection–Mass Spectrometry for the Discovery of Pharmacologically Active Compounds from Combinatorial Libraries and Natural Products. J. Mass Spectrom 56, (2021) 10.1002/jms.4647 [DOI] [PubMed] [Google Scholar]
- 13.Rush MD, Walker EM, Prehna G, Burton T, van Breemen RB: Development of a Magnetic Microbead Affinity Selection Screen (MagMASS) Using Mass Spectrometry for Ligands to the Retinoid X Receptor-α. J. Am. Soc. Mass. Spectrom 28, 479–485 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safarik I, Safarikova M: Magnetic Techniques for the Isolation and Purification of Proteins and Peptides. BioMagnetic Res. Technol 2, 1–17 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Almeida FG; Vanzolini KL; Cass QB: Angiotensin Converting Enzyme Immobilized on Magnetic Beads as a Tool for Ligand Fishing. J. Pharm. Biomed. Anal 132, 159–164 (2017) [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Chen Y, Xiao C, Chen D, Xiao Y, Mei Z: Rapid Screening and Identification of α-Amylase Inhibitors from Garcinia xanthochymus Using Enzyme-Immobilized Magnetic Nanoparticles Coupled with HPLC and MS. J. Chromatogr. B 960, 166–173 (2014) [DOI] [PubMed] [Google Scholar]
- 17.Pochet L, Heus F, Jonker N, Lingeman H, Smit AB, Niessen WM, Kool J: Online Magnetic Bead Based Dynamic Protein Affinity Selection Coupled to LC–MS for the Screening of Acetylcholine Binding Protein Ligands. J. Chromatogr. B 879, 1781–1788 (2011) [DOI] [PubMed] [Google Scholar]
- 18.Vanzolini KL, Jiang Z, Zhang X, Vieira LCC, Corrêa AG, Cardoso CL, Cass QB, Moaddel R: Acetylcholinesterase Immobilized Capillary Reactors Coupled to Protein Coated Magnetic Beads: A New Tool for Plant Extract Ligand Screening. Talanta 116, 647–652 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salvatori G, Luberto L, Maffei M, Aurisicchio L, Roscilli G, Palombo F, Marra E: SARS-CoV-2 Spike Protein: An Optimal Immunological Target for Vaccines. J. Transl. Med 18, 1–3 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierri CL: SARS-CoV-2 Spike Protein: Flexibility as a New Target for Fighting Infection. Signal Transduct. Targeted Ther 5, 1–3 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D: Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292.e286 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajirahimkhan A, Simmler C, Yuan Y, Anderson JR, Chen S-N, Nikolić D, Dietz BM, Pauli GF, van Breemen RB, Bolton JL: Evaluation of Estrogenic Activity of Licorice Species in Comparison with Hops Used in Botanicals for Menopausal Symptoms. PLoS One 8: e67947 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G: XCMS Online: A Web-based Platform to Process Untargeted Metabolomic Data. Anal. Chem 84, 5035–5039 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O, Olson AJ: AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem 31, 455–461 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, Wang Q, Zhou H, Yan J, Qi J: Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 181, 894–904.e899 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S: Identification of SARS-CoV-2 Inhibitors Using Lung and Colonic Organoids. Nature 589, 270–275 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, Zhu Y, Xu J, Yao G, Zhang P, Wang M, Zhao Y, Lin G, Chen H, Chen L, Zhang J: Glycyrrhizic Acid Exerts Inhibitory Activity Against the Spike Protein of SARS-CoV-2. Phytomedicine 85, 153364 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomplun S, Jbara M, Quartararo AJ, Zhang G, Brown JS, Lee YC, Ye X, Hanna S, Pentelute BL: De Novo Discovery of High-Affinity Peptide Binders for the SARS-CoV-2 Spike Protein. ACS Cent. Sci 7, 156–163 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman DJ, Cragg GM: Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod 79, 629–661 (2016) [DOI] [PubMed] [Google Scholar]
- 30.Kanjanasirirat P, Suksatu A, Manopwisedjaroen S, Munyoo B, Tuchinda P, Jearawuttanakul K, Seemakhan S, Charoensutthivarakul S, Wongtrakoongate P, Rangkasenee N: High-content Screening of Thai Medicinal Plants Reveals Boesenbergia rotunda Extract and its Component Panduratin A as anti-SARS-CoV-2 Agents. Sci. Rep 10, 1–12 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]


