ABSTRACT
Urinary tract infection (UTI) is among the most common infections treated worldwide each year and is caused primarily by uropathogenic Escherichia coli (UPEC). Rising rates of antibiotic resistance among uropathogens have spurred a consideration of alternative treatment strategies, such as bacteriophage (phage) therapy; however, phage-bacterial interactions within the urinary environment are poorly defined. Here, we assess the activity of two phages, namely, HP3 and ES17, against clinical UPEC isolates using in vitro and in vivo models of UTI. In both bacteriologic medium and pooled human urine, we identified phage resistance arising within the first 6 to 8 h of coincubation. Whole-genome sequencing revealed that UPEC strains resistant to HP3 and ES17 harbored mutations in genes involved in lipopolysaccharide (LPS) biosynthesis. Phage-resistant strains displayed several in vitro phenotypes, including alterations to adherence to and invasion of human bladder epithelial HTB-9 cells and increased biofilm formation in some isolates. Interestingly, these phage-resistant UPEC isolates demonstrated reduced growth in pooled human urine, which could be partially rescued by nutrient supplementation and were more sensitive to several outer membrane-targeting antibiotics than parental strains. Additionally, phage-resistant UPEC isolates were attenuated in bladder colonization in a murine UTI model. In total, our findings suggest that while resistance to phages, such as HP3 and ES17, may arise readily in the urinary environment, phage resistance is accompanied by fitness costs which may render UPEC more susceptible to host immunity or antibiotics.
IMPORTANCE UTI is one of the most common causes of outpatient antibiotic use, and rising antibiotic resistance threatens the ability to control UTI unless alternative treatments are developed. Bacteriophage (phage) therapy is gaining renewed interest; however, much like with antibiotics, bacteria can readily become resistant to phages. For successful UTI treatment, we must predict how bacteria will evade killing by phage and identify the downstream consequences of phage resistance during bacterial infection. In our current study, we found that while phage-resistant bacteria quickly emerged in vitro, these bacteria were less capable of growing in human urine and colonizing the murine bladder. These results suggest that phage therapy poses a viable UTI treatment if phage resistance confers fitness costs for the uropathogen. These results have implications for developing cocktails of phage with multiple different bacterial targets, of which each is evaded only at the cost of bacterial fitness.
KEYWORDS: antimicrobial resistance, bacteriophage therapy, urinary tract infection, uropathogenic E. coli
INTRODUCTION
Urinary tract infection (UTI) is an extremely common bacterial infection, causing nearly 10 million cases in the United States alone each year (1, 2). These infections disproportionately affect women, with approximately half of women experiencing at least one UTI during their lifetime (3). Uropathogenic Escherichia coli (UPEC) is the leading cause of UTI worldwide, causing upward of 75% of infections. UTI is one of the most common causes of outpatient antibiotic prescriptions (4, 5), and the rise of antibiotic resistance among UPEC isolates threatens existing treatments for UTI (6, 7). Current technological and economic challenges limit the development of novel antibiotics, and antibiotic resistance develops rapidly once antibiotics are introduced (8–10). Because of these challenges, several new nonantibiotic alternatives to treat UPEC UTI have been proposed (11–17). One such alternative are bacteriophages (phages), which are viruses that use bacteria as their natural host.
Soon after the discovery of phages in the early 1900s, phage therapy was applied to bacterial infections (18), including UTI caused by UPEC (19). Despite this early interest, phage therapy was abandoned largely in favor of antibiotics, but with the modern rise in antibiotic-resistant infections, interest in phage therapy is resurging (20, 21). Phage therapy holds several appealing properties. Bacteriophages outnumber bacteria by an estimated 10:1 ratio (22), and phages that target human pathogens are isolated readily from environmental and human sources (23–25). Additionally, phages replicate within the bacterial host, generating a source of new phages for the duration of the pathogen presence (self-dosing). Moreover, phages may have fewer off-target impacts on the host microbiota (26). To date, phage therapy for UTI has been confined to compassionate care use (27–31) and has shown generally favorable results. Clinical trials testing dosing and administration methods for UTI phage therapy are currently in the early stages of development (32, 33). A randomized clinical trial conducted on UPEC UTI described phage therapy as noninferior to standard-of-care antibiotics but also nonsuperior to placebo treatment (34). Factors specific to the urinary environment or to uropathogens, such as UPEC, may impact phage efficacy, and thus, understanding phage-bacterial interactions in the urinary tract is critical to drive UTI phage therapies.
A key challenge to phage therapy efficacy is the emergence of bacterial resistance toward phage killing. Similar to evolving resistance to antibiotics, bacteria can rapidly develop resistance to phage. Resistance mechanisms include blocking of phage adsorption through the mutation of phage receptors, masking of phage targets, and producing competitive inhibitors (35, 36). Additionally, even after phage internalization, bacteria can resist phage infection by blocking DNA entry or degrading viral DNA intracellularly (37). Phage resistance is also associated with reduced bacterial fitness, particularly related to phage receptor modification (35, 38). Fitness costs include reduced virulence and increased susceptibility to antimicrobials (35, 39–43) or immune clearance (44). Phage resistance paired with reduced virulence suggests that “steering” bacteria toward these phenotypes may be a viable approach for treating infections (44). Despite the current knowledge on phage resistance and potential fitness costs, the effect of phage resistance in uropathogens within the urinary tract has not been assessed.
In this study, we evaluate mechanisms of UPEC phage resistance to two genetically distinct bacteriophages (24) and test the hypothesis that phage resistance may bring associated bacterial fitness costs within the urinary environment. Through unbiased screening and sequencing, we found that UPEC resistance to phages HP3 and ES17 is associated with mutations in the lipopolysaccharide (LPS) biosynthesis pathway. Phage resistance mutations negatively impact UPEC growth in urine and resistance to membrane-targeting antibiotics and result in reduced colonization of the urinary tract in vivo. Together, these findings provide insight into mechanisms and associated impacts of phage resistance during uropathogenesis and suggest phage-associated mutations reduce UPEC fitness in the urinary tract. These findings provide a critical knowledge base for the future development of UTI phage therapy.
RESULTS
UPEC strains resistant to phage emerge rapidly in vitro in LB and human urine.
Conventionally, bacterium-phage interactions are studied in bacteriologic medium; however, recent work has shifted to assess these interactions in the context of the host, including ex vivo blood or in vivo tissues (44–46). To assess how phage activity is influenced by the urinary environment, we compared the susceptibility of four UPEC strains to two distinct phages, namely, HP3 and ES17 (24, 31) either in LB medium or human urine. These phage target extraintestinal pathogenic E. coli (ExPEC), including UPEC (24, 31, 35, 47, 48). For UPEC strains, we selected well-characterized UPEC cystitis and pyelonephritis isolates (UTI89 and CFT073, respectively) and DS515 and DS566, which are recent isolates from patients with neurogenic bladders from spinal cord injury.
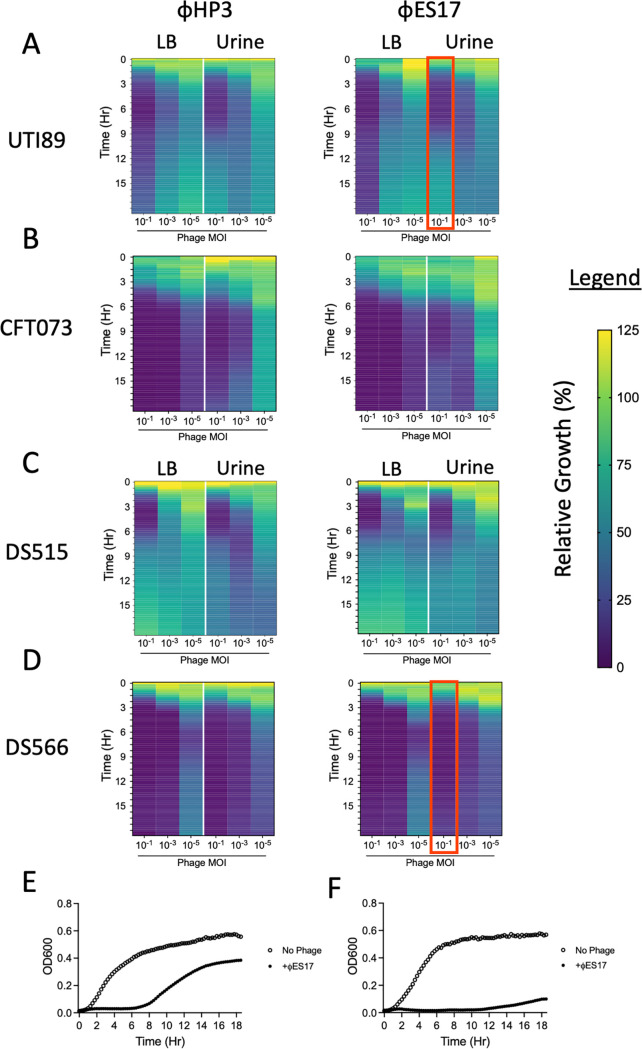
Approximately 107 CFU of UPEC were challenged with 106, 104, or 102 PFU of phage (multiplicity of infection [MOI] of 10−1, 10−3, and 10−5, respectively) in 96-well microtiter plates. Within 1 to 2 h of phage challenge, the relative growth of phage-treated wells declined rapidly. Minimal growth (<25% growth of optical density at 600 nm [OD600] in treated versus untreated wells) was observed for the first ~6 h in the presence of the two higher MOIs for both phages (Fig. 1A to D; see Fig. S1 to S3 in the supplemental material). After this time, many of the cultures began to “rebound” their relative growth, suggesting bacterial resistance toward the phage. This activity was observed most strongly in strains UTI89 (Fig. 1A) and DS515 (Fig. 1C). There were no clear differences in relative growth based on the medium used (LB versus urine), although a decreasing phage MOI reduced the relative growth suppression. UTI89 and DS566 were selected for further investigation into the mechanisms of phage resistance based on the differential growth kinetics observed during phage challenge. While relative growth rebounded quickly in phage-challenged strains (Fig. 1E), DS566 relative growth was slower, reaching only ~20% of no-phage control growth after 18 h (Fig. 1F).
FIG 1.
Phage-bacterial dynamics are similar in LB and pooled human urine with resistance developing under both conditions. Heatmaps of relative bacterial growth (OD600 of phage treated well/OD600 of untreated well) of bacteria challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All bacteria were challenged at multiplicities of infection (MOIs) of 10−1, 10−3, and 10−5. (E) Representative growth curve of UTI89 challenged with ES17 at a MOI of 10−1 in urine highlighted with red box in A. (F) Representative growth curve of DS566 challenged with ES17 at MOI of 10−1 in urine highlighted with red box in D. All heatmaps are representative of three independent experiments performed in at least technical duplicates. Growth curves for these data appear in Fig. S1 and S3.
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−1. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S1, JPG file, 0.3 MB (300KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−3. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S2, JPG file, 0.3 MB (299.3KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−5. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S3, JPG file, 0.3 MB (301.5KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phage resistance is associated with mutations in lipopolysaccharide (LPS) biosynthesis.
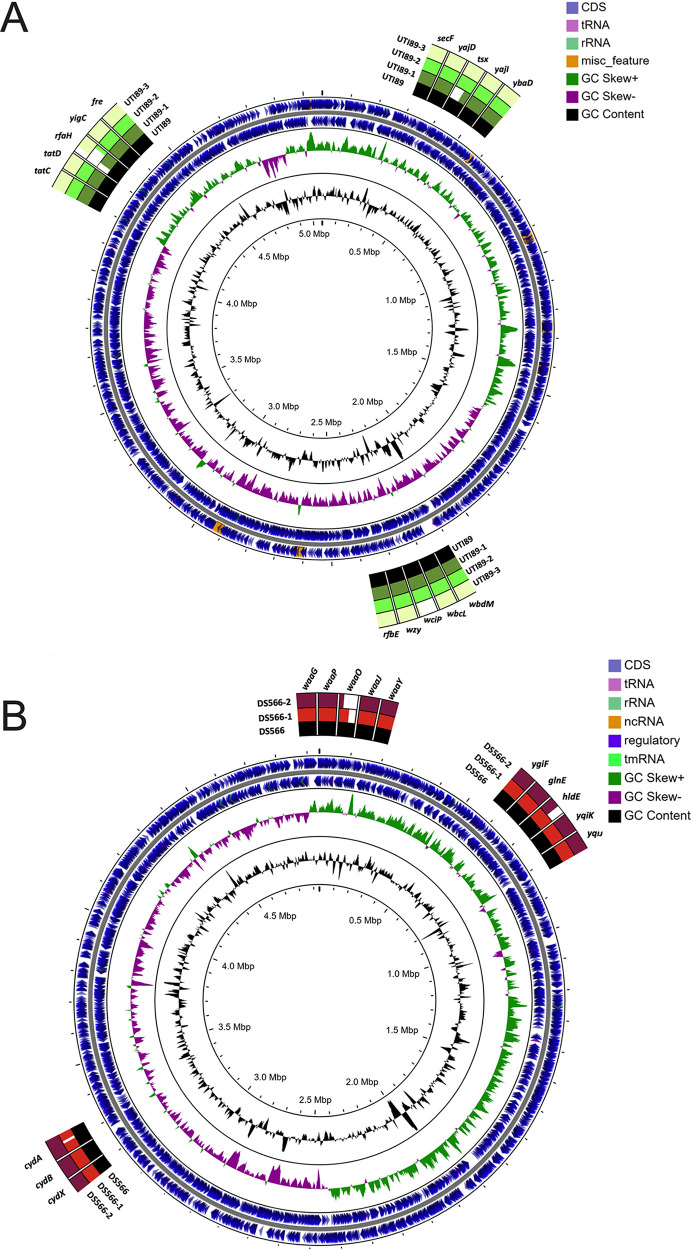
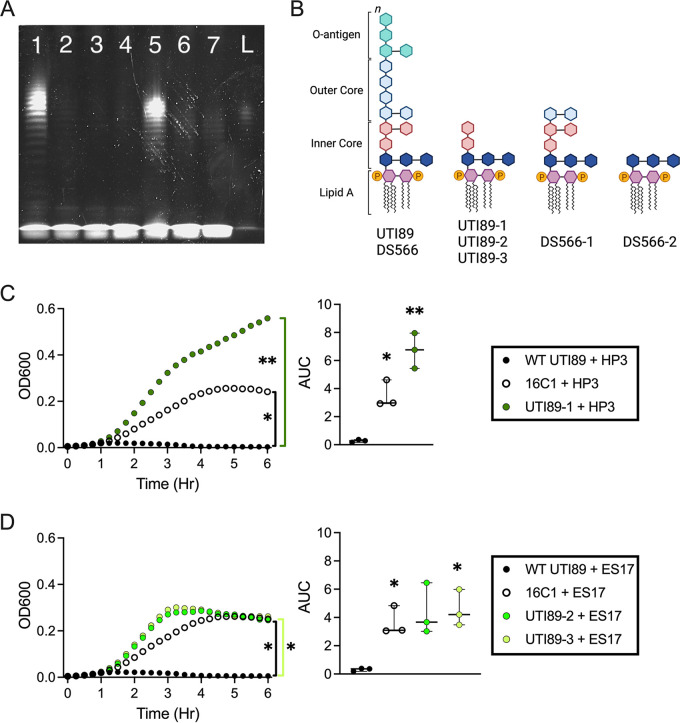
To isolate phage-resistant UPEC, bacteria were passaged a total of four times in the presence of phage in liquid culture and on agar containing phage. Three UTI89 isolates resistant to HP3 or ES17 and two DS566 isolates resistant to ES17 were subjected to whole-genome sequencing. All UTI89 isolates resistant to phage, regardless of which phage, had mutations in the transcription factor rfaH (locus tag b3842) (Fig. 2A, Table 1). DS566 strains had mutations in galactotransferase rfaI/waaO (locus tag b3627). DS566-2 had an additional mutation in rfaE/hldE (locus tag b3052) (Fig. 2B; Table 1). Several non-LPS-related mutations were also observed (see Table S1 in the supplemental material). To visualize changes to LPS in phage-resistant strains, LPS was isolated via hot aqueous-phenol extraction and subjected to SDS-PAGE. Loss of the LPS O-antigen was apparent in all phage-resistant strains compared with the parental strains (Fig. 3A). These results agree with predicted LPS structures in rfaH and rfaE/hldE mutants (truncated inner core), while rfaI/waaO is likely to result in outer core truncation (49) (Fig. 3B). Phage-resistant strain UTI89-2 was serially passaged three times, and no sensitivity to phage ES17 was regained (see Fig. S4 in the supplemental material), suggesting that these mutations are stable even in the absence of selective pressure. To test whether a specific mutation of LPS biosynthesis could confer resistance to phages HP3 or ES17, we challenged UTI89 16C1, which harbors a transposon insertion in the rfa operon, with phage. Strain 16C1, as well as phage-resistant strain UTI89-1, grew significantly better than wild-type UTI89 in the presence of HP3 (P = 0.023 and P = 0.0046, respectively) (Fig. 3C). Interestingly, UTI89-1 growth was enhanced compared with 16C1 when challenged with HP3, suggesting a mutation of the promoter rfaH may confer more resistance than a mutation to the rfa operon itself. In the presence of ES17, 16C1 also grew significantly better than wild-type UTI89 (P = 0.041) (Fig. 3D). Similarly, phage-resistant strains UTI89-2 and UTI89-3 achieved greater growth than wild-type UTI89 (P = 0.092 and P = 0.029, respectively) (Fig. 3D).
FIG 2.
Whole-genome sequencing of phage-resistant bacteria identifies multiple mutations in UPEC strains UTI89 and DS566 exposed to phages HP3 and ES17. Alignment of whole-genome sequences depicting mutations identified in UTI89 phage-resistant mutants (A) and DS566 phage-resistant mutants (B) compared with that of parental wild-type (WT) strains.
TABLE 1.
LPS biosynthesis-associated mutations observed in phage-resistant UPECa
| Strain | Phage resistant against | Gene(s) mutated | Earliest LPS mutation | Predicted effect |
|---|---|---|---|---|
| UTI89-1 | HP3 | rfaH | rfaH | Truncated LPS inner core |
| UTI89-2 | ES17 | rfaH | rfaH | Truncated LPS inner core |
| UTI89-3 | ES17 | rfaH, wbbL | rfaH | Truncated LPS inner core |
| DS566-1 | ES17 | rfaI/waaO | rfaI/waaO | Truncated LPS outer core |
| DS566-2 | ES17 | rfaI/waaO, rfaE/hldE | rfaE/hldE | Truncated LPS inner core |
Mutations associated with LPS biosynthesis in phage-resistant UPEC were identified through whole-genome sequencing. The site and type of LPS biosynthesis-associated mutations were observed in phage-resistant strains arising from in vitro screening and were identified by whole-genome sequencing.
FIG 3.
Phage resistance to HP3 and ES17 is associated with LPS deficiency. (A) Representative SDS-PAGE image of WT and phage-resistant UPEC LPS isolated through hot aqueous-phenol extraction and run on a 4% to 12% SDS-polyacrylamide gel loaded with 15 μL of isolated product. Lane 1, WT UTI89; lane 2, UTI89-1; lane 3, UTI89-2; lane 4, UTI89-3; lane 5, WT DS566; lane 6, DS566-1; lane 7, DS566-2; L, LPS standard. Isolation and visualization of LPS was performed in two independent experiments with comparable results. (B) Predicted LPS structures based on gene mutations noted in the sequencing analysis. (C and D) Growth of WT UTI89, rfa transposon mutant 16C1, or phage-resistant UTI89 in the presence of HP3 (C) or ES17 (D) over 6 h. Growth curves of bacteria in the presence of phage are representative of the mean value of three independent experiments, with each performed with four technical replicates. Bars represent median and 95% confidence intervals (CIs). Growth curves were analyzed by repeated measures two-way ANOVA with Geisser-Greenhouse correction and Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01.
Serially passaged phage-resistant UPEC strains do not readily regain sensitivity to ES17. Three independent cultures of phage-resistant strain UTI89-2 were serially passaged three times in LB media. Each passage, the parental UTI89-2 strain and wild-type UTI89 were challenged with ES17 for 6 h. Data are grouped by passage number 0 (A), 1 (B), 2 (C), and 3 (D). Each growth curve represents the mean value of three technical replicates. Download FIG S4, JPG file, 0.3 MB (297.7KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Non-LPS-associated mutations identified during sequencing of phage-resistant isolates. Site and type of non-LPS biosynthesis-associated mutations observed in phage-resistant strains arising from in vitro screening and identified by whole-genome sequencing. Download Table S1, DOCX file, 0.1 MB (54.2KB, docx) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phage resistance is associated with altered bacterial biofilm formation and adherence and invasion of bladder cells.
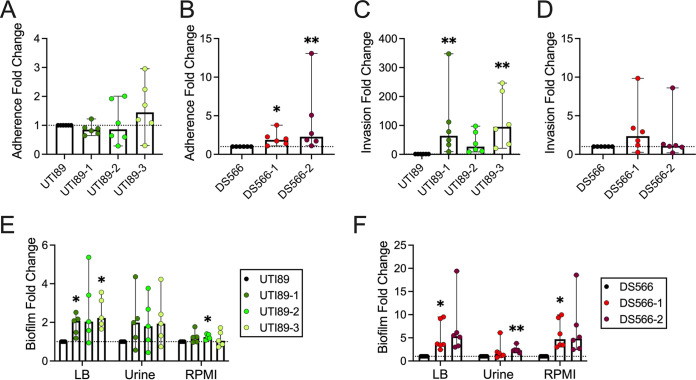
UPEC adheres to and invades the bladder epithelium to form intracellular bacterial reservoirs capable of reseeding infection (50, 51); thus, we assessed if phage resistance alters UPEC interactions with HTB-9 bladder cells. No change in adherence was seen in any of the UTI89 phage-resistant strains (Fig. 4A); however, both DS566 strains had modestly increased adherence relative to the wild type, with a 1.9-fold increase for DS566-1 (P = 0.014) and 2.3-fold increase for DS566-2 (P = 0.0023) (Fig. 4B). Invasion of HTB-9 cells was increased 50- to 100-fold in 2 UTI89 strains, namely, UTI89-1 and UTI89-3 (P = 0.0053 and P = 0.0012, respectively) (Fig. 4C), while no change in invasion was detected in the DS566 strains (Fig. 4D).
FIG 4.
Adherence and invasion of HTB-9 cells as well as biofilm formation are altered in phage-resistant UPEC. (A and B) UTI89 (A) and DS566 (B) and respective phage-resistant strain adherence to HTB-9 cells after 30 min of infection; MOI of 1. HTB-9 cells were infected with UTI89 (C) or DS566 (D) and respective phage-resistant mutants (MOI of 1) for 2 h before the medium was changed to medium containing antibiotics to kill extracellular bacteria. After 2 h of antibiotic treatment, cells were lysed and intracellular bacteria enumerated. Biofilm formation of UTI89 (E) or DS566 (F) and respective phage-resistant mutants in LB, urine, or RPMI 1640 quantified by crystal violet uptake. All adherence, invasion, and biofilm assays were performed in at least technical duplicate in five to six independent experiments, with points on graphs representing mean values of each independent experiment. Bars represent median and 95% confidence intervals. Data were analyzed by Kruskal-Wallis test using Dunn’s multiple-comparison test (A to D) or repeated-measures two-way ANOVA with Geisser-Greenhouse correction and Dunnett’s multiple-comparison test (E, F). *, P < 0.05; **, P < 0.01.
As LPS modifications may change bacterial surface properties (52, 53), we evaluated the ability of phage-resistant strains to form biofilms in LB and under host mimetic conditions (pooled human urine and RPMI 1640). In general, biofilm formation by phage-resistant UPEC was increased relative to that of parental strains, although not all conditions achieved statistical significance as indicated in figure panels. UTI89-1 (1.9-fold, P = 0.049) and UTI89-3 (2.5-fold, P = 0.033), demonstrated significantly increased biofilm formation in LB medium, while UTI89-2 demonstrated enhanced biofilm formation in RPMI 1640 (1.2-fold, P = 0.032) (Fig. 4E). DS566-1 displayed increased biofilm formation in both LB and RPMI 1640 compared with its wild-type strain (3.6-fold, P = 0.036, and 4.7-fold, P = 0.021, respectively), whereas DS566-2 had increased biofilm formation in pooled human urine (2.3-fold, P = 0.0065) (Fig. 4F).
Phage resistance is associated with increased antibiotic susceptibility in urine.
Since LPS truncation may allow antimicrobials to access the bacterial surface more easily, we compared the susceptibility of wild-type and phage-resistant strains to antibiotics colistin (polymyxin E) and polymyxin B, which both interact with the bacterial outer membrane, in pooled human urine. We observed an approximately 2-fold decrease in the colistin MIC for strains UTI89-1, UTI89-2, and UTI89-3 (Fig. 5A) (P = 0.0096, P = 0.0040, and P = 0.014, respectively) and a nonsignificant change in MIC versus polymyxin B for these same strains (Fig. 5B). In DS566, we observed a 2-fold reduction in MIC for DS566-1 to both colistin and polymyxin B, while DS566-2 had colistin and polymyxin B MICs that were 8- to 16-fold lower than the wild-type strain (0.19 μg/mL and 0.09 μg/mL, respectively) (P = 0.0001 and P = 0.0001) (Fig. 5C and D). In LB medium, differences between phage-resistant derivatives and parental strains were largely absent (see Fig. S5 in the supplemental material).
FIG 5.
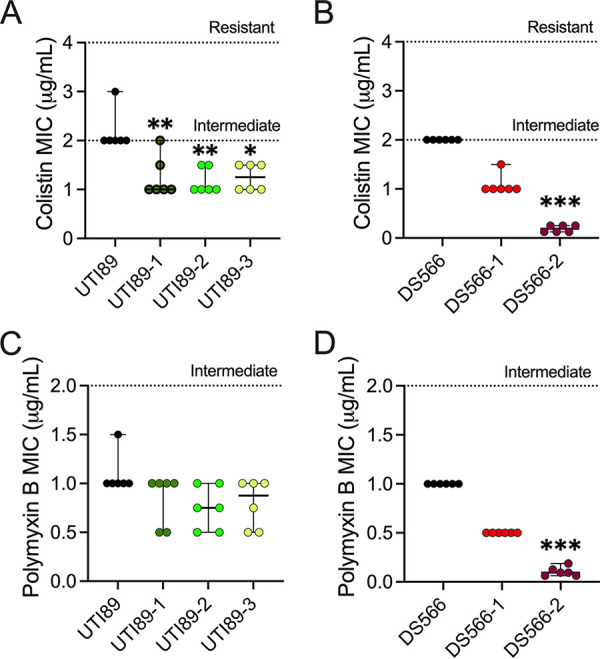
Phage resistance renders UPEC more susceptible to antibiotics that target the bacterial outer membrane. Colistin MICs of UTI89 (A) and DS566 (B) and respective phage-resistant strains in pooled human urine. Polymyxin B MICs of UTI89 (C) and DS566 (D) and respective phage-resistant mutants in pooled human urine. Assays were performed in technical duplicate in six independent experiments, with points on graphs representing mean values of each independent experiment. Bars represent median and 95% confidence intervals. Data were analyzed by Kruskal-Wallis test using Dunn’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MICs for antibiotics targeting the bacterial outer membrane are not altered when phage-resistant bacteria are grown in LB media. Colistin MICs of UTI89 (A) and DS566 (B) and isogenic phage-resistant mutants. Polymyxin B MICs of UTI89 (C) and DS566 (D) and isogenic phage-resistant mutants. Individual points are representative of independent experiments performed in duplicate. Bars represent median and 95% confidence intervals. Download FIG S5, TIF file, 0.3 MB (346.8KB, tif) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phage-resistant UPEC growth is attenuated in urine.
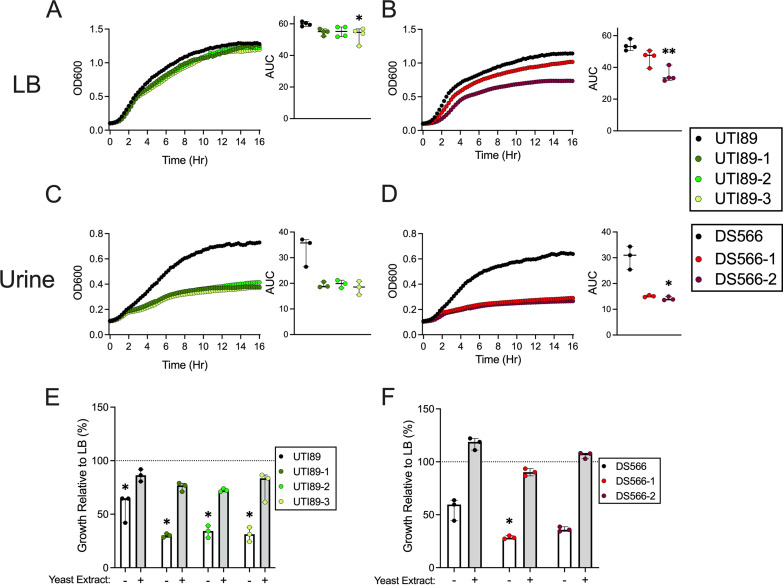
To evaluate the impact of phage resistance and associated LPS modifications on bacterial growth, bacteria were grown in either LB medium or pooled human urine for 16 h. While the growth of most phage-resistant bacteria was similar to that of wild-type strains in LB medium (Fig. 6A and B), bacterial growth was severely attenuated in urine across strains regardless of the specific mutations (Fig. 6C and D). Defective growth in urine suggests the possibility that phage-resistant UPEC strains either (i) are more susceptible to active inhibitory factors or (ii) are less fit to grow under nutrient-poor conditions than parental strains. To delineate between these possibilities, we supplemented urine with yeast extract, a primary nutrient source in LB medium. Yeast extract supplementation rescued phage-resistant strain growth to levels resembling growth in LB (Fig. 6E and F). These results suggest that consequences of phage resistance (e.g., LPS deficiency) may directly or indirectly limit nutrient acquisition under nutrient-deplete conditions, such as human urine; however, deciphering this mechanism further is beyond the scope of this study.
FIG 6.
Phage-resistant UPEC growth is attenuated in urine but can be partially restored with nutrient supplementation. WT and phage-resistant strains were grown for 16 h with optical density (left panels) and area under curve (AUC; right panels) displayed. Phage-resistant mutants of UTI89 (A) and DS566 (B) grow similar to parental strains in LB media, but growth is attenuated in pooled human urine (C and D, respectively). UTI89 (E) and DS566 (F) and respective phage-resistant mutants were grown in pooled human urine with or without yeast extract supplementation or LB media for 16 h. Percent growth relative to LB for urine (white bar) and yeast extract-supplemented urine (gray bar) are displayed. Assays were performed in at least technical triplicate in 3 to 4 independent experiments, with points on graphs representing mean values of each independent experiment. Bars represent median with 95% CIs. All data were analyzed by Kruskal-Wallis test using Dunn’s multiple-comparison test. Growth in urine and in urine supplemented with yeast extract was compared with that of the LB control for each strain. *, P < 0.05; **, P < 0.01.
Phage-resistant UPEC strains exhibit decreased bladder colonization.
Because phage-resistant strains had comparable or increased biofilm formation, adherence, and internalization to their parental strains in vitro, we evaluated the ability of two phage-resistant strains (UTI89-2 and DS566-2) to colonize the mouse urinary tract compared with their parental strains. These strains were selected because they harbor distinct LPS biosynthesis pathway mutations with different degrees of LPS truncation limited to the outer core (UTI89-2) or outer and inner cores (DS566-2). Mice received transurethral bacteria (108 CFU), and at 24 h postinfection, bladder UPEC burdens were quantified. UTI89-2 was significantly attenuated in its colonization compared with wild-type UTI89 (249-fold decrease, P = 0.0007) (Fig. 7A). Although wild-type DS566 achieved lower bacterial burdens in the bladder than wild-type UTI89, DS566-2 displayed a potent colonization defect, with bacterial burdens below the limit of detection in all bladders at 24 h postinfection (P < 0.0001) (Fig. 7B).
FIG 7.
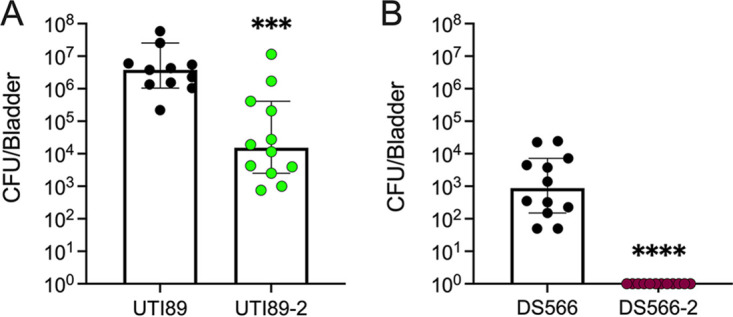
Phage-resistant UPEC strains display decreased bladder colonization in a murine model of UTI. Female C57BL/6J mice were infected transurethrally with 108 CFU of UTI89 or UTI89-2 (A) or DS566 or DS566-2 (B). After 24 h, bladders were removed, and bladder bacterial burdens were assessed. Points represent individual mice (biological replicates) assayed over two separate experiments. (A) UTI89, n = 11; UTI89-2, n = 12. (B) DS566, n = 12; DS566-2, n = 12. Lines represent median and 95% confidence intervals. Data were analyzed by Mann-Whitney test. ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
Although well-characterized in bacteriologic media, phage-bacterial dynamics are not well-defined in the urinary environment. Here, we identified that (i) in both LB and human urine, resistance arises to two genetically distinct phages with mutations converging on LPS biosynthesis; (ii) phage resistance attenuates UPEC growth in urine, but defective growth is partially rescued by nutrient supplementation; (iii) phage-resistant bacteria are sensitized to membrane-interacting antibiotics in human urine; and finally, (iv) while phage resistance may enhance adherence, invasion, and biofilm formation in vitro, it does not translate to heightened bladder colonization in vivo. In total, these findings suggest that phage-resistant bacteria may arise during phage therapy for UTI but that the resulting bacteria may be less capable of causing disease and/or sensitized to other treatment options.
Phage resistance is a well-documented phenomenon; however, few studies have assessed phage resistance outside bacteriologic media and in environments relevant to human medicine. Our results demonstrate that UPEC-phage dynamics are similar across environmental conditions but may be more highly influenced by bacterial host genetics as revealed by differences in phage killing kinetics among UPEC strains. In both LB and urine, phage resistance developed within 16 h, and mutations observed in response to HP3 and ES17 phages shared roles in LPS synthesis. In agreement with our study, ExPEC strains have been shown previously to evade HP3 killing by mutating LPS synthesis (35). Additionally, 16C1, an independently derived LPS mutant in UTI89 (54), was also resistant to HP3 and ES17. LPS is a major component of the Gram-negative outer membrane and provides stability and anchoring for other outer membrane proteins (55). LPS mutation in E. coli attenuates several cellular processes, including cellular membrane integrity, resistance to antibiotics, growth in low pH and high detergent conditions, and importantly, resistance to bacteriophage (35, 52, 56–60). Here, we demonstrate that these same detrimental effects occur in the urinary environment among phage-resistant strains. Some of our strains harbor a mutation in transcription factor rfaH, which is involved in the assembly of the LPS (61) but also regulates capsule and alpha-hemolysin among other factors (62). The contributions of these other pathways to phage resistance and associated phenotypes were not examined in the current study.
Urine composition displays dynamic nutrient and solute concentrations throughout the day and between individuals (63–65). Phage-resistant UPEC strains were equally capable of growth in LB as parental strains but grew poorly in urine. This result echoes recent work by García et al. who demonstrated that several genes involved in LPS biosynthesis are important for bacterial growth in urine (66). The addition of yeast extract to urine rescued bacterial growth to levels resembling growth in LB, suggesting that this phenotype may be due ineffective nutrient acquisition or utilization under deplete conditions. In human urine, amino acid and carbohydrate metabolic pathways are upregulated in UPEC compared with iron-limited LB, and mutations in these central metabolic pathways attenuate UPEC fitness in vivo (67). Outer membrane proteins, used to transport nutrients through the outer membrane, often form complexes with LPS for assembly and insertion into the membrane (52, 55, 68). Decreased efficacy or number of outer membrane proteins in phage-resistant strains may explain how nutrient supplementation overcomes growth defects in urine; however, the exact mechanisms underlying these results are outside the scope of this study.
Increased biofilm formation occurs in E. coli harboring LPS “deep-rough” mutations in vitro (69). Since enhanced biofilms could lead to worse outcomes in patients treated with phages, we assessed if the phage-resistant UPEC displayed this same increased biofilm formation. Partially aligning with previous literature, we observed in vitro biofilm increases in UTI89 strains containing rfaH mutations, although increased biofilm formation was not observed uniformly across all conditions. Increased biofilm formation was reported in E. coli containing rfaH mutations, although differing E. coli strains, biofilm generation, and enumeration methods were used (70). One strain, UTI89-1, also possessed a frameshift mutation in tsx, encoding an outer membrane protein and receptor to a T6-like phage (71); however, given the phenotypic similarities between UTI89-1 and the other two phage-resistant UTI89 strains not harboring tsx mutations, we hypothesize tsx is unlikely to significantly contribute to the observed in vitro phenotypes under our experimental conditions. Additionally, we observed increased biofilm formation in phage-resistant DS566 strains that was most pronounced in DS566-2, the most severely attenuated LPS structure based on genomic predictions. Although in vitro biofilm assays may not fully reflect the capacity to form biofilms in vivo, our results corroborate observations by Nakao et al. who demonstrated enhanced biofilm production by E. coli defective in LPS heptose biosynthesis (69). A limitation of our study is that all phage-resistant strains identified were isolated in vitro and yet previous work by our lab suggests that resistance is likely to develop in vivo as well (35).
UPEC interactions with the bladder epithelium are crucial for UTI pathogenesis. We observed a minimal impact of phage resistance to HTB-9 cell adherence in the UTI89 background, although adherence was significantly increased in DS566 phage-resistant strains. These results echo work done by Nagy et al. who found no role for rfaH in adherence to intestinal cells (72). In contrast, two of three UTI89 phage-resistant strains showed an increased invasion of HTB-9 cells. Similarly, avian pathogenic E. coli strains lacking rfaH are more readily engulfed by chicken macrophages than the wild type (73). The mechanism for this increased uptake was not further investigated but could explain the increased invasion of HTB-9 cells.
Since our results suggested that phage resistance could alter host-UPEC interactions, we used a mouse model of UTI to investigate in vivo infection outcomes. Phage-resistant strains in both UTI89 and DS566 backgrounds were worse at colonizing the murine bladder than their wild-type counterparts. Similarly, targeted inactivation of rfaH dramatically lowers the recovery of UPEC strain 536 from the urinary tract at 21 days postinfection (62). Additionally, Aguiniga et al. used targeted gene deletions to identify LPS domains essential for colonization of the bladder using UPEC strain NU14 (74). While the LPS mutations assessed by this group are not identical to those observed in our study, the functional consequences (e.g., outer membrane truncation and inner membrane truncation) are likely similar. In addition, García et al. have shown recently that rfaG, another gene involved in LPS biosynthesis, is important for colonization of the murine bladder (66). Of note, we did not investigate all phage-resistant strains isolated from this study in vivo, and it remains possible that alternative phenotypes may be present in other phage-resistant UPEC strains. Although our study does not investigate the immune response to phage-resistant UPEC with LPS modifications, others have observed the “rough” LPS phenotype in several asymptomatic bacteriuria strains, suggesting that these pathogens may be less virulent in humans (75). In fact, asymptomatic bacteriuria isolates with truncated LPS, among other virulence gene mutations, have been suggested as possible competitors which could be intentionally introduced in patients to prevent recurrent UPEC infection (76–79). In agreement with our study, bladder colonization using these UPEC strains is not always achieved despite frequent bacterial inoculations, suggesting a possible colonization defect of these strains (79).
Phage and antibiotics have the potential to work together synergistically (43, 80). Indeed, we observed that phage-resistant UPEC strains were more sensitive to antibiotics targeting the bacterial membrane in urine, a condition that was not investigated previously. Colistin and polymyxin B both permeabilize the bacterial outer membrane by interacting with lipid A (81). As observed in the biofilm and adherence and invasion assays, enhanced susceptibility to antibiotics was most pronounced in DS566-2, the strain with the most significant predicted LPS truncation. Although Gram-negative pathogens lacking the lipid A portion of LPS are resistant to colistin (82, 83), phage-driven mutations in our strains are predicted to retain lipid A.
In summary, bacteria quickly become resistant to phage under urinary tract conditions; however, mutations providing phage resistance may come at the cost of bacterial fitness and ultimately reduce pathogenesis in the urinary environment. Many other targets for phage exist in addition to LPS (38) raising the appealing possibility that combining phages with distinct targets could both reduce bacterial burden and attenuate bacterial virulence of emerging antibiotic-resistant bacteria. These findings expand our knowledge of phage resistance of UPEC in the context of the urinary tract and support its continued development as a target for controlling UTI.
MATERIALS AND METHODS
Bacterial strains and mammalian cell lines.
Uropathogenic E. coli strains UTI89 (84), CFT073 (ATCC 700928) (85), DS515, and DS566 (accession CP092534) were used to isolate bacteriophage-resistant bacteria. DS515 and DS566 are urinary isolates from spinal cord injury patients with neurogenic bladders at the Michael E. DeBakey VA Medical Center (Houston, TX). UTI89 strain 16C1 harbors a transposon insertion within the rfa operon and has been described previously (54). All bacterial strains were grown overnight, shaking, at 37°C prior to experiments. Phages HP3 (accession KY608967) and ES17 (accession MN508615) were isolated from environmental sources and wastewater, respectively (24, 31). A human bladder epithelium carcinoma cell line (ATCC HTB-9) was grown in RPMI 1640 (Corning) containing 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% CO2 and was passaged every 3 to 5 days.
Bacteriophage preparation.
Purified HP3 and ES17 stocks were prepared as described previously (47), their titers were determined, and they were stored in phage buffer (35) at 4°C until use.
Human urine pool preparation.
Urine samples were collected from six healthy male and female volunteers, 20 to 50 years old, under approval of the Baylor College of Medicine (BCM) institutional review board (IRB) (protocol H-47537). Following collection, urine was warmed to 37°C and filtered (0.22 μm) before storage at 4°C.
UPEC growth in the presence of phage.
Overnight UPEC cultures were diluted 1:100 in LB or pooled human urine and added to 96-well microtiter plates. For phage challenge, bacteriophage preparations diluted in phosphate-buffered saline (PBS) were added at an MOI of 0.1, 0.001, or 0.00001 to reach a final volume of 150 μL. Control bacterial wells were treated with PBS alone. Growth at 37°C was measured by OD600 every 15 min for 18 h under aerobic shaking conditions using a Tecan Infinite 200 plate reader. Relative bacterial growth was determined by calculating the percent OD600 of a given well compared with the mean of bacterium-only wells at that same time point.
Isolation of phage-resistant bacteria.
Overnight bacterial cultures were diluted 1:100 in fresh LB or pooled human urine and challenged with phage (MOI, 0.1) in a 150-μL total volume. OD600 was measured every 15 min as described above. Buffer (PBS) without phage was a control for noninfected bacterial growth. After 18 h, phage-treated wells with bacterial growth were streaked onto soft agar overlay plates containing phage. This process was repeated once for colonies which grew on phage top agar to isolate clonal populations. Phage-resistant isolates were confirmed by phage spot assay onto lawns of UPEC.
Purification and visualization of LPS.
UPEC LPS was isolated through hot aqueous-phenol extraction following methods described previously (86). Extracted LPS samples (15 μL) were run at 120 V for 1.5 h on 4% to 12% SDS-polyacrylamide gels and stained using the Pro-Q Emerald 300 lipopolysaccharide gel stain kit (Molecular Probes) following the manufacturer’s directions. Gels were visualized on a ProteinSimple AlphaImager HP system.
Generating bacterial growth curves for phage-resistant bacteria in LB and urine.
For growth assessments of phage-resistant bacteria, overnight cultures were diluted 1:100 in LB media or pooled human urine and added to 96-well microtiter plates (100 μL). Growth at 37°C was measured by OD600 every 15 min for 16 h under aerobic shaking conditions using a BioTek Cytation 5 plate reader (Gen5 v3.10).
Biofilm assays.
Bacterial biofilms were quantified as described previously with minor adaptations (87). Overnight cultures were diluted to an OD600 of 0.1 in LB, urine, or RPMI 1640 before 200 μL of it was added to 96-well tissue culture plates. Plates were incubated under stationary, aerobic conditions at 37°C for 24 h, and the OD600 was measured to quantify overall bacterial growth. Nonadherent bacteria were removed, and biofilms were washed three times with PBS and dried at 55°C for 1 h. Crystal violet (0.2%, 200 μL) was added, and plates were incubated at room temperature for 30 min. Crystal violet was removed, and biofilms were washed five times with PBS. To release the crystal violet, 200 μL of an 80:20 mixture of ethanol and acetone was added to each well. The released crystal violet solution (100 μL) was transferred to a new 96-well plate and absorbance measured at OD595 on a BioTek Cytation 5 instrument.
Adherence and invasion assays.
Adherence and invasion assays were performed as described previously (88, 89). Briefly, HTB-9 cell confluent monolayers were washed, and 400 μL of fresh RPMI 1640 medium was added. Mid-log-phase bacterial cultures (OD600, 0.4) were diluted 1:10 in PBS, and 100 μL of the bacterial dilution was added to wells. To facilitate bacterial-cell contact, plates were spun at 200 × g for 2 min and incubated at 37°C in 5% CO2. For adherence assays, after 30 min, cells were washed six times with PBS before the addition of 100 μL of 0.025% Trypsin-EDTA. Cells were incubated at 37°C for 7 min, after which time, 400 μL of 0.025% Triton X-100 was added to each well to lyse HTB-9 cells. The contents of the wells were pipetted up and down 25 times before being diluted and plated onto LB agar. For invasion assays, infected HTB-9 cells were incubated with bacteria for 2 h before the medium was removed and replaced with 500 μL of RPMI 1640 medium containing 50 μg/mL gentamicin. The cells were again incubated for 2 h before being washed and lysed as described for adherence assays.
MIC assays.
MIC assays were conducted as described previously (90). Briefly, overnight cultures of bacteria were subcultured to mid-log phase (OD600, 0.4 to 0.6) before being pelleted and resuspended 1:10 in PBS. Antibiotics were diluted in LB or urine and added to 96-well plates. Bacteria were added at a 1:10 dilution to each well before being incubated under stationary, aerobic conditions at 37°C overnight. To measure metabolic activity, resazurin (Sigma-Aldrich) was added at a concentration of 6.75 μg/mL to each well and plates were incubated for an additional 3 h at 37°C. Fluorescence, indicated by a resazurin-to-resofurin conversion, was measured using an excitation/emission of 550 nm/600 nm on a BioTek Cytation 5 instrument. MIC was determined as the lowest antibiotic concentration at which a >90% reduction in fluorescent signal was observed compared with no-antibiotic controls. MIC breakpoints were obtained from the Clinical and Laboratory Standards Institute (CLSI) (91).
Murine UTI model experiments.
All animal experiments were approved by the Baylor College of Medicine (BCM) Institutional Animal Care and Use Committee (protocol AN-8233) and were performed under accepted veterinary standards. Female C57BL/6J mice (strain number 000664) were purchased from Jackson Laboratories or from BCM vivarium stock, and all experiments were conducted when mice were aged 8 to 12 weeks. Animals were allowed to eat and drink ad libitum throughout the duration of experiments. An established murine UTI model was used as described previously (89, 90). Mice were anesthetized with inhaled isoflurane, and approximately 108 CFU of bacteria suspended in PBS was instilled transurethrally into the bladders of mice in a 50-μL volume. At 24 h postinfection, bladders were removed and homogenized in tubes containing 1.0-mm-diameter zirconia/silica beads (Biospec Products; catalog number 11079110z) using a MagNA Lyser instrument (Roche Diagnostics). Serial dilutions of homogenized organs were plated on LB agar and enumerated the following day.
Sequencing and analysis of phage-resistant UPEC.
DNA was isolated from overnight bacterial cultures using the E.Z.N.A. bacterial DNA kit (Omega Bio-Tek) following the manufacturer’s instructions. Sequencing was performed by Novogene using the Illumina platform. Reads were trimmed to Q30 and a minimum length of 50 bp using BBDuk (v38.84). Phage-resistant isolates were compared with wild-type strains using three methods, as follows: (i) reads were independently assembled de novo using Geneious assembler (Geneious 2022.0.1). Contigs of resistant bacteria were then compared with parental contigs using progresssiveMauve (version 26 February 2015) and disagreements extracted (92); (ii) Reads from phage-resistant bacteria were mapped to wild-type bacteria from method 1, followed by variant analysis using Geneious 2022.0.1 variant finder. (iii) Resistant bacteria were compared with parental bacteria using snippy-multi script in Snippy (93). Snippy results agreeing with methods 1 and 2 are presented. For reference selection in Snippy, bacterial reads were validated through the EDGE bioinformatic (v2.4.0) software phylogenetic analysis module, using RAxML and a prebuilt E. coli single nucleotide polymorphism (SNP) database (94, 95). The published UTI89 genome (accession CP000243.1) was used as a reference. Since DS566 did not cluster with another strain closely enough to be used as a reference, it was further categorized using Center for Genomic Epidemiology MLST 2.0 software (v2.0.4; database version 18 October 2021) which predicted it belonged to sequence type 1193 (ST1193) (96, 97). E. coli MCJCHV-1 (accession CP030111.1) was chosen as a reference for DS566. Variants present in both parental strains and resistant progenies were discarded. Circular diagrams were made using CGView Server and modified using Microsoft PowerPoint (98).
Statistics.
In vitro experiments were performed at least three times independently with at least one technical duplicate. Mean values of independent experiments were used to represent biological replicates for statistical analyses. In vivo experiments were conducted at least twice independently with individual mice serving as biological replicates. Experimental data were combined prior to statistical analyses. Mann-Whitney tests were used to compare murine bladder colonization (Fig. 7A and B). The Kruskal-Wallis test with Dunn’s multiple comparisons was used to compare adherence and invasion differences between the strains (Fig. 4A to D), to assess changes in MIC values (Fig. 5A to D), and to analyze bacterial growth in LB and urine (Fig. 6A to F). Two-way repeated-measures analysis of variance (ANOVA) with Geisser-Greenhouse correction and Dunnett’s multiple comparisons was used for biofilm experiments (Fig. 4E and F) and for analysis of bacterial growth in the presence of phage (Fig. 3C and D). Statistical analyses were performed using Prism, v9.2.0 (GraphPad Software Inc., La Jolla, CA). P values of <0.05 were considered statistically significant.
Data availability.
Assembled wild-type DS566 (accession CP092534) and phage-resistant UTI89-1 (accession CP092531), UTI89-2 (accession CP092530), UTI89-3 (accession CP092529), DS566-1 (accession CP092533), and DS566-2 (accession CP092532) sequencing data generated from this study are available in NCBI GenBank under project number PRJNA808067.
ACKNOWLEDGMENTS
We are grateful to Sabrina Green for providing the phage for experiments and to Christina Collins and David Hunstad for supplying UTI89 mutant 16C1. J.J.Z. and M.E.M. were supported by an NIH T32 award (T32GM136554), B.C.S. was supported by an NIH T32 award (T32AI055413), and V.M.-E. was supported by a scholarship from Baylor Research Advocates for Student Scientists (BRASS) and an NIH Kirschstein-NRSA F31 (F31AI167547). Studies were supported by an NIH NIAID U19 award (U19AI157981) to A.W.M. and K.A.P. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Anthony W. Maresso, Email: maresso@bcm.edu.
Kathryn A. Patras, Email: katy.patras@bcm.edu.
Sarah E. F. D'Orazio, University of Kentucky
REFERENCES
- 1.Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 13:1–38. [PubMed] [Google Scholar]
- 3.Foxman B, Brown P. 2003. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am 17:227–241. doi: 10.1016/s0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 4.Ronald A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med 113:14–19. doi: 10.1016/s0002-9343(02)01055-0. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro DJ, Hicks LA, Pavia AT, Hersh AL. 2014. Antibiotic prescribing for adults in ambulatory care in the USA, 2007–09. J Antimicrob Chemother 69:234–240. doi: 10.1093/jac/dkt301. [DOI] [PubMed] [Google Scholar]
- 6.Kot B. 2019. Antibiotic resistance among uropathogenic Escherichia coli. Pol J Microbiol 68:403–415. doi: 10.33073/pjm-2019-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunduki GK, Heinz E, Phiri VS, Noah P, Feasey N, Musaya J. 2021. Virulence factors and antimicrobial resistance of uropathogenic Escherichia coli (UPEC) isolated from urinary tract infections: a systematic review and meta-analysis. BMC Infect Dis 21:753. doi: 10.1186/s12879-021-06435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conly J, Johnston B. 2005. Where are all the new antibiotics? The new antibiotic paradox. Can J Infect Dis Med Microbiol 16:159–160. doi: 10.1155/2005/892058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher HW. 2020. Bad bugs, no drugs 2002–2020: progress, challenges, and call to action. Trans Am Clin Climatol Assoc 131:65–71. [PMC free article] [PubMed] [Google Scholar]
- 10.Wagenlehner FME, Bjerklund Johansen TE, Cai T, Koves B, Kranz J, Pilatz A, Tandogdu Z. 2020. Epidemiology, definition and treatment of complicated urinary tract infections. Nat Rev Urol 17:586–600. doi: 10.1038/s41585-020-0362-4. [DOI] [PubMed] [Google Scholar]
- 11.Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. 2016. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh C, Sarkar P, Issa R, Haldar J. 2019. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol 27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 14.Lemire JA, Harrison JJ, Turner RJ. 2013. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 11:371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 15.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Gordillo Altamirano FL, Barr JJ. 2019. Phage therapy in the postantibiotic era. Clin Microbiol Rev 32:e00066-18. doi: 10.1128/CMR.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene SE, Pinkner JS, Chorell E, Dodson KW, Shaffer CL, Conover MS, Livny J, Hadjifrangiskou M, Almqvist F, Hultgren SJ. 2014. Pilicide ec240 disrupts virulence circuits in uropathogenic Escherichia coli. mBio 5:e02038. doi: 10.1128/mBio.02038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. 2011. Phage treatment of human infections. Bacteriophage 1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldwell JA. 1928. Bacteriophagy in urinary infections following the administration of the bacteriophage therapeutically. Arch Intern Med 41:189–197. doi: 10.1001/archinte.1928.00130140051002. [DOI] [Google Scholar]
- 20.Pires DP, Costa AR, Pinto G, Meneses L, Azeredo J. 2020. Current challenges and future opportunities of phage therapy. FEMS Microbiol Rev 44:684–700. doi: 10.1093/femsre/fuaa017. [DOI] [PubMed] [Google Scholar]
- 21.Melo LDR, Oliveira H, Pires DP, Dabrowska K, Azeredo J. 2020. Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit Rev Microbiol 46:78–99. doi: 10.1080/1040841X.2020.1729695. [DOI] [PubMed] [Google Scholar]
- 22.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. 2004. Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhary N, Narayan C, Mohan B, Taneja N. 2021. Characterization and in vitro activity of a lytic phage RDN37 isolated from community sewage water active against MDR uropathogenic E. coli. Indian J Med Microbiol 39:343–348. doi: 10.1016/j.ijmmb.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Gibson SB, Green SI, Liu CG, Salazar KC, Clark JR, Terwilliger AL, Kaplan HB, Maresso AW, Trautner BW, Ramig RF. 2019. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front Microbiol 10:2537. doi: 10.3389/fmicb.2019.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chibeu A, Lingohr EJ, Masson L, Manges A, Harel J, Ackermann HW, Kropinski AM, Boerlin P. 2012. Bacteriophages with the ability to degrade uropathogenic Escherichia coli biofilms. Viruses 4:471–487. doi: 10.3390/v4040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarker SA, Berger B, Deng Y, Kieser S, Foata F, Moine D, Descombes P, Sultana S, Huq S, Bardhan PK, Vuillet V, Praplan F, Brussow H. 2017. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environ Microbiol 19:237–250. doi: 10.1111/1462-2920.13574. [DOI] [PubMed] [Google Scholar]
- 27.Rostkowska OM, Międzybrodzki R, Miszewska-Szyszkowska D, Górski A, Durlik M. 2021. Treatment of recurrent urinary tract infections in a 60-year-old kidney transplant recipient. The use of phage therapy. Transpl Infect Dis 23:e13391. doi: 10.1111/tid.13391. [DOI] [PubMed] [Google Scholar]
- 28.Ujmajuridze A, Chanishvili N, Goderdzishvili M, Leitner L, Mehnert U, Chkhotua A, Kessler TM, Sybesma W. 2018. Adapted bacteriophages for treating urinary tract infections. Front Microbiol 9:1832. doi: 10.3389/fmicb.2018.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aslam S, Lampley E, Wooten D, Karris M, Benson C, Strathdee S, Schooley RT. 2020. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect Dis 7:ofaa389. doi: 10.1093/ofid/ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khawaldeh A, Morales S, Dillon B, Alavidze Z, Ginn AN, Thomas L, Chapman SJ, Dublanchet A, Smithyman A, Iredell JR. 2011. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J Med Microbiol 60:1697–1700. doi: 10.1099/jmm.0.029744-0. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger A, Clark J, Karris M, Hernandez-Santos H, Green S, Aslam S, Maresso A. 2021. Phage therapy related microbial succession associated with successful clinical outcome for a recurrent urinary tract infection. Viruses 13:2049. doi: 10.3390/v13102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ClinicalTrials.gov. Safety, tolerability, and PK of LBP-EC01 in patients with lower urinary tract colonization caused by E. coli. https://ClinicalTrials.gov/show/NCT04191148.
- 33.ClinicalTrials.gov. Bacteriophage therapy in patients with urinary tract infections. https://ClinicalTrials.gov/show/NCT04287478.
- 34.Leitner L, Ujmajuridze A, Chanishvili N, Goderdzishvili M, Chkonia I, Rigvava S, Chkhotua A, Changashvili G, McCallin S, Schneider MP, Liechti MD, Mehnert U, Bachmann LM, Sybesma W, Kessler TM. 2021. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomised, placebo-controlled, double-blind clinical trial. Lancet Infect Dis 21:427–436. doi: 10.1016/S1473-3099(20)30330-3. [DOI] [PubMed] [Google Scholar]
- 35.Salazar KC, Ma L, Green SI, Zulk JJ, Trautner BW, Ramig RF, Clark JR, Terwilliger AL, Maresso AW. 2021. Antiviral resistance and phage counter adaptation to antibiotic-resistant extraintestinal pathogenic Escherichia coli. mBio 12:e00211-21. doi: 10.1128/mBio.00211-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 37.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 38.Leon M, Bastias R. 2015. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 6:343. doi: 10.3389/fmicb.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chart H, Row B, Threlfall EJ, Ward LR. 1989. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol Lett 51:37–40. doi: 10.1016/0378-1097(89)90073-6. [DOI] [PubMed] [Google Scholar]
- 40.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, Camilli A. 2014. Evolutionary consequences of intra-patient phage predation on microbial populations. Elife 3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. 2016. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 6:26717. doi: 10.1038/srep26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordillo Altamirano F, Forsyth JH, Patwa R, Kostoulias X, Trim M, Subedi D, Archer SK, Morris FC, Oliveira C, Kielty L, Korneev D, O'Bryan MK, Lithgow TJ, Peleg AY, Barr JJ. 2021. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol 6:157–161. doi: 10.1038/s41564-020-00830-7. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Loh B, Gordillo Altamirano F, Yu Y, Hua X, Leptihn S. 2021. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg Microbes Infect 10:2205–2219. doi: 10.1080/22221751.2021.2002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, Debarbieux L. 2017. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22:38–47.e4. doi: 10.1016/j.chom.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 45.Ma L, Green SI, Trautner BW, Ramig RF, Maresso AW. 2018. Metals enhance the killing of bacteria by bacteriophage in human blood. Sci Rep 8:2326. doi: 10.1038/s41598-018-20698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moses S, Vagima Y, Tidhar A, Aftalion M, Mamroud E, Rotem S, Steinberger-Levy I. 2021. Characterization of Yersinia pestis phage lytic activity in human whole blood for the selection of efficient therapeutic phages. Viruses 13:89. doi: 10.3390/v13010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green SI, Kaelber JT, Ma L, Trautner BW, Ramig RF, Maresso AW. 2017. Bacteriophages from ExPEC reservoirs kill pandemic multidrug-resistant strains of clonal group ST131 in animal models of bacteremia. Sci Rep 7:46151. doi: 10.1038/srep46151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green SI, Gu Liu C, Yu X, Gibson S, Salmen W, Rajan A, Carter HE, Clark JR, Song X, Ramig RF, Trautner BW, Kaplan HB, Maresso AW. 2021. Targeting of mammalian glycans enhances phage predation in the gastrointestinal tract. mBio 12:e03474-20. doi: 10.1128/mBio.03474-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnaitman CA, Klena JD. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev 57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. 2006. Maturation of intracellular Escherichia coli communities requires SurA. Infect Immun 74:4793–4800. doi: 10.1128/IAI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagnout C, Sohm B, Razafitianamaharavo A, Caillet C, Offroy M, Leduc M, Gendre H, Jomini S, Beaussart A, Bauda P, Duval JFL. 2019. Pleiotropic effects of rfa-gene mutations on Escherichia coli envelope properties. Sci Rep 9:9696. doi: 10.1038/s41598-019-46100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang G, Meredith TC, Kahne D. 2013. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol 16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunstad DA, Justice SS, Hung CS, Lauer SR, Hultgren SJ. 2005. Suppression of bladder epithelial cytokine responses by uropathogenic Escherichia coli. Infect Immun 73:3999–4006. doi: 10.1128/IAI.73.7.3999-4006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vivijs B, Aertsen A, Michiels CW. 2016. Identification of genes required for growth of Escherichia coli MG1655 at moderately low pH. Front Microbiol 7:1672. doi: 10.3389/fmicb.2016.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB. 2018. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front Microbiol 9:2153. doi: 10.3389/fmicb.2018.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem 273:26310–26316. doi: 10.1074/jbc.273.41.26310. [DOI] [PubMed] [Google Scholar]
- 59.Huss P, Meger A, Leander M, Nishikawa K, Raman S. 2021. Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. Elife 10:e63775. doi: 10.7554/eLife.63775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qimron U, Marintcheva B, Tabor S, Richardson CC. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci USA 103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein G, Stupak A, Biernacka D, Wojtkiewicz P, Lindner B, Raina S. 2016. Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J Biol Chem 291:22999–23019. doi: 10.1074/jbc.M116.748954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J, Emody L. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect Immun 70:4406–4413. doi: 10.1128/IAI.70.8.4406-4413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koopman MG, Krediet RT, Koomen GC, Strackee J, Arisz L. 1989. Circadian rhythm of proteinuria: consequences of the use of urinary protein:creatinine ratios. Nephrol Dial Transplant 4:9–14. [PubMed] [Google Scholar]
- 64.Zhang T, Chang X, Liu W, Li X, Wang F, Huang L, Liao S, Liu X, Zhang Y, Zhao Y. 2017. Comparison of sodium, potassium, calcium, magnesium, zinc, copper and iron concentrations of elements in 24-h urine and spot urine in hypertensive patients with healthy renal function. J Trace Elem Med Biol 44:104–108. doi: 10.1016/j.jtemb.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 65.Taylor EN, Curhan GC. 2007. Differences in 24-hour urine composition between black and white women. J Am Soc Nephrol 18:654–659. doi: 10.1681/ASN.2006080854. [DOI] [PubMed] [Google Scholar]
- 66.García V, Gronnemose RB, Torres-Puig S, Kudirkiene E, Piantelli M, Ahmed S, Andersen TE, Moller-Jensen J, Olsen JE, Herrero-Fresno A. 2021. Genome-wide analysis of fitness-factors in uropathogenic Escherichia coli during growth in laboratory media and during urinary tract infections. Microb Genom 7:000719. doi: 10.1099/mgen.0.000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arunmanee W, Pathania M, Solovyova AS, Le Brun AP, Ridley H, Basle A, van den Berg B, Lakey JH. 2016. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc Natl Acad Sci USA 113:E5034–E5043. doi: 10.1073/pnas.1602382113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakao R, Ramstedt M, Wai SN, Uhlin BE. 2012. Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PLoS One 7:e51241. doi: 10.1371/journal.pone.0051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beloin C, Michaelis K, Lindner K, Landini P, Hacker J, Ghigo JM, Dobrindt U. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J Bacteriol 188:1316–1331. doi: 10.1128/JB.188.4.1316-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manning PA, Reeves P. 1976. Outer membrane of Escherichia coli K-12: TSX mutants (resistant to bacteriophage T6 and colicin K) lack an outer membrane protein. Biochem Biophys Res Commun 71:466–471. doi: 10.1016/0006-291x(76)90810-x. [DOI] [PubMed] [Google Scholar]
- 72.Nagy G, Dobrindt U, Grozdanov L, Hacker J, Emody L. 2005. Transcriptional regulation through RfaH contributes to intestinal colonization by Escherichia coli. FEMS Microbiol Lett 244:173–180. doi: 10.1016/j.femsle.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 73.Gao Q, Xu H, Wang X, Zhang D, Ye Z, Gao S, Liu X. 2013. RfaH promotes the ability of the avian pathogenic Escherichia coli O2 strain E058 to cause avian colibacillosis. J Bacteriol 195:2474–2480. doi: 10.1128/JB.02074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aguiniga LM, Yaggie RE, Schaeffer AJ, Klumpp DJ. 2016. Lipopolysaccharide domains modulate urovirulence. Infect Immun 84:3131–3140. doi: 10.1128/IAI.00315-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect Immun 76:695–703. doi: 10.1128/IAI.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hull R, Rudy D, Donovan W, Svanborg C, Wieser I, Stewart C, Darouiche R. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J Urol 163:872–877. doi: 10.1016/S0022-5347(05)67823-8. [DOI] [PubMed] [Google Scholar]
- 77.Darouiche RO, Donovan WH, Del Terzo M, Thornby JI, Rudy DC, Hull RA. 2001. Pilot trial of bacterial interference for preventing urinary tract infection. Urology 58:339–344. doi: 10.1016/S0090-4295(01)01271-7. [DOI] [PubMed] [Google Scholar]
- 78.Trautner BW, Hull RA, Thornby JI, Darouiche RO. 2007. Coating urinary catheters with an avirulent strain of Escherichia coli as a means to establish asymptomatic colonization. Infect Control Hosp Epidemiol 28:92–94. doi: 10.1086/510872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darouiche RO, Green BG, Donovan WH, Chen D, Schwartz M, Merritt J, Mendez M, Hull RA. 2011. Multicenter randomized controlled trial of bacterial interference for prevention of urinary tract infection in patients with neurogenic bladder. Urology 78:341–346. doi: 10.1016/j.urology.2011.03.062. [DOI] [PubMed] [Google Scholar]
- 80.Tagliaferri TL, Jansen M, Horz HP. 2019. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol 9:22. doi: 10.3389/fcimb.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure–activity relationships of polymyxin antibiotics. J Med Chem 53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trebosc V, Gartenmann S, Totzl M, Lucchini V, Schellhorn B, Pieren M, Lociuro S, Gitzinger M, Tigges M, Bumann D, Kemmer C. 2019. Dissecting colistin resistance mechanisms in extensively drug-resistant acinetobacter baumannii clinical isolates. mBio 10:e01083-19. doi: 10.1128/mBio.01083-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moosavian M, Emam N, Pletzer D, Savari M. 2020. Rough-type and loss of the LPS due to lpx genes deletions are associated with colistin resistance in multidrug-resistant clinical Escherichia coli isolates not harbouring mcr genes. PLoS One 15:e0233518. doi: 10.1371/journal.pone.0233518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis MR, Jr, Goldberg JB. 2012. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J Vis Exp 28:3916. doi: 10.3791/3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patras KA, Derieux J, Al-Bassam MM, Adiletta N, Vrbanac A, Lapek JD, Zengler K, Gonzalez DJ, Nizet V. 2018. Group B Streptococcus biofilm regulatory protein A contributes to bacterial physiology and innate immune resistance. J Infect Dis 218:1641–1652. doi: 10.1093/infdis/jiy341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patras KA, Wescombe PA, Rosler B, Hale JD, Tagg JR, Doran KS. 2015. Streptococcus salivarius K12 limits group B Streptococcus vaginal colonization. Infect Immun 83:3438–3444. doi: 10.1128/IAI.00409-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patras KA, Ha AD, Rooholfada E, Olson J, Ramachandra Rao SP, Lin AE, Nizet V. 2019. Augmentation of urinary lactoferrin enhances host innate immune clearance of uropathogenic Escherichia coli. J Innate Immun 11:481–495. doi: 10.1159/000499342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patras KA, Coady A, Babu P, Shing SR, Ha AD, Rooholfada E, Brandt SL, Geriak M, Gallo RL, Nizet V. 2020. Host cathelicidin exacerbates group B Streptococcus urinary tract infection. mSphere 5:e00932-19. doi: 10.1128/mSphere.00932-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.CLSI. 2019. Performance standards for antimicrobial susceptibility testing, M100. CLSI, Wayne, PA. [Google Scholar]
- 92.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seemann T. Rapid haploid variant calling and core genome alignment. https://github.com/tseemann/snippy. Accessed 12 September.
- 94.Li PE, Lo CC, Anderson JJ, Davenport KW, Bishop-Lilly KA, Xu Y, Ahmed S, Feng S, Mokashi VP, Chain PS. 2017. Enabling the democratization of the genomics revolution with a fully integrated Web-based bioinformatics platform. Nucleic Acids Res 45:67–80. doi: 10.1093/nar/gkw1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−1. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S1, JPG file, 0.3 MB (300KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−3. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S2, JPG file, 0.3 MB (299.3KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of UPEC in the presence of phages HP3 and ES17 at an MOI of 10−5. Bacteria were challenged with HP3 (left column) or ES17 (right column) in LB media and pooled human urine during 18 h of growth. Data are grouped by UPEC strain UTI89 (A), CFT073 (B), DS515 (C), and DS566 (D). All curves represent the mean value of three independent experiments performed in at least technical duplicate. Download FIG S3, JPG file, 0.3 MB (301.5KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Serially passaged phage-resistant UPEC strains do not readily regain sensitivity to ES17. Three independent cultures of phage-resistant strain UTI89-2 were serially passaged three times in LB media. Each passage, the parental UTI89-2 strain and wild-type UTI89 were challenged with ES17 for 6 h. Data are grouped by passage number 0 (A), 1 (B), 2 (C), and 3 (D). Each growth curve represents the mean value of three technical replicates. Download FIG S4, JPG file, 0.3 MB (297.7KB, jpg) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Non-LPS-associated mutations identified during sequencing of phage-resistant isolates. Site and type of non-LPS biosynthesis-associated mutations observed in phage-resistant strains arising from in vitro screening and identified by whole-genome sequencing. Download Table S1, DOCX file, 0.1 MB (54.2KB, docx) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MICs for antibiotics targeting the bacterial outer membrane are not altered when phage-resistant bacteria are grown in LB media. Colistin MICs of UTI89 (A) and DS566 (B) and isogenic phage-resistant mutants. Polymyxin B MICs of UTI89 (C) and DS566 (D) and isogenic phage-resistant mutants. Individual points are representative of independent experiments performed in duplicate. Bars represent median and 95% confidence intervals. Download FIG S5, TIF file, 0.3 MB (346.8KB, tif) .
Copyright © 2022 Zulk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Assembled wild-type DS566 (accession CP092534) and phage-resistant UTI89-1 (accession CP092531), UTI89-2 (accession CP092530), UTI89-3 (accession CP092529), DS566-1 (accession CP092533), and DS566-2 (accession CP092532) sequencing data generated from this study are available in NCBI GenBank under project number PRJNA808067.