ABSTRACT
Human papillomaviruses (HPVs) are highly infectious and cause the most common sexually transmitted viral infections. They induce hyperproliferation of squamous epithelial tissue, often forming warts. Virally encoded proteins reprogram gene expression and cell growth to create an optimal environment for viral replication. In addition to their normal roles in infection, functional alterations induced by viral proteins establish conditions that frequently contribute to human carcinogenesis. In fact, ~5% of human cancers are caused by HPVs, with virtually all cervical squamous cell carcinomas (CESC) and an increasing number of head and neck squamous cell carcinomas (HNSC) attributed to HPV infection. The Cancer Genome Atlas (TCGA) molecularly characterized thousands of primary human cancer samples in many cancer types, including CESC and HNSC, and created a comprehensive atlas of genomic, epigenomic, and transcriptomic data. This publicly available genome-wide information provides an unprecedented opportunity to expand the knowledge of the role that HPV plays in human carcinogenesis. While many tools exist to mine these data, few, if any, focus on the comparison of HPV-positive cancers with their HPV-negative counterparts or adjacent normal control tissue. We have constructed a suite of web-based tools, The HPV Induced Cancer Resource (THInCR), to utilize TCGA data for research related to HPV-induced CESC and HNSC. These tools allow investigators to gain greater biological and medical insights by exploring the impacts of HPV on cellular gene expression (mRNA and microRNA), altered gene methylation, and associations with patient survival and immune landscape features. These tools are accessible at https://thincr.ca/.
IMPORTANCE The suite of analytical tools of THInCR provides the opportunity to investigate the roles that candidate target genes identified in cell lines or other model systems contribute to in actual HPV-dependent human cancers and is based on large-scale TCGA data sets. Expression of target genes, including both mRNA and microRNA, can be correlated with HPV gene expression, epigenetic changes in DNA methylation, patient survival, and numerous immune features, like leukocyte infiltration, interferon gamma response, T cell response, etc. Data from these analyses may immediately provide evidence to validate in vitro observations, reveal insights into mechanisms of virus-mediated alterations in cell growth, behavior, gene expression, and innate and adaptive immunity and may help hypothesis generation for further investigations.
KEYWORDS: human papillomavirus, cervical cancer, head and neck cancer, TCGA, gene expression, survival, methylation, correlation, oncogene, DNA methylation, analysis resource, cancer, database, HPV
INTRODUCTION
There are currently over 400 known human papillomavirus (HPV) types (1). These viruses, as exemplified by HPV type 16 (HPV16), contain a small, circular, double-stranded DNA genome of about 8 kbp which encodes 8 or 9 genes. There are 30 HPV types that preferentially infect the anogenital mucosa, causing papillomas (warts) (2). These epithelium-specific viruses are highly transmissible, with peak incidence of infection occurring with the onset of sexual activity (3). Indeed, mucosal HPVs are the most common sexually transmitted infection in North America (4). Furthermore, mucosal HPV types are classified as high risk (HR) or low risk based on their oncogenic potential (2, 5). HR HPV-induced lesions frequently initiate carcinogenesis, commonly as a result of damage to the viral genome, leading to random integration into the host genome. This often leads to a constitutively elevated level of expression of the viral E6 and E7 oncogenes (2). Persistent infection by HR HPVs causes virtually all cervical and other anogenital cancers (2). HR HPV is associated with 85 to 90% of cervical cancers (CESC), with HPV16 predominating via its association with ~50% of all CESC (6, 7). Prognosis for patients with HPV-positive (HPV+) CESC is better than for their HPV-negative (HPV−) counterparts (8, 9), and it is clear that HPV+ and HPV− diseases are clinically and pathologically distinct diseases (10, 11). Worldwide, CESC is the 3rd most prevalent cancer in women, with 570,000 new cases in 2018 (https://gco.iarc.fr/). HPV-induced CESC remains a leading cause of cancer death in younger women in economically disadvantaged countries (2).
In addition to anogenital cancers, HR HPV infection of the oral mucosa is an independent etiological agent of head and neck squamous cell carcinoma (HNSC), which is the 6th most common cancer worldwide, with ~890,000 cases recorded per year (https://gco.iarc.fr/) (12). HNSC include malignant squamous lesions arising in the oral cavity, larynx, pharynx, and oropharynx. HPV infection now accounts for ~25% of all HNSC cases; most HPV+ HNSC cases are caused by HPV16, and the remainder are caused by other HR types, with all cases expressing the viral E6 and E7 oncogenes (13–15).
Over the last 30 years, the frequency of HPV-induced oral cancers has increased to epidemic proportions, likely due to changes in sexual behaviors (16, 17). In the United States, more than 70% of oropharyngeal cancers are HPV+ (18). Indeed, the incidence of oropharyngeal cancer has been increasing steadily since 1973 in the United States, with base-of-tongue and tonsillar cancers increasing by 2.1% and 3.9% per year, respectively, among white men and women ages 20 to 44 years (19). Similarly, this increase in oropharyngeal cancers was nearly 5% per year between 1970 and 2016 in Sweden (Swedish National Board of Health and Welfare, https://www.socialstyrelsen.se/en/). HPV+ HNSC often occurs in younger people, nonsmokers, and nondrinkers compared to their HPV− counterparts (20). Although HPV+ tumors generally exhibit favorable clinical outcomes (21, 22), ~15% of these patients do not survive their disease (23, 24). In summary, HPV+ HNSC is considered a distinct epidemiological, molecular, and clinical entity (25–27) that is caused by a sexually transmitted virus and is increasing at an epidemic rate (15, 28–30).
HPV+ cancers constitutively retain expression of viral genes, primarily the E6 and E7 oncogenes (2, 15), but transcripts for other viral genes are detected in some cancers (31, 32). How these HPV proteins reprogram the infected cell in terms of gene expression, immune response, and metabolism relevant to the development and progression of HPV-induced cancer is a highly active area of investigation (33–36). These studies are complicated by the fact that HPV does not replicate in animal systems, necessitating in vitro studies using primary human epithelial cells or human cancer cell lines in culture, in vivo studies in transgenic animals, or studies of related animal papillomaviruses as surrogate models of carcinogenesis (37–39). Although tremendously valuable insights into HPV infection and oncogenesis have been made in these surrogate systems, the ability to validate these observations in primary human cancers remains the gold standard. Such validation is often hampered by the lack of human tissue, clinical data, and power to generate statistically robust insights.
The Cancer Genome Atlas (TCGA) represents a large-scale effort by the National Cancer Institute and the National Human Genome Research Institute to create a comprehensive atlas of genomic, epigenomic, and transcriptomic data from primary, surgically resected human cancers (https://www.cancer.gov/tcga). This effort has generated publicly available, comprehensive, multidimensional maps of genomic changes in more than 11,000 tumor samples from 33 distinct types of human tumors. All TCGA samples were processed via a uniform data analysis pipeline that included mRNA sequencing, microRNA (miRNA) sequencing, and DNA methylation profiling. Substantial clinical data are available for many of the TCGA samples, which allows the comparison of many clinical variables, including patient outcome assessment (40).
In terms of HPV-induced cancers, the TCGA CESC data sets are derived from nearly 300 cancer patients (41, 42), while the TCGA HNSC data set is derived from over 500 cancer patients (25, 42). All samples were surgically obtained from treatment-naive individuals, thus avoiding the potentially confounding effects of chemotherapy or radiotherapy on these molecular studies. Initial annotations of tumor HPV positivity and the HPV type present have been expanded and reported in ancillary publications (43, 44), providing the opportunity to make direct comparisons between HPV+ tumors and their corresponding HPV− counterparts for many molecular and genetic features. Additionally, viral mRNA read counts for those samples that are positive for HPV16, -33, and -35 (HPV16/33/35+) are available (32), as is a systematic analysis of the clinical follow-up intervals for patient survival (40) and estimates of numerous specific immune landscape markers (45). Despite the plethora of useful data in the TCGA and many tools to access it programmatically (46), it remains relatively difficult to analyze this large-scale data primarily by HPV status, particularly for researchers without strong bioinformatic skill sets.
Here we introduce a suite of intuitive web-based tools, The HPV Induced Cancer Resource (THInCR), to assist in querying and visualization of gene expression, methylation, survival, and immune landscape data for both the CESC and HNSC TCGA cohorts. These analysis and visualization tools are intended specifically as a resource for those working in the field of HPV-dependent cancers and should not require any significant computational or bioinformatics expertise. THInCR was developed to allow investigators to rapidly gain greater biological and medical insights by exploring the impact of HPV on cellular gene expression (mRNA and miRNA), altered gene methylation, and associations with patient survival and immune landscape features from primary human cervical and head and neck cancers. The tools can be accessed at https://thincr.ca/, and the stand-alone version can be downloaded at https://github.com/msaland/THInCR-Suite.
RESULTS
The THInCR suite is a collection of 5 unique tools created for the purpose of exploring a multitude of factors that may be impacted by HPV status in patients with CESC or HNSC. Such factors include differential mRNA and miRNA expression, gene loci methylation, overall patient survival, and changes in immune landscape features. Using the tools present in the THInCR suite, each of these factors can also be correlated with HPV status, genomic location, or gene expression. Analyses are focused on samples positive for the closely related HPV16, -33, and -35 types, which represent the most prevalent infections in both the CESC and HNSC cohorts. This was done to reduce potential variability related to intrinsic functional differences between distantly related HPVs. Table 1 lists the number of samples used in the calculations for each THInCR tool. Note that the CESC cohort has relatively few HPV− or normal control samples, which impacts the ability to define statistically significant differences compared to the more balanced HNSC cohort.
TABLE 1.
Number of patient samples analyzed from the TCGA CESC and HNSC cohorts for each THInCR tool
| Cohort | Patient subset | No. of patient samples analyzed with TCGA tool for: |
||||||
|---|---|---|---|---|---|---|---|---|
| mRNA-seq | miRNA-seq | mRNA vs viral mRNA | miRNA vs viral mRNA | Methylation | Immune comparisons | Survival | ||
| CESC | HPV16/33/35+ | 180 | 180 | 91 | 91 | 180 | 176 | 180 |
| HPV− | 19 | 19 | NAa | NA | 19 | 18 | 19 | |
| Normal control | 3 | 3 | NA | NA | 3 | NA | NA | |
| HNSC | HPV16/33/35+ | 72 | 64 | 65 | 65 | 72 | 71 | 72 |
| HPV− | 442 | 409 | NA | NA | 442 | 437 | 442 | |
| Normal control | 40 | 40 | NA | NA | 50 | NA | NA | |
NA, not applicable.
Differentially expressed gene analysis.
As obligate intracellular parasites, HPVs dramatically reprogram gene expression in the infected cell to provide a more conducive state for the virus replicative cycle (47). As HPV+ CESC and HNSC consistently retain expression of a subset of viral genes, including the critical E6 and E7 oncogenes, at least some of these changes in cellular gene expression will likely persist in these cancers.
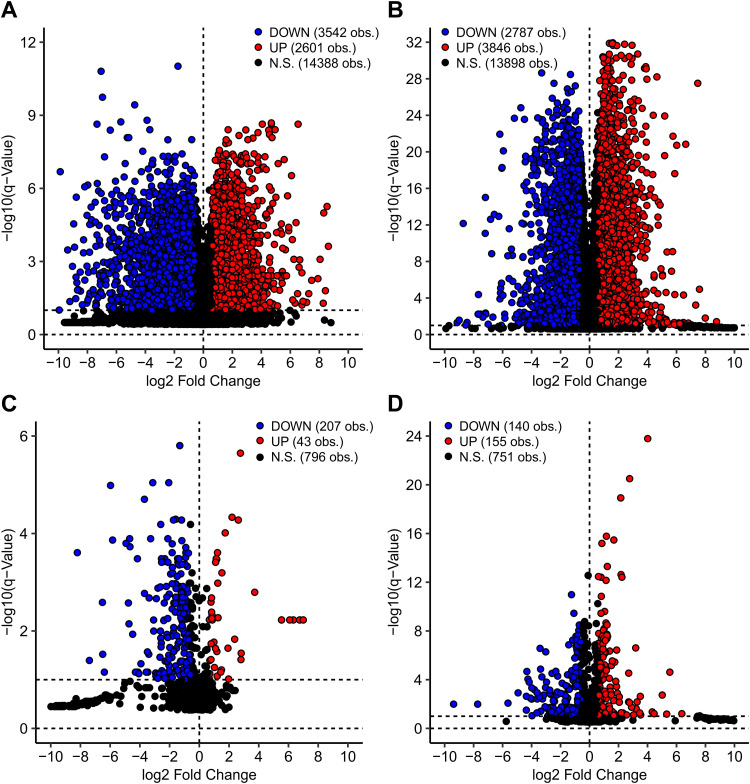
Thousands of cellular mRNAs and miRNAs are differentially expressed between HPV+ cancers and their HPV− counterparts, many of which may be important in the process of carcinogenesis, the maintenance of the cancer phenotype, immune evasion, treatment response, etc. (Fig. 1). The first tool in the THInCR suite is the differential gene expression analysis tool, which allows a user to select a gene of interest (GOI) to determine if it is differentially expressed between HPV+, HPV−, or normal control tissue, providing easily digestible summaries of this information. This tool can be used to determine if a GOI identified as differentially regulated by HPV in experimental systems is similarly dysregulated in actual primary human cancers from different anatomical sites. Such a validation could indicate that the GOI is worth further pursuit via additional experimentation, allowing the rapid exploration of expression of related genes, and may promote hypothesis generation. Examples of exactly this type of confirmation are already present in the literature (48, 49), but with THInCR these analyses can be performed by anyone in the field. The tool in question is subdivided into two different tools: one for miRNA and the other for mRNA expression patterns, each featuring the CESC and HNSC data sets. These data are restricted to only the HPV16+ and closely related HPV33/35+ samples from the TCGA CESC and HNSC cohorts.
FIG 1.
Volcano plots of differentially expressed genes (DEGs) between HPV+ and HPV− patients for CESC mRNA (A), HNSC mRNA (B), CESC miRNA (C), and HNSC miRNA (D) TCGA data sets. Each dot represents an individual gene. Genes shaded in blue exhibited a statistically decreased level of expression in HPV16/33/35+ cancers. Genes shaded in red exhibited a statistically increased level of expression in HPV+ cancers, whereas expression of those indicated in black was not significantly different. Calculations were performed with a false discovery rate (FDR) of 10%.
For each tool, users have the option of selecting a GOI, upon which two different boxplots are generated, one for each featured data set, showing the distribution of expression levels of the selected gene for HPV+, HPV−, and normal control tissue. Pairwise P values and associated q values are also calculated, with significant differences highlighted with bright colors, drawing the viewers' attention to statistically significant changes. A table containing statistical information, such as minimum, maximum, and quartile values, for each of the subsets within the boxplots is also generated. The boxplots can be downloaded as .png files, whereas the table with the requisite information to redraw the boxplots can be downloaded as a .csv file, allowing for local data storage for future reference and figure creation. Downloadable tables of all differentially expressed cellular mRNAs and miRNAs are also available through a link provided on the tool’s webpage, to assist users in identifying differentially expressed genes (DEGs) that might merit future investigation.
Correlation of cellular gene expression with HPV genes.
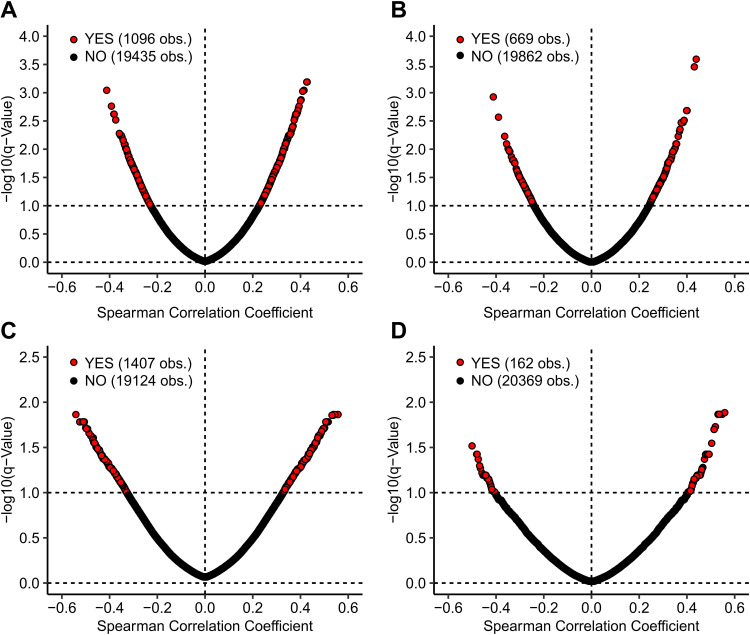
The presence of genes that are differentially expressed between HPV+ and HPV− cancers suggests that they could be directly or indirectly related to the expression of viral factors retained in these cancers. Furthermore, a causal relationship between a viral gene and altered expression of a cellular gene might be reflected in a proportional or inversely proportional relationship between their expression levels. To investigate this, we utilized the mRNA expression data for each viral gene (E1, E2, E4, E5, E6, E7, L1, and L2) previously reported (32). These data are restricted to only the HPV16+ and closely related HPV33/35+ samples from the TCGA CESC and HNSC cohorts. The expression of many cellular mRNAs in both CESC and HNSC are significantly correlated with HPV E6 or E7 expression (Fig. 2). The presence of these correlations suggests a causal relationship, which can be investigated as a mechanism or used in hypothesis generation.
FIG 2.
Spearman correlation coefficient versus negative log of significance for CESC cellular mRNA versus E6 (A) or E7 (B) mRNA levels and for HNSC cellular mRNA versus E6 (C) or E7 (D) mRNA. Only HPV16/33/35+ samples were included in these analyses. Genes shaded in red exhibited a statistically increased level of expression in HPV+ cancers, whereas expression of those indicated in black was not significantly different. Calculations were performed with an FDR of 10%.
The tool in question is subdivided into two different tools: one for miRNA and the other for mRNA expression patterns, each featuring the CESC and HNSC data sets. For each tool, users have the option of selecting a GOI, upon which two different heatmaps are generated, one for each featured data set, showing the correlation between the 8 HPV16/33/35 genes and the selected GOI, with the Spearman correlation score and significance shown for each set of correlations. Correlation scores and pairwise P values and associated q values are also shown in a separate table, and significant values are highlighted with bright colors, drawing the viewers' attention to statistically significant correlations. The heatmaps can be downloaded as .png files, whereas the table with the heatmap P values and q values can be downloaded as a .csv file, allowing for local data storage for future reference and figure creation. Downloadable tables of all correlations are also available through the tool’s webpage, via a provided link to assist users in identifying highly correlated genes that might merit future investigation.
Analysis of the impact of HPV status on gene methylation.
In addition to targeting specific transcriptional regulators like p53 and Rb, HPV-encoded proteins can alter transcription in infected or virally transformed cells via epigenetic mechanisms (33, 50). In particular, genome-wide changes in DNA methylation are clearly observed in both HPV+ CESC and HNSC (51, 52). Genome-wide methylation data obtained using the Infinium HumanMethylation450 BeadChip array are available for both the TCGA CESC and HNSC cohorts, allowing the interrogation of the methylation status of ~450,000 CpG sites located throughout the genome (53). Using these data, comparison of the methylation status of HPV+, HPV−, and normal control tissues revealed a strong HPV-dependent dysregulation of methylation at the CDKN2A gene, which encodes p16(INK4A), in CESC and HNSC (54). Thus, these data can be used to provide some insight into the mechanism by which HPV infection induces p16 expression, a surrogate marker historically used for clinical HPV status (55, 56).
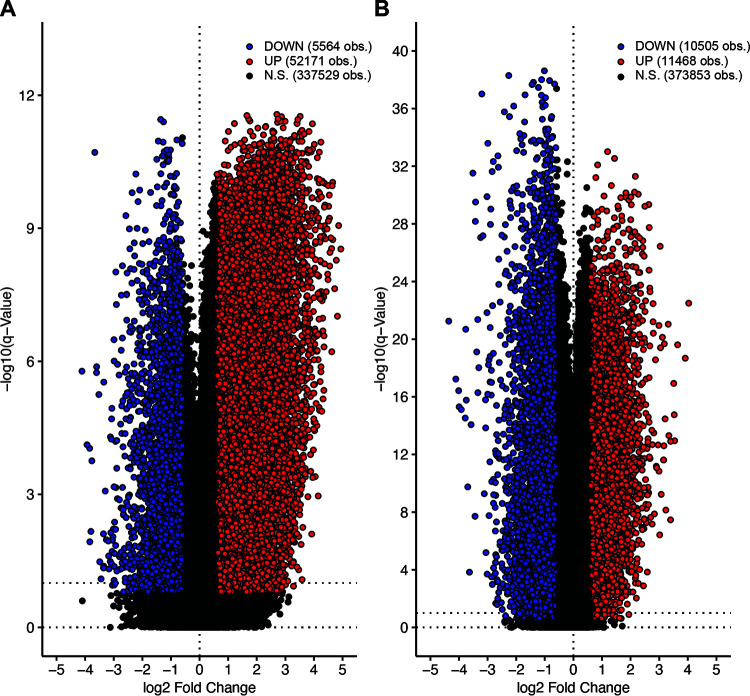
Both the TCGA CESC and HNSC cohorts exhibited many genomic probes that are differentially methylated between HPV+ cancers and their HPV− counterparts (Fig. 3). Many of these methylation sites may be important in the process of carcinogenesis, the maintenance of the cancer phenotype, immune evasion, treatment response, etc. The third tool in the THInCR suite is the differential CpG methylation tool, which allows a user to select a GOI to determine if any probes within it, plus 15% the gene length on either side, are differentially methylated between HPV+, HPV−, and normal control tissue, providing easily digestible summaries of this information. This tool can be used to determine if a GOI identified as differentially methylated by HPV in experimental systems is similarly dysregulated in actual primary human cancers from different anatomical sites. Such a validation could indicate that the GOI is worth further pursuit via further experimentation, allows the rapid exploration of expression of related genes, and may promote hypothesis generation. Examples of exactly this type of analysis are already present in the literature (49, 54, 57), but with THInCR, this can be performed by anyone in the field. Note that these data are restricted to only the HPV16+ and closely related HPV33/35+ samples from the TCGA CESC and HNSC cohorts.
FIG 3.
Volcano plots of differentially methylated sites between HPV+ and HPV− patients for the CESC (A) and HNSC (B) TCGA data sets. Each dot represents an individual methylation probe from the Infinium HumanMethylation450 BeadChip array. Probes shaded in blue exhibited a statistically decreased level of expression in HPV16/33/35+ cancers. Probes shaded in red exhibited a statistically increased level of expression in HPV+ cancers, whereas expression of those indicated in black was not significantly different. Calculations were performed with an FDR of 10%.
This tool encompasses both the miRNA and mRNA expression patterns observed in the CESC and HNSC data sets. For each tool, users have the option of selecting a GOI, along with the genomic region to focus on, which generates line plots for the CESC and HNSC cohorts. These show the average methylation beta value for each genomic probe within the selected region, with average methylation values shown for HPV+, HPV−, and normal control at a specific probe. The line plot also displays the coding strand for the selected GOI, with a left-to-right arrow representing a forward or Watson strand, whereas the reverse represents the reverse or Crick orientation. A comparison table with P values and q values is also generated for differences in methylation beta values by HPV status. Significant values are highlighted with bright colors, drawing the viewers' attention to statistically significant differences. Additionally, users can select a probe within the selected region by using a pulldown menu, which generates two boxplots, one for each featured data set. The line plots and boxplots can be downloaded as .png files, whereas the table with the average methylation beta values, P values, and q values can be downloaded as a .csv file, allowing for local data storage for future reference and figure creation. Downloadable tables of all differentially methylated probes are also available through a link provided on the tool’s webpage, to assist users in identifying areas of methylation that might merit future investigation.
Impact of gene expression on patient survival.
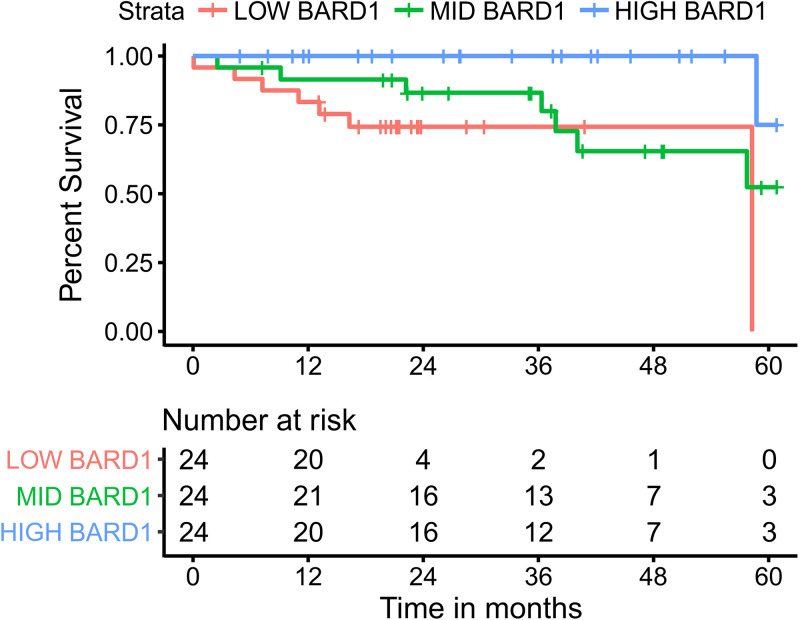
High-quality data on patient survival are available for both the CESC and HNSC TCGA cohorts (40), providing the opportunity to determine if altered expression of a GOI is associated with patient outcomes. Many existing studies have exploited these data to identify numerous individual prognostic genes or gene signatures associated with patient survival in the various TCGA, including studies analyzing all 33 cohorts (58). For example, low expression of BARD1 in HNSC shows a significant correlation with patient survival, suggesting that high expression functions in some way as a positive factor for cancer, whether promoting growth, evasion of the immune system, or resistance to genotoxic agents (Fig. 4).
FIG 4.
Example survival curve with 3 comparison groups. Analysis was based on mRNA expression levels of BRCA1-associated RING domain 1 mRNA (BARD1; Gene ID 580), with the gene being differentially regulated between HPV+ and HPV− samples for HNSC. This figure was generated natively as part of the THInCR suite, as an example of data output. The HPV16/33/35+ samples from the TCGA HNSC cohort were divided into high-, middle-, and low-expressing subsets for Kaplan-Meier survival analysis.
The fourth tool in the THInCR suite allows Kaplan-Meier analysis of patient survival based on expression levels of an individual mRNA or miRNA in both the CESC and HNSC data sets. Once again, these data are restricted to only the HPV16+ and closely related HPV33/35+ samples from the TCGA CESC and HNSC cohorts. For each tool, users have the option of selecting a GOI, as well as the number of comparison groups, which ranges between 2 and 4. In this way, the cohort is divided into 2, 3, or 4 equally sized subsets based on expression of the target gene for survival comparisons between subsets. Four different survival plots are generated, each representing survival in HPV+ CESC, HPV− CESC, HPV+ HNSC, and HPV− HNSC. In this way, genes specifically affecting survival in HPV+ cancers can be identified, as well as genes associated with outcome independent of HPV status. Pairwise P values and associated q values are also calculated, with the values shown in a separate region and significant values highlighted with bright colors, drawing the viewers' attention to statistically significant correlations. The survival plots can be downloaded as .png files, allowing for local data storage for future reference and figure creation.
Impact of gene expression on the immune landscape.
The field of tumor immunology has exploded in the last decade, highlighting the critical role that the immune system plays in tumor development and patient outcomes. The tumor immune landscape is complex, and it has become increasingly clear that it varies widely between different cancer types and even individual tumors of the same type, leading to vastly different treatment responses and patient outcomes (59). Importantly, HPV+ HNSC are immune “hot” tumors, with markedly more immune infiltration and higher levels of CD8+ T-cell activation than HPV− HNSC (60, 61). Although molecular comparisons of the immune landscape between HPV+ and HPV− CESC are scarce, these are pathologically distinct tumors with different clinical outcomes, suggesting that they will also exhibit immunological differences (10, 11). As a “stealth” virus, HPV encodes many distinct immunomodulatory functions that help HPV+ tumors evade adaptive immunity (62). Thus, detailed comparisons of the immune landscape between HPV+ and HPV− cancers provide an opportunity to identify immunological determinants and the underlying mechanisms by which they are achieved, which may translate to improved treatment of HPV+ CESC and HNSC.
A recent study entailed an extensive immunogenomic analysis of more than 10,000 tumors from the TCGA (45). This analysis was used to construct the final tool in the THInCR suite, which allows a user to investigate the impact of expression of a GOI, either mRNA or miRNA, on numerous immune landscape features (Table 2). These analyses are restricted to only the HPV16+ and closely related HPV33/35+ samples from the TCGA CESC and HNSC cohorts. Users have the option of selecting a GOI, upon which a table featuring 53 immune landscape features is generated, showing correlation and significance for each of the 4 categories (CESC HPV+, CESC HPV−, HNSC HPV+, HNSC HPV−), with significant values highlighted with bright colors, drawing the viewers' attention to statistically significant correlations. Users then have the option of selecting an immune landscape feature, upon which two scatterplots and two boxplots are generated, one for each featured data set (see the example analysis in Fig. 5). The line plots and boxplots can be downloaded as .png files, whereas the table with the correlation and significance scores can be downloaded as a .csv file, allowing for local data storage for future reference and figure creation.
TABLE 2.
The 53 immune landscape features available for analysis in the THInCR suite of tools
| Immune landscape featurea | |
|---|---|
| Aneuploidy score | Monocytes |
| B cells memory | Neutrophils |
| B cells naive | NK cells activated |
| BCR evenness | NK cells resting |
| BCR richness | Nonsilent mutation rate |
| BCR Shannon | No. of segments |
| CTA score | Plasma cells |
| Dendritic cells | Proliferation |
| Dendritic cells activated | Silent mutation rate |
| Dendritic cells resting | SNV neoantigens |
| Eosinophils | Stromal fraction |
| Fraction altered | T cells CD4 memory activated |
| Homologous recombination defects | T cells CD4 memory resting |
| IFN gamma response | T cells CD4 Naive |
| Indel neoantigens | T cells CD8 |
| Intratumor heterogeneity | T cells follicular helper |
| Leukocyte fraction | T cells gamma delta |
| Lymphocyte infiltration signature score | T cells regulatory Tregs |
| Lymphocytes | TCR evenness |
| Macrophage regulation | TCR richness |
| Macrophages | TCR Shannon |
| Macrophages M0 | TGF beta response |
| Macrophages M1 | Th1 cells |
| Macrophages M2 | Th17 cells |
| Mast cells | Th2 cells |
| Mast cells activated | Wound healing |
| Mast cells resting | |
BCR, B cell receptor; CTA, cancer testis antigens; IFN, interferon; SNV, single nucleotide variant; TCR, T cell receptor; TGF, transforming growth factor.
FIG 5.
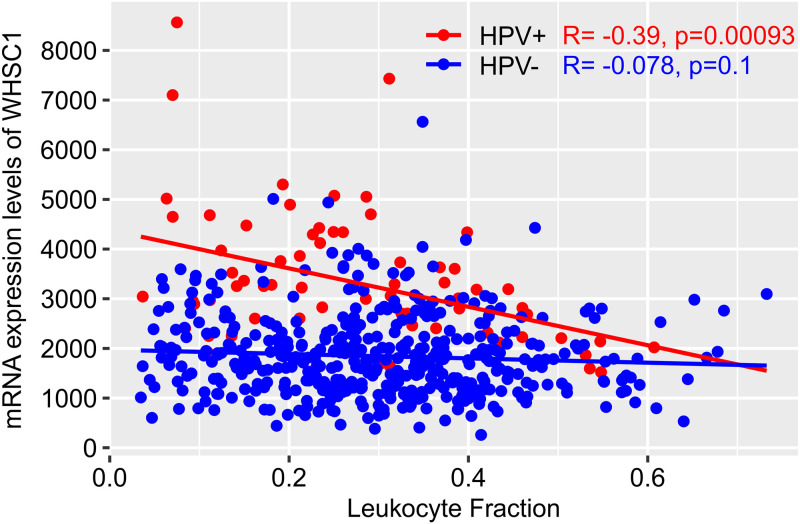
Example of a correlation plot between NSD2/WHSC1 mRNA expression levels and the leukocyte fraction immune landscape feature for the HNSC data set. The figure was generated natively as part of the THInCR suite. Red dots represent HPV16/33/35+ HNSC samples, while blue dots represent HPV− HNSC samples. For HPV+, R = −0.39, P = 9.3e−4; for HPV−, R = −0.078, P = 0.1.
DISCUSSION
High-risk HPV infection continues to be responsible for ~5% of human cancers, despite the availability of subunit vaccines against HPV since 2006. These vaccines induce neutralizing antibodies that are predominantly type specific (63). Importantly, these vaccines are prophylactic, rather than therapeutic, as they target the L1 virion protein, which is not often expressed in HPV+ cancers. While vaccination will reduce HPV-induced cancer in the long term, there would still be many infections caused by HPV types not neutralized by these vaccines, which include the nonavalent vaccine (64). In addition, these vaccines do not benefit the millions of people with existing HPV infections and are prohibitively expensive in many areas with high HPV infection rates. Thus, despite excellent vaccines, HPV will likely remain an important human pathogen, and a major cause of cancer, over the long term (65). As a result, studies aimed at understanding the molecular basis by which HPV reprograms the infected cell, predisposing it to oncogenic transformation, remain an important area of tumor virus research that is directly relevant to human health.
The large-scale multimodal data generated by the TCGA remain a tremendous resource for cancer research, including tumor virus research. Although level 3 preanalyzed high-level data are freely available to any user via the Broad GDAC Firehose (https://gdac.broadinstitute.org/), their utility to wet lab researchers and clinicians with limited computational skills is limited without a significant investment of time and effort. This has spawned a plethora of web-based utilities that can greatly assist most areas of investigation (46, 66). THInCR was created specifically with the needs of the HPV research community in mind and using the CESC and HNSC TCGA cohorts. It provides access to a gene-centered approach to study the impact of HPV status on cellular mRNA and miRNA gene expression and DNA methylation. Furthermore, it allows the user to explore correlations with expression of individual HPV genes, patient survival, and immune landscape features. While there are notable advantages to using these large-scale data, an important limitation is that these tools are almost entirely based on RNA expression levels, which may not accurately reflect actual protein expression and/or activity, as described in detail by others (45). Importantly, THInCR uses actual molecular annotation to define true HPV status, rather than p16 status as a surrogate marker, as originally reported for the TCGA HNSC cohort (26). The simple, intuitive, and interactive interface facilitates the visualization, interpretation, and acquisition of otherwise-complex information relevant to studies of HPV oncogenesis that are not reachable in other packages. Basic and clinical researchers working in this field can readily explore the features of their favorite gene in two different anatomical sites, while comparing expression, methylation, survival, and immune landscape changes between HPV+ and HPV− disease.
Another important feature provided by THInCR is the ability to download graphical interpretations of each selected analysis suitable for documenting the analysis or presenting it informally. Importantly, at the press of a button, sets of tabular data in a format easily amenable for advanced customized graphical rendering for publication can also be downloaded. Additionally, master files of DEGs, differentially methylated probes, and gene expression correlations with viral genes can also be downloaded for further analysis or reference by the user.
In conclusion, THInCR is an intuitive interface to explore and interpret HPV-dependent changes in gene expression and DNA methylation using molecularly annotated CESC and HNSC data sets from the TCGA. HPV+ versus HPV− versus normal control tissue comparisons are possible in the form of graphs and comprehensive tables. These data can provide evidence to validate the tumor relevance of existing experimental observations in model systems and/or facilitate novel hypothesis generation. THInCR is freely accessible at https://thincr.ca/.
MATERIALS AND METHODS
Implementation and software.
The web server has been deployed on Amazon Web Services (AWS) running Ubuntu version 22.04, Shiny Server version 1.5.18.987, and R version 4.2.0. The following packages, along with required dependencies, have been installed: Shiny version 1.7.1, dplyr version 1.0.9, shinythemes version 1.2.0, data.table version 1.14.2, ggplot2 version 3.3.6, gdata version 2.18.0.1, reshape2 version 1.4.4, ggpubr version 0.4.0, DT version 0.23, plotly version 4.10.0, tidyverse version 1.3.1, survival version 3.3-1, survminer 0.4.9, Hmisc version 4.7-0, and scales version 1.2.0. Server setup was performed according to Bordet’s instructions at https://www.charlesbordet.com/en/guide-shiny-aws/. The service is available 24 h/day, 7 days/week at https://thincr.ca/, barring downtime for maintenance. Tutorial videos for tools are available through the home page under the section “Video Guides.” A stand-alone version of THInCR for Windows can be downloaded at https://github.com/msaland/THInCR-Suite.
Sample collection and ethics.
All data were downloaded from the Cancer Genome Atlas (TCGA) via the Broad Genome Data Analysis Center’s Firehose server (https://gdac.broadinstitute.org/) or other publicly available sources as noted below; therefore, no ethical approval was needed. Table 1 lists the number of available samples used in the calculations for each THInCR tool.
Data sources for mRNA and miRNA expression levels, patient cohort composition, and analysis workflow.
Level 3 mRNA and miRNA expression data for the TCGA HNSC and CESC data sets were sourced from Broad Genome Data Analysis Center’s Firehose server (https://gdac.broadinstitute.org/), with the data sets manually annotated as described by Gameiro et al. (67). The mRNA data sets feature expression patterns of 20,533 unique genes, whereas the miRNA data sets feature 1,048 unique genes. The CESC mRNA and miRNA data sets are each comprised of 165 HPV16+, 9 HPV33+, and 6 HPV35+ samples. The HNSC mRNA data set is comprised of 61 HPV16+, 8 HPV33+, and 3 HPV35+ samples. The miRNA data set is comprised of 54 HPV16+, 7 HPV33+, and 3 HPV35+ samples. Boxplots were generated using the ggplot2 package (version 3.3.5). Maximum and minimum boxplot values are represented as 1.5× upper and lower quartile ranges, respectively. The correlation of cellular gene mRNA or miRNA expression and HPV status was performed via sorting the data set into HPV16/33/35+, HPV−, or normal (noncancerous) subsets, with subsequent calculations performed with R’s built-in wilcox.test function with the conf.level parameter set to 0.95. Patient cellular gene mRNA and miRNA expression with >50% zero or null values were marked as nonsignificant regardless of calculated P value. q values were calculated for each comparison group with a false-discovery rate (FDR) of 10%.
Data sources for viral mRNA expression levels, patient cohort composition, and analysis workflow.
The HPV16/33/35 viral mRNA expression data sets were sourced from Ren et al. (32), with the data sets manually annotated as described by Gameiro et al. (67). These data sets feature expression levels of the E1, E2, E4, E5, E6, E7, L1, and L2 genes expressed by a subset of HPV16+, HPV33+, and HPV35+ TCGA CESC and HNSC patients, with expression being summed into a single value for individual genes with multiple mRNAs. The CESC data set features 91 patient observations, with 88 HPV16+, 2 HPV33+, and 1 HPV35+ sample. The HNSC data set features 65 patient observations, with 54 HPV16+, 8 HPV33+, and 3 HPV35+ samples. The correlation of HPV16/33/35 mRNA and cellular gene mRNA or miRNA expression was performed via R’s built-in cor.test function, with the function being run with the linear relationship and Spearman correlation coefficient arguments. Patient cellular gene mRNA or miRNA and HPV mRNA expression with >50% zero or null values were marked as nonsignificant regardless of calculated P value. q values were calculated for each comparison group with an FDR of 10%. Boxplots and scatterplots were generated using the ggplot2 package (version 3.3.5). Maximum and minimum boxplot values are represented as 1.5× upper and lower quartile ranges, respectively.
Data sources for DNA methylation levels, patient cohort composition, and analysis workflow.
Level 3 Infinium HumanMethylation450 BeadChip array data sets for the TCGA HNSC and CESC cohorts were sourced from the Broad Genome Data Analysis Centers Firehose server (https://gdac.broadinstitute.org/), with the data sets manually annotated for HPV status as described by Gameiro et al. (67). The data sets feature the methylation beta values for 395,329 different probes, along with chromosome number and genomic coordinates. The CESC data set features 303 patient observations, with 180 HPV16/33/35+, 19 HPV−, and 3 normal controls. The HNSC data set features 558 patient observations, with 72 HPV16/33/35+, 442 HPV−, and 50 normal controls. The correlation of probe methylation beta value and genomic loci was performed via sorting the data set into HPV16/33/35+ and HPV− subsets, with subsequent calculations performed via R’s built-in wilcox.test function with the conf.level parameter set to 0.95. Probe methylation beta with >50% zero or null values were marked as nonsignificant regardless of calculated P values. q values were calculated for each comparison group with an FDR of 10%. Boxplots and line plots were generated using the ggplot2 package (version 3.3.5). Maximum and minimum boxplot values are represented with 1.5× upper and lower quartile ranges, respectively.
Data sources for patient survival, patient cohort composition, and analysis workflow.
The TCGA HNSC and CESC overall survival (OS) data sets were sourced from Liu et al. (40), with the data sets manually annotated for HPV status as described by Gameiro et al. (67). The CESC data set has 180 HPV16/33/35+ and 19 HPV− patient observations. The HNSC data set has 72 HPV16/33/35+ and 442 HPV− patient observations. The correlation of survival and cellular gene mRNA and miRNA expression was performed via sorting the data set into HPV16/33/35+ and HPV− subsets, with subsequent calculations performed via the pairwise_survdiff and Surv functions, available via survminer and survival packages, respectively. Users have the option of selecting the number of comparison groups, upon which the subsets are broken down by the selected quantile. Patient cellular gene mRNA and miRNA expression with >50% zero or null values were marked as nonsignificant regardless of calculated P values. q values were calculated for each comparison group with an FDR of 10%. Kaplan-Meier survival plots were generated using the ggsurvplot function available through the survminer package (version 0.4.9).
Data sources for immune landscape features, patient cohort composition, and analysis workflow.
The immune landscape features for the TCGA HNSC and CESC data sets were sourced from Thorsson et al. (45), with the data sets manually annotated for HPV status as described by Gameiro et al. (67). The data sets included 53 immune landscape features as listed in Table 2. The correlation of immune landscape features and cellular gene mRNA and miRNA expression was performed via sorting the data set into HPV+ and HPV− subsets, with subsequent calculations performed via R’s built-in cor.test function, with the function being run with the linear relationship and Spearman correlation coefficient arguments. Immune landscape features and patient cellular gene mRNA and miRNA expression with >50% zero or null values were marked as nonsignificant regardless of calculated P values. q values were calculated for each comparison group with an FDR of 10%.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Institutes of Health Research (PJT-173496). We thank Greg Fonseca, Bart Kolendowski, Tanner Tessier, Haider Hassan, Trevor Shepherd, and Farhad Ghasemi for helpful advice and discussion during the implementation of this project.
Contributor Information
Joe S. Mymryk, Email: jmymryk@uwo.ca.
Laimonis A. Laimins, Northwestern University
REFERENCES
- 1.Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, Sun Q, Kaur R, Huyen Y, McBride AA. 2017. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zur Hausen H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim Biophys Acta Rev Cancer 1288:F55–F78. doi: 10.1016/0304-419X(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 3.Cutts FT, Franceschi S, Goldie S, Castellsague X, de Sanjose S, Garnett G, Edmunds WJ, Claeys P, Goldenthal KL, Harper DM, Markowitz L. 2007. Human papillomavirus and HPV vaccines: a review. Bull World Health Organ 85:719–726. doi: 10.2471/blt.06.038414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstock H, Berman S, Cates W. 2004. Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspect Sex Reprod Health 36:6–10. doi: 10.1363/psrh.36.6.04. [DOI] [PubMed] [Google Scholar]
- 5.Münger K, Basile JR, Duensing S, Eichten A, Gonzalez SL, Grace M, Zacny VL. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888–7898. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 6.Guan P, Howell-Jones R, Li N, Bruni L, De Sanjosé S, Franceschi S, Clifford GM. 2012. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 131:2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 7.de Sanjose S, Quint WGV, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GAH, Lombardi LE, Banjo A, Menéndez C, Domingo EJ, Velasco J, Nessa A, Chichareon SCB, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJLM, Al-Jassar WF, Cruz E, Wright TC, Puras A, et al. 2010. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 8.Riou G, Bourhis J, Favre M, Orth G, Jeannel D, Le Doussal V. 1990. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA. Lancet 335:1171–1174. doi: 10.1016/0140-6736(90)92693-C. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Carunchio L, Soveral I, Steenbergen RDM, Torné A, Martinez S, Fusté P, Pahisa J, Marimon L, Ordi J, Del Pino M. 2015. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG 122:119–127. doi: 10.1111/1471-0528.13071. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H, Shiraishi K, Kato T. 2021. Molecular pathology of human papilloma virus-negative cervical cancers. Cancers 13:6351. doi: 10.3390/cancers13246351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing B, Guo J, Sheng Y, Wu G, Zhao Y. 2020. Human papillomavirus-negative cervical cancer: a comprehensive review. Front Oncol 10:606335. doi: 10.3389/fonc.2020.606335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 13.Kreimer AR, Clifford GM, Boyle P, Franceschi S. 2005. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 14.Dayyani F, Etzel CJ, Liu M, Ho CH, Lippman SM, Tsao AS. 2010. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC). Head Neck Oncol 2:15. doi: 10.1186/1758-3284-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomaić V. 2016. Functional roles of E6 and E7 oncoproteins in HPV-induced malignancies at diverse anatomical sites. Cancers 8:95. doi: 10.3390/cancers8100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramqvist T, Dalianis T. 2010. Oropharyngeal cancer epidemic and human papillomavirus. Emerg Infect Dis 16:1671–1677. doi: 10.3201/eid1611.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marur S, D'Souza G, Westra WH, Forastiere AA. 2010. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol 11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. 2011. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiboski CH, Schmidt BL, Jordan RCK. 2005. Tongue and tonsil carcinoma: increasing trends in the U.S. population ages 20-44 years. Cancer 103:1843–1849. doi: 10.1002/cncr.20998. [DOI] [PubMed] [Google Scholar]
- 20.Leemans CR, Braakhuis BJM, Brakenhoff RH. 2011. The molecular biology of head and neck cancer. Nat Rev Cancer 11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Torabi SJ, Yarbrough WG, Mehra S, Osborn HA, Judson B. 2018. Association of human papillomavirus status at head and neck carcinoma subsites with overall survival. JAMA Otolaryngol Head Neck Surg 144:519–525. doi: 10.1001/jamaoto.2018.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. 2008. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 23.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. 2010. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Sullivan B, Huang SH, Siu LL, Waldron J, Zhao H, Perez-Ordonez B, Weinreb I, Kim J, Ringash J, Bayley A, Dawson LA, Hope A, Cho J, Irish J, Gilbert R, Gullane P, Hui A, Liu F-F, Chen E, Xu W. 2013. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J Clin Oncol 31:543–550. doi: 10.1200/JCO.2012.44.0164. [DOI] [PubMed] [Google Scholar]
- 25.Gillison ML. 2004. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol 31:744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network. 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, Brägelmann J, DeBoer R, Weichselbaum RR, Langerman A, Portugal L, Blair E, Stenson K, Lingen MW, Cohen EEW, Vokes EE, White KP, Hammerman PS. 2015. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols AC, Palma DA, Dhaliwal SS, Tan S, Theuer J, Chow W, Rajakumar C, Um S, Mundi N, Berk S, Zhou R, Basmaji J, Rizzo G, Franklin JH, Fung K, Kwan K, Wehrli B, Salvadori MI, Winquist E, Ernst S, Kuruvilla S, Read N, Venkatesan V, Todorovic B, Hammond JA, Koropatnick J, Mymryk JS, Yoo J, Barrett JW. 2013. The epidemic of human papillomavirus and oropharyngeal cancer in a Canadian population. Curr Oncol 20:212–219. doi: 10.3747/co.20.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jégu J, Binder-Foucard F, Borel C, Velten M. 2013. Trends over three decades of the risk of second primary cancer among patients with head and neck cancer. Oral Oncol 49:9–14. doi: 10.1016/j.oraloncology.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Price KAR, Cohen EE. 2012. Current treatment options for metastatic head and neck cancer. Curr Treat Options Oncol 13:35–46. doi: 10.1007/s11864-011-0176-y. [DOI] [PubMed] [Google Scholar]
- 31.Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, Lee S, Hadjipanayis AG, Ivanova EV, Wilkerson MD, Protopopov A, Yang L, Seth S, Song X, Tang J, Ren X, Zhang J, Pantazi A, Santoso N, Xu AW, Mahadeshwar H, Wheeler DA, Haddad RI, Jung J, Ojesina AI, Issaeva N, Yarbrough WG, Hayes DN, Grandis JR, El-Naggar AK, Meyerson M, Park PJ, Chin L, Seidman JG, Hammerman PS, Kucherlapati R, Cancer Genome Atlas Network . 2014. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA 111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren S, Gaykalova DA, Guo T, Favorov AV, Fertig EJ, Tamayo P, Callejas-Valera JL, Allevato M, Gilardi M, Santos J, Fukusumi T, Sakai A, Ando M, Sadat S, Liu C, Xu G, Fisch KM, Wang Z, Molinolo AA, Gutkind JS, Ideker T, Koch WM, Califano JA. 2020. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene 39:6327–6339. doi: 10.1038/s41388-020-01431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basukala O, Banks L. 2021. The not-so-good, the bad and the ugly: HPV E5, E6 and E7 oncoproteins in the orchestration of carcinogenesis. Viruses 13:1892. doi: 10.3390/v13101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J Virol 78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehoux M, D'Abramo CM, Archambault J. 2009. Molecular mechanisms of human papillomavirus-induced carcinogenesis. Public Health Genomics 12:268–280. doi: 10.1159/000214918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarth JA, Patterson MR, Morgan EL, Macdonald A. 2021. The human papillomavirus oncoproteins: a review of the host pathways targeted on the road to transformation. J Gen Virol 102:001540. doi: 10.1099/jgv.0.001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doorbar J. 2016. Model systems of human papillomavirus-associated disease. J Pathol 238:166–179. doi: 10.1002/path.4656. [DOI] [PubMed] [Google Scholar]
- 38.Lambert PF. 2016. Transgenic mouse models of tumor virus action. Annu Rev Virol 3:473–489. doi: 10.1146/annurev-virology-100114-054908. [DOI] [PubMed] [Google Scholar]
- 39.Chow LT. 2015. Model systems to study the life cycle of human papillomaviruses and HPV-associated cancers. Virol Sin 30:92–100. doi: 10.1007/s12250-015-3600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J, Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, et al. 2018. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research Network. 2017. Integrated genomic and molecular characterization of cervical cancer. Nature 543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M, Desai N, Sültmann H, Moch H, Cooper CS, Eils R, Ferretti V, Lichter P, Kleinheinz K, Nakagawa H, Ojesina AI, Pedamallu CS, Schlesner M, Su X, PCAWG Consortium . 2020. The landscape of viral associations in human cancers. Nat Genet 52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bratman SV, Bruce JP, O'Sullivan B, Pugh TJ, Xu W, Yip KW, Liu F-F. 2016. Human papillomavirus genotype association with survival in head and neck squamous cell carcinoma. JAMA Oncol 2:823–826. doi: 10.1001/jamaoncol.2015.6587. [DOI] [PubMed] [Google Scholar]
- 44.Banister CE, Liu C, Pirisi L, Creek KE, Buckhaults PJ. 2017. Identification and characterization of HPV-independent cervical cancers. Oncotarget 8:13375–13386. doi: 10.18632/oncotarget.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt R, et al. 2018. The immune landscape of cancer. Immunity 48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Li H, Jiang S, Li R, Li W, Chen H, Bo X. 2019. A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Brief Bioinform 20:1524–1541. doi: 10.1093/bib/bby023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee S, Do Kang S, Alam S, Salzberg AC, Milici J, van der Burg SH, Freeman W, Meyers C. 2019. Tissue-specific gene expression during productive human papillomavirus 16 infection of cervical, foreskin, and tonsil epithelium. J Virol 93:e00915-19. doi: 10.1128/JVI.00915-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sitz J, Blanchet SA, Gameiro SF, Biquand E, Morgan TM, Galloy M, Dessapt J, Lavoie EG, Blondeau A, Smith BC, Mymryk JS, Moody CA, Fradet-Turcotte A. 2019. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc Natl Acad Sci USA 116:19552–19562. doi: 10.1073/pnas.1906102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cicchini L, Blumhagen RZ, Westrich JA, Myers ME, Warren CJ, Siska C, Raben D, Kechris KJ, Pyeon D. 2017. High-risk human papillomavirus E7 alters host DNA methylome and represses HLA-E expression in human keratinocytes. Sci Rep 7:3633. doi: 10.1038/s41598-017-03295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soto D, Song C, McLaughlin-Drubin ME. 2017. Epigenetic alterations in human papillomavirus-associated cancers. Viruses 9:248. doi: 10.3390/v9090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K. 2016. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie 121:298–311. doi: 10.1016/j.biochi.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa T, Kurokawa T, Mima M, Imamoto S, Mizokami H, Kondo S, Okamoto Y, Misawa K, Hanazawa T, Kaneda A. 2021. DNA methylation and HPV-associated head and neck cancer. Microorganisms 9:801. doi: 10.3390/microorganisms9040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. 2011. High density DNA methylation array with single CpG site resolution. Genomics 98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Gameiro SF, Kolendowski B, Zhang A, Barrett JW, Nichols AC, Torchia J, Mymryk JS. 2017. Human papillomavirus dysregulates the cellular apparatus controlling the methylation status of H3K27 in different human cancers to consistently alter gene expression regardless of tissue of origin. Oncotarget 8:72564–72576. doi: 10.18632/oncotarget.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsoumpou I, Arbyn M, Kyrgiou M, Wentzensen N, Koliopoulos G, Martin-Hirsch P, Malamou-Mitsi V, Paraskevaidis E. 2009. p16INK4a immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev 35:210–220. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larsen G, Gyldenløve M, Jensen DH, Therkildsen MH, Kiss K, Norrild B, Konge L, Von Buchwald C. 2014. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Br J Cancer 110:1587–1594. doi: 10.1038/bjc.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loick SM, Fröhlich A, Gabrielpillai J, Franzen A, Vogt TJ, Dietrich J, Wiek C, Scheckenbach K, Strieth S, Landsberg J, Dietrich D. 2022. DNA methylation and mRNA expression of OX40 (TNFRSF4) and GITR (TNFRSF18, AITR) in head and neck squamous cell carcinoma correlates with HPV status, mutational load, an interferon-γ signature, signatures of immune infiltrates, and survival. J Immunother 45:194–206. doi: 10.1097/CJI.0000000000000407. [DOI] [PubMed] [Google Scholar]
- 58.Nagy Á, Munkácsy G, Győrffy B. 2021. Pancancer survival analysis of cancer hallmark genes. Sci Rep 11:6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. 2017. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 60.Mandal R, Şenbabaoğlu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee K-W, Ganly I, Hakimi AA, Chan TA, Morris LGT. 2016. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 1:e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gameiro SF, Ghasemi F, Barrett JW, Koropatnick J, Nichols AC, Mymryk JS, Maleki Vareki S. 2018. Treatment-naïve HPV+ head and neck cancers display a T-cell-inflamed phenotype distinct from their HPV- counterparts that has implications for immunotherapy. Oncoimmunology 7:e1498439. doi: 10.1080/2162402X.2018.1498439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shamseddine AA, Burman B, Lee NY, Zamarin D, Riaz N. 2021. Tumor immunity and immunotherapy for HPV-related cancers. Cancer Discov 11:1896–1912. doi: 10.1158/2159-8290.CD-20-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roden RB, Hubbert NL, Kirnbauer R, Christensen ND, Lowy DR, Schiller JT. 1996. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J Virol 70:3298–3301. doi: 10.1128/JVI.70.5.3298-3301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joura EA, Giuliano AR, Iversen O-E, Bouchard C, Mao C, Mehlsen J, Moreira ED, Ngan Y, Petersen LK, Lazcano-Ponce E, Pitisuttithum P, Restrepo JA, Stuart G, Woelber L, Yang YC, Cuzick J, Garland SM, Huh W, Kjaer SK, Bautista OM, Chan ISF, Chen J, Gesser R, Moeller E, Ritter M, Vuocolo S, Luxembourg A, Broad Spectrum HPV Vaccine Study . 2015. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N Engl J Med 372:711–723. doi: 10.1056/NEJMoa1405044. [DOI] [PubMed] [Google Scholar]
- 65.Schiller JT, Lowy DR. 2012. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol 10:681–692. doi: 10.1038/nrmicro2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng H, Zhang G, Zhang L, Wang Q, Li H, Han Y, Xie L, Yan Z, Li Y, An Y, Dong H, Zhu W, Guo X. 2020. Comprehensive review of web servers and bioinformatics tools for cancer prognosis analysis. Front Oncol 10:68. doi: 10.3389/fonc.2020.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gameiro SF, Zhang A, Ghasemi F, Barrett JW, Nichols AC, Mymryk JS. 2017. Analysis of class I major histocompatibility complex gene transcription in human tumors caused by human papillomavirus infection. Viruses 9:252. doi: 10.3390/v9090252. [DOI] [PMC free article] [PubMed] [Google Scholar]