Abstract
In vivo MRS is a non-invasive measurement technique used not only in humans, but also in animal models using high-field magnets. MRS enables the measurement of metabolite concentrations as well as metabolic rates and their modifications in healthy animals and disease models. Such data open the way to a deeper understanding of the underlying biochemistry, related disturbances and mechanisms taking place during or prior to symptoms and tissue changes. In this work, we focus on the main preclinical 1H, 31P and 13C MRS approaches to study brain metabolism in rodent models, with the aim of providing general experts’ consensus recommendations (animal models, anesthesia, data acquisition protocols). An overview of the main practical differences in preclinical compared with clinical MRS studies is presented, as well as the additional biochemical information that can be obtained in animal models in terms of metabolite concentrations and metabolic flux measurements. The properties of high-field preclinical MRS and the technical limitations are also described.
Keywords: anesthesia, brain metabolism, consensus review, dynamic MRS, neurochemical profile, preclinical MRS
Graphical Abstract
1 |. INTRODUCTION
The use of rodents as experimental models provides a great opportunity to increase our understanding of human tissue development, function and metabolism, which is relevant to better understand pathologies and to develop treatment strategies. While basic cellular or metabolic questions can be studied using in vitro models (e.g. cell cultures of different levels of complexity, from monotypic monolayers to complex organotypic pluricellular cultures), understanding more complex traits of the living organism (e.g. interactions between different cell types, organ systems and behavior) requires the use of in vivo experimental models, for which mice and rats have been the most widely used for many reasons such as their proximity to the human genome, their short generation time and their small size.
In vivo MRI/MRS have become tools of choice to study and better understand the central nervous system (CNS) activity and metabolism as well as human brain diseases, both in human subjects and through the use of animal models, including mice or rats. MRI and MRS have the great advantages of being non-invasive and allowing an easy longitudinal follow-up of brain development and activity, disease evolution and treatment efficacy.1–3 While patients are usually followed in clinics using magnetic fields of 1.5 or 3 T, the development of new technologies, allowing MRI/MRS at magnetic field strength of 9.4 T and higher, enables the study of the CNS in rodent models with unprecedented image/spectral resolution, allowing for example the deciphering of up to 20 different neurochemicals by 1H MRS (Table 1).4–6 Moreover, combining MRS of various nuclei (e.g. 1H, 13C, 31P) broadens the range of brain metabolic and neurotransmission pathways available to study, including the time course of metabolic activities, particularly with 13C MRS.7–9
TABLE 1.
| Neurotransmission | Energy metabolism | Antioxidants | Osmolytes | Membrane metabolism |
|---|---|---|---|---|
| glutamate | creatine | glutathione | taurine | phosphoethanolamine |
| glutamine | PCr | ascorbate | myo-inositol | phosphocholine |
| GABA | glucose | scyllo-inositol | glycerophosphocholine | |
| N-acetylaspartylglutamate | lactate | NAA | ||
| aspartate | alanine | |||
| glycine |
This consensus recommendation paper aims at providing specific technical recommendations for MRS in preclinical studies of brain function, metabolism and diseases using animal models, including a broad overview of technical specificities of 13C, 31P and 1H MRS and the characteristics of the corresponding in vivo MRS data. For MRS aspects and challenges common to clinical and preclinical studies, the reader is referred to the other consensus reviews of this special issue.
2 |. ANIMAL PHYSIOLOGY AND ANESTHESIA
For several reasons discussed below, rats have been considered for decades as better models than mice to tackle human pathology, and have been used for a long time as models for surgery, exposure to toxins or pathological vectors, or development of treatments by exposure to various agents (e.g. drugs, vaccines, viral vectors).10 In contrast, since 1980 and the availability of genetically modified mice (and particularly homologous recombination technology),11 the mouse has been the most used preclinical model to investigate normal gene functions as well as human genetic diseases. While generating genetically modified rats was much more difficult than for mice historically, the recent technologies (in particular Crispr/CAS9)12–15 make the development of genetically modified rats easier and provide new and valuable rat models to better understand both basic gene functions and human diseases.
Important differences exist between mice and rats, the two most used in vivo experimental models.16 While mice have advantages over rats in their body size for reduced housing costs and in readily available genetic modification techniques, they are less used for brain metabolic imaging studies due to the small size of the brain, and lower similarity to humans in terms of CNS metabolism and circuitry as well as behavior. In contrast, while rats have higher costs for purchase, housing and consumables, they have long been recognized, especially in neuroscience and behavioral research, as better models to study basic functions and metabolism of the CNS as well as human brain diseases because of their larger brain size, their similarities to humans in terms of brain metabolism and more reliable behavioral tests as described below. In particular, while rat and mouse brains develop in a very similar manner, several major differences have been identified in terms of circuitry and brain function, as well as of their respective behaviors, making rats often better models for preclinical studies of human brain diseases. For examples, rats show a more social behavior and are generally preferred to mice in cognitive tests, making them an attractive model for the study of autistic spectrum disorders; rats and humans, in contrast to mice, show very similar levels and spatial distributions of 5-HT6 serotonin receptors in the CNS, making rats very interesting models for the study of drug addictions, psychiatric disorders such as schizophrenia and attention deficit hyperactivity disorders; some neurodegenerative diseases, such as Parkinson’s and Huntington’s, are also better modeled in rats than mice (see Reference 17 and references therein for more examples and details). On the practical side, however, one disadvantage of working with rats over mice is the large change in body size over their lifetime, which could require RF coil geometry adaption over lifetime. Mice are easier in this respect for MRS experiments.
Effective and standardized mouse MRI/MRS studies require attention to many aspects of the experimental design. One of the important aspects of the experimental design is the anesthesia. Anesthesia is critical for in vivo preclinical MRI and MRS. It decreases the stress of the animals, potential pain in case of surgical intervention, and their biological motions, e.g. physical activity and head movement, as well as respiratory and cardiac activities.18–20 This section provides a brief overview of pre-anesthetic considerations, a review of the existing literature regarding the effects of anesthesia on the neurochemical profile of rodents, i.e. rats and mice, and a practical guideline for selecting an appropriate anesthetic protocol for MRS studies of small animals.
2.1 |. Pre-anesthetic considerations for in vivo MRS studies
The effect of anesthesia depends on a variety of factors including stress, strain, sex, circadian cycles, weight and age of the animals.20,33 These factors not only may change the effectiveness of anesthesia and required dosage but also may have a direct impact on MRS measurements of the neurochemical profile in the rodent brain.34
2.1.1 |. Stress
Transporting, handling, restraining and injection of anesthetics may cause acute stress, which leads to alterations in physiological parameters, such as corticosteroid and epinephrine levels, glucose levels, respiration and heart rate.33 Some period of acclimatization, which takes in general between 24 h and 48 h (up to 7 days following ethics legal guidelines), is necessary after transporting the animals to the imaging facility to decrease the stress level.35 The personnel working with animals should be well trained to acclimatize and handle the animals properly before anesthetizing the animals to reduce potential acute stress before MRS experiments. Stress-induced alterations in physiological parameters such as corticosteroids may increase the required dosage for proper anesthesia and alter the concentration of neurochemicals. A few MRS studies have successfully demonstrated the effect of stress on the neurochemical profile in the rodent brain.36,37 For example, female adolescent rats exposed to early life stress demonstrated reduced glutamate, glutamine and N-acetylaspartate (NAA) compared with controls in the prefrontal cortex.36 In a chronic unpredictable stress rat model, increased gamma-aminobutyric acid (GABA)/glutamine and glutamate/glutamine ratios have been positively correlated with plasma corticosterone levels.37
2.1.2 |. Strain
Genetic variation, i.e. genetic background, among mice is well documented. Earlier studies demonstrated that the genetic background of the mouse strain can have a substantial effect on physiological parameters such as the sleep time of anesthesia and stress.33,38–40 For example, Lovell reported that there were variations in pentobarbitone sleeping time between mice from different strains.38 Strain-specific differences in neurochemical concentrations have also been demonstrated in C57BL/6 compared with BALB/c and NMRI mice.41 The concentrations of NAA, creatine and phosphocreatine (PCr), choline-containing compounds, glucose and lactate were different between C57BL/6 and BALB/c mice,41 and glucose and lactate levels were different between C57BL/6 and NMRI mice.
2.1.3 |. Weight
The effective dosage of anesthesia depends on the body weight of the animal. Obese rodents may react differently to anesthesia. Obesity is a heterogenic condition, which may arise due to different factors, such as genetics and diet. Obesity should be considered as an important confounder when accounting for the effect of anesthesia on animal physiology and MRS findings. Alteration in body composition may affect the endocrine response, cardiovascular and respiratory function, and pharmacological response of the animal to the anesthesia. It is important to take into account the possible variations in drug absorption related to obesity. For example, obese rodents exhibit altered biodistribution of lipophilic agents, and they have a low metabolic rate compared with lean animals. Lipophilic molecules, such as anesthetics and analgesics, may cause deposition of drugs in the adipose tissue of obese rodents, and eventually delay the time onset of anesthetics.40,41 Overall, the body composition of the rodents is an essential confounding factor when accounting for effects of anesthesia on the animal physiology, which may affect MRS findings.
2.1.4 |. Sex
The sex effect should be taken into account in collecting and analyzing MRS data. Few rodent studies reported a substantial effect of sex differences on the neurochemical profile of rodents.42,43 The presence of menstrual cycle effects on the neurochemical profile of the human brain has been demonstrated.44 However, the estrous cycle effect on the neurochemical concentrations of female rodents has not been thoroughly studied. The sex effect should be taken into consideration when evaluating anesthetic effects. A variety of articles have discussed the effect of sex on anesthetic dosages, metabolism and pharmacokinetics.22,45–48 Due to differences in sexual hormones, plasma corticosteroids and hepatic enzymes between female and male rodents, the effects of anesthesia on animals’ physiological parameters may differ.33 Therefore, the dosage of the anesthetic should be adjusted if the type of anesthesia is affected by the sex difference. For example, the suggested dosage for ketamine anesthesia is higher for female mice compared with male mice.22
2.1.5 |. Circadian cycles
Rodents have circadian rhythms, which provide rhythmic variations in their many physiological functions, including hormones.49 A recent in vivo MRS study reported that diurnal changes occurred in the neurochemical profile in rats when MRS data were collected after light onset or offset.50 In some animal facilities light onset and offset time points are reversed. Therefore, reporting the time of the experiment and the time schedule for light onset and offset of animal facilities is an important parameter while assessing the data collected from randomly assigned experimental groups. The timing of the experiments and treatment should be controlled and reported in order to increase the reproducibility between MRS studies.33
2.1.6 |. Age
The age of the animals has an important effect on the neurochemical profile of the rodent brain51,52 and also on anesthetic variability. For example, young mice (<8 weeks) cannot metabolize anesthetics as effectively as adult mice can.22 Therefore, the impact of the same levels of anesthetics on cerebral metabolites might be different in young mice relative to mature mice (>3 months). In a longitudinal setting, the brain size of rodents may also change due to age or progression of a disease. For example, atrophy in the region of interest may occur due to a neurodegenerative disease.53 The MRS voxel size may, therefore, need to be adjusted according to age or disease-dependent changes of the brain. If absolute metabolite concentration is derived using unsuppressed tissue water, edema or age-related changes in water content of the brain should be taken into consideration as well.
2.2 |. Guidelines and recommendations for anesthesia protocols
A variety of inhaled and injectable anesthetics are available for MRS studies of rodents.21 The appropriate choice of the anesthetic procedure is essential, as anesthetics may have variable effects on the neurochemical profile in rodents, as well as other side effects, as summarized in Table 2 and described in more detail in Supplementary Materials. A detailed discussion of the influence of inhaled and injectable anesthetics on the physiology of animals and more information about the properties of these agents can be found elsewhere.18,19,22,54,55
TABLE 2.
Characteristics of commonly used anesthetics and their impact on brain metabolites
| Physiological effects | Side effects | Effect on brain metabolites↓↑* statistically significant changes(p < 0.05) | Type | References | |
|---|---|---|---|---|---|
| Propofol | Rapid and short-acting anesthesia effect, fast recovery time | Muscle twitching, apnea, hypotension, decreased cardiac output | Lactate ↓, glutamate ↓* (compared with isoflurane) | Injectable | 20,21 |
| Halothanes (e.g. isoflurane, sevoflurane) | Rapid and short-acting anesthesia effect, fast recovery time | Respiratory depression, dose dependent hypotension, increased cerebral blood flow, immune suppression | Lactate ↑, GABA ↑, choline-containing compounds ↑, myo-inositol ↑, glucose ↓, NAA ↑, total creatine ↑, creatine ↑, glutamate ↑, glutamine ↓, alanine ↑ * (compared with without isoflurane) | Inhaled | 22,23 |
| Thiopental | Ultra-short acting | Severe tissue necrosis (if administered via non-i.v. routes), prolonged recovery if the animal has low body fat, myocardial depression, decreased cardiac output, hypotension | Glucose ↑* (compared with light alpha-chloralose) | Injectable | 20,24 |
| Pentobarbitone | Poor analgesia characteristics (more reliable for rats than for mice) | Hyperexcitability, significant cardiovascular depression in mice, hypotension in rats | GABA ↓ glucose ↓, taurine ↓, propylene glycol ↑* (compared with isoflurane), glucose ↑ (compared with light alpha-chloralose) | Injectable | 20,25–27 |
| Ketamine | Rapid analgesia but less muscle relaxation | Respiratory depression, pain in injection side (due to low pH), increased cardiac output, heart rate, blood pressure | Glutamate ↑* (1H-13C NMR study; 80 mg per kg ketamine treated group compared with saline treated group) | Injectable | 18,20,28 |
| Xylazine/ketamine | The synergistic effect causes anesthesia with extended analgesia | Body temperature may decrease, increased urination, defecation, salivation, ocular lesions, hypoglycemia | Alanine ↓, ascorbate (or vitamin C) ↑, aspartate ↑, GABA ↑, glycine ↑, PCr ↑ (compared with isoflurane) | Injectable | 27,29 |
| Urethane | Provides long-lasting anesthesia | Mutagenic and carcinogenic in experimental animals | Lactate ↑ (compared with no urethane group) | Injectable | 30,31 |
| Alpha-chloralose | Provides long-lasting light anesthesia | Poor analgesic properties, prolonged and poor recovery | Unknown | Injectable | 32 |
The type, administration method and duration of anesthesia, as well as the dosage of the anesthetic, should be optimized carefully according to the aims of each experiment.21,24–26,56 Overall, an inhaled anesthetic is easier to handle for MRI/MRS experiments due to its means of administration, the possibility to adapt the dose inside the magnet during the experiment, its fast kinetics and ease of use for longitudinal studies. Information about the drug doses of anesthetics that are commonly used in rodents in MRI and MRS studies can be found elsewhere.20 The correct dosage of the anesthetic should provide adequate sedation but also adequate analgesia and less variability in physiological parameters during MRI/MRS experiments.20 Monitoring and recording the respiration rate and temperature of animals under anesthesia is essential (if available, pulse oximetry and electrocardiography can provide further control). If the experiment requires repeated exposure to the anesthetic, one having a quicker recovery phase and fewer side effects should be chosen. Providing an adequate environment for the animal not only during exposure to anesthesia but also during the recovery phase is vital to prevent complications, such as hypothermia, stress and respiratory arrest (i.e., recovery on a heating pad or under a heating lamp should be common practice). Ocular protection should likewise be provided, as rodents may keep their eyes open under anesthesia. The correct application of anesthesia is essential, as inappropriate use of these agents may cause physiological instability and deleterious effects, including pain, fear, distress, hypothermia and hypoxia.18
2.3 |. Physiological parameters and physiological monitoring
A main limitation for animal subjects undergoing in vivo MRS under anesthesia is the impact of anesthesia on measurements, especially in studies on brain metabolism. Although anesthesia helps acquire signals with minimal motion, minimal stress and maximal reproducibility, all anesthetic drugs alter normal physiology in some way and may confound results.
An essential step to minimize these effects is to monitor physiological parameters during the animal preparation and during the scan.20,57–59 Anesthesia typically induces hypothermia, which can impact energy metabolism. Moreover, a consistent anesthesia level among the analyzed groups is required to avoid biased results or to artificially increase the variability of the measured cerebral metabolic parameters. If anesthesia is too light, this could lead in the worst case to partial awakening of the animal, potentially inducing stress, pain and motion. A careful monitoring of the animal respiration frequency is a very good way to follow and adapt the anesthesia level.
It is recommended to monitor the body temperature and keep it stable (with the help of MR-compatible heating systems, such as a hot air stream in the bore or a hot water pipe circuit). The temperature should be kept in the range of 36.5–37.5 °C for mice and 37.5–38.5 °C for rats. The normal undisturbed respiration rate is ~100–180/min in mice and ~70–120/min in rats, and a decrease by 50% is acceptable during anesthesia. If the breathing rate is too low, the animal will gasp and not oxygenate properly. It is recommended to first test the anesthesia protocol on the chosen animal model in bench experiments through visual inspection of the animal. Further parameters such as blood parameters (pH, pO2, pCO2,…) and heart rate will help to monitor the physiology, but require more equipment and blood sampling, which is not always achievable in the center of the MR scanner, especially considering the small blood volume of mice.
2.4 |. MRS in awake rodents
Performing MRS with awake rodents is challenging and requires a relatively long training period for the animals to stay still during scans. Restraining awake animals without proper training may induce stress and affect MRS results. For awake-rodent MRS studies, monitoring serum cortisone levels and heart rate of the animals is recommended.60 There are a variety of methodologies for training and acclimating the animals to the MRI environment.61–65 These methodologies vary among different research centers and related ethical committees.61
3 |. HARDWARE
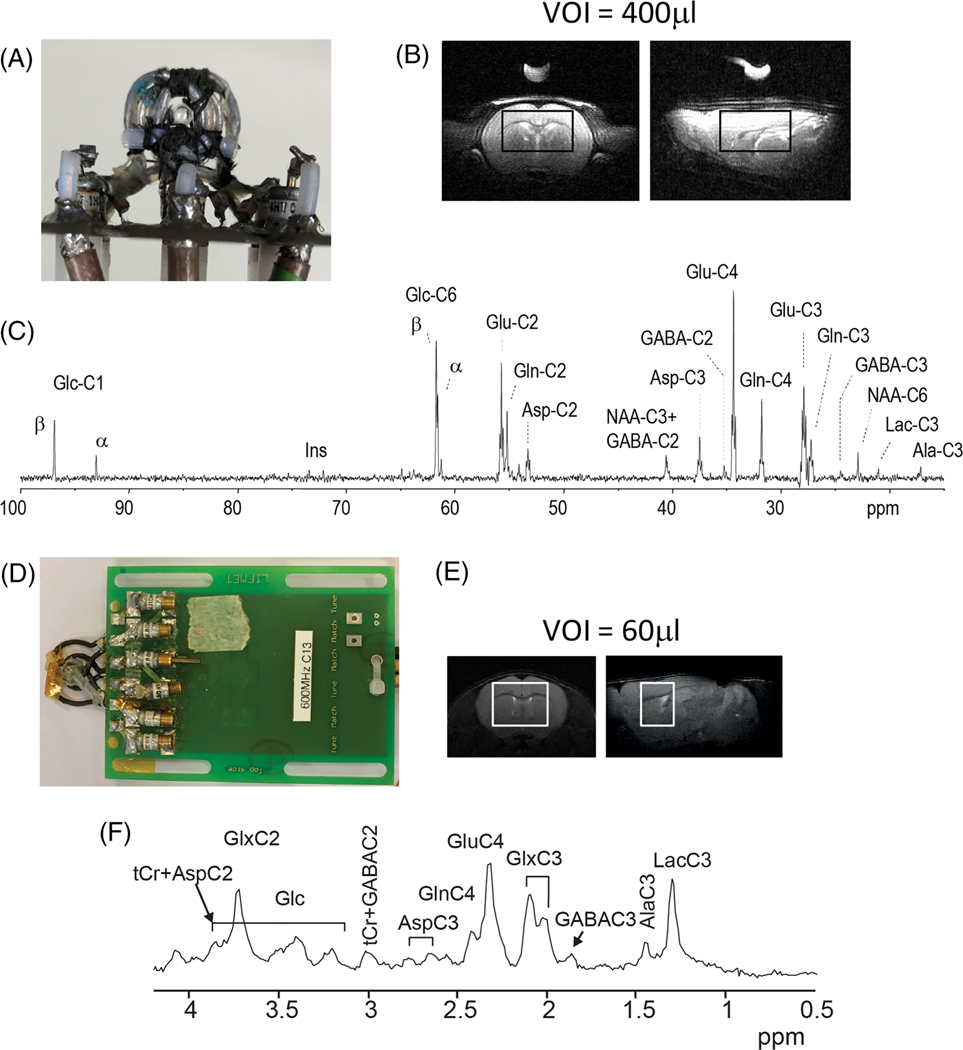
The small size of the brain and strong B0 inhomogeneity induced in the brain by the air/tissue interface are two major differences distinguishing preclinical studies of small animals from clinical studies and leading to different hardware requirements. The small volumes of interest (VOIs) (rats 50–150 μL, mice 2–15 μL) necessitated by the small size of the brain and significant regional differences in neurochemical concentrations benefit from ultra-high magnetic field strength (≥9.4 T), where the increased sensitivity compensates for the reduced signal-to-noise ratio (SNR) due to the small size of the VOI. The small VOIs at ultra-high fields put higher requirements on gradient strength (ideally ≥400 mT/m) compared with that for human systems, which is typically 70 mT/m for 7 T clinical MR systems. It is known that rapid switching of magnetic field gradients stimulates impulses in peripheral nerves, known as peripheral nerve stimulation, though these effects have hitherto not been reported to be of concern in preclinical research. Another advantage of using ultra-high fields is the increased chemical shift dispersion, which helps to resolve overlapped resonances and simplifies strongly coupled spin systems. However, to take advantage of the increased chemical shift dispersion and spectral resolution, B0 inhomogeneity needs to be minimized by using an efficient shimming method and shim system, strong enough to compensate for the field gradient induced in the brain. Stronger shims are required for the mouse brain than for the rat brain. In addition, the required strength of the shims in the mouse brain is region dependent.66 At 9.4 T, shim strengths of up to 47 μT/cm2 for XZ, YZ, Z2 and 23.5 μT/cm2 for XY and X2Y2 are needed for mouse brain spectroscopy,66 while the strength of the shims scales linearly with the field strength, since the amplitude of susceptibility-induced B0 inhomogeneity scales with the B0 field strength.67,68 Automatic shimming methods such as FAST (EST)MAP69,70 or 3D B0 mapping71 can be used efficiently for shimming on preclinical systems. Regarding the B0 shim quality expressed as the full-width at half-maximum72 of water linewidths in a specific brain region, the lowest linewidths can be achieved from more homogeneous regions, such as hippocampus and striatum (i.e. 9–12 Hz in the rat brain at 9.4 T for a voxel of 2 × 2.8 × 2 mm3 for hippocampus and 2.2 × 2 × 2.5 mm3 for striatum using FAST (EST)MAP). In the cerebellum, the water linewidth is broader (i.e. 14–17 Hz at 9.4 T in the rat brain for a voxel of 2.5 × 2.5 × 2.5 mm3) due to intrinsic properties of the tissue (i.e. microscopic heterogeneity).68
The power of the RF amplifiers is lower in preclinical than in human systems, since much smaller and more efficient coils (either volume or surface coils) are used on preclinical systems. Volume RF coils provide uniform images of the whole brain due to their homogeneous B1 field. However, when used for signal reception in MRS, they can lead to increased contamination from areas outside the VOI and collect more thermal noise from the measured object due to their larger field of view. Surface coils provide much higher SNR from regions close to the RF coil and higher B1 efficiency than volume coils, but the B1 field is spatially inhomogeneous. The usage of adiabatic RF pulses can mitigate B1 inhomogeneity. It is also worth mentioning that, in contrast to human studies, legally unlimited B1 and strong gradients enable the optimum RF coil (or combination of RF coils) to be chosen for a specific experiment from the point of view of SNR and chemical shift displacement (CSD) error. Combinations of volume coils for RF transmission and receive loops or receiver arrays have been used in a few recent pre-clinical applications,73–76 but rarely for brain studies. Such coil combinations can potentially improve the measurement in deeper brain regions and for 1H magnetic resonance spectroscopic imaging (MRSI), but are technically more complicated and experimentally challenging.
To maximize sensitivity, most 13C and 31P studies have used surface coils. The most commonly used arrangement is a combination of a single-loop coil for 13C or 31P and quadrature coil for 1H (Reference 77) or vice versa.78–82 Alternate coil arrangements have been proposed to further increase sensitivity with quadrature detection.74,83,84
Cryogenically cooled RF coils can be used for further noise reduction. For small sample volumes, the thermal noise in the coil and the receive pathway is the dominant noise source. By cooling the respective components,85 in direct comparison with a room-temperature coil, a reduction of the overall noise by a factor of 2–3 has been reported86 (Figure 1). This enables a remarkable reduction of the acquisition time or acquisitions from smaller volumes within a reasonable acquisition time. A limiting factor for the general usage of cryogenically cooled coils results from the requirement of dominating coil noise, which restricts its application to small animals.87,88
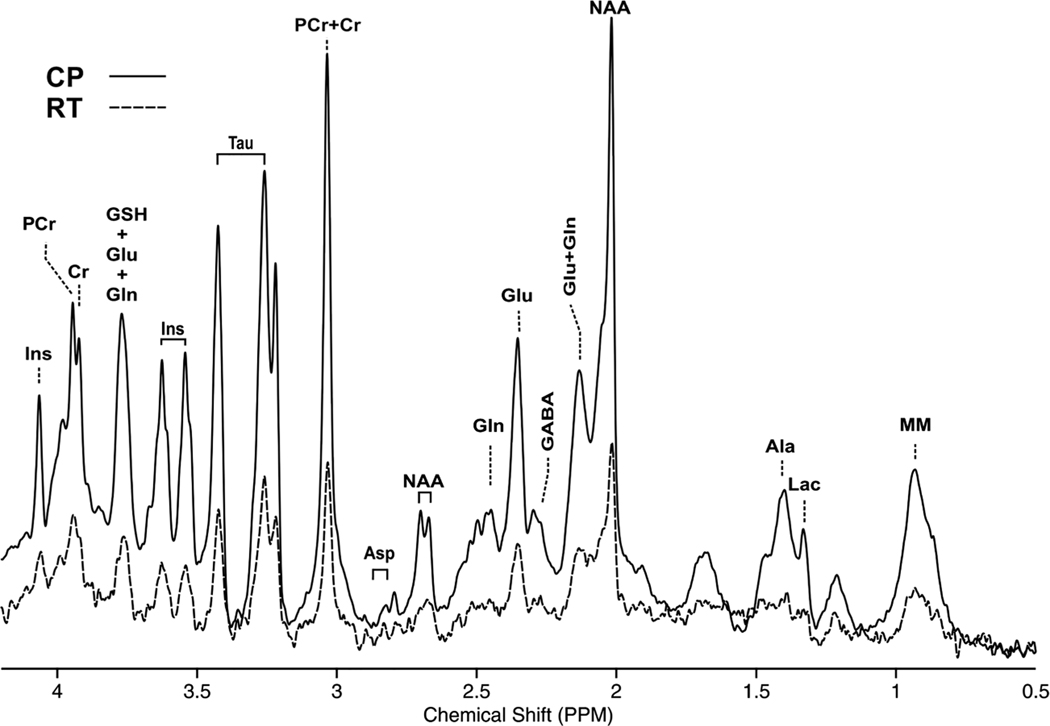
FIGURE 1.
Spectra acquired with the STEAM sequence (TR/TE = 5000/3.5 ms, 384 averages) in a 2.0 × 1.1 × 2.0 mm3 VOI located in the mouse frontal cortex. A cryogenically cooled 1H two-element phased-array transmit/receive coil was employed for excitation and signal reception (solid line). As a comparison, a 72 mm diameter birdcage quadrature volume resonator was used for excitation and a 1H receive-only 2 × 2 surface array coil was used for signal reception (dotted line). A 5.2-fold higher SNR was obtained with the cryoprobe (CP) compared with the room-temperature probe (RT)
4 |. SEQUENCES AND ACQUISITION PROTOCOLS: 1H MRS
The methodology of preclinical localized 1H MRS is very similar to that of clinical 1H MRS. In preclinical studies (in contrast to TE = 20–30 ms in ‘short-TE” MRS protocols provided by manufacturers of human scanners), ultra-short-TE (≤10 ms) spectroscopic localization sequences are usually possible to achieve and preferentially used because they provide the most accurate quantitative information from a 1H MR spectrum by minimizing the J evolution in coupled spin systems and reducing T2 losses. With the wide availability of ultra-high-field (9.4 T and above) preclinical MR scanners, minimal J-modulation 1H MRS studies in rodents also benefit from the high spectral dispersion that enables the measurement of a large number of metabolites including those (such as GABA, glutathione and lactate) that generally require spectral editing at lower magnet fields in clinical studies.89,90
The use of longTR minimizes signal attenuation due toT1 weighting at the expense of a long acquisition time. The length of the acquisition time, however, is not as critical an issue in preclinical studies (where rodents are carefully anesthetized and immobilized) as it is in clinical studies. T1 relaxation times of metabolites in the rat brain are ~1.5 s at 9.4 T and similar beyond 9.4 T.91 Therefore, a TR of 4–5s would result in signal reduction of 3.6–7.0%, while a 20% change inT1 will only lead to 2.6–3.9% signal difference for group comparison studies.
The localization performance of a 1H MRS sequence is very important and the following properties should be considered when choosing the localization sequence: (1) an ability to detect signals originating from the VOI; (2) an ability to suppress signals from outside of the VOI; (3) minimal CSD error related to the bandwidth of the localization pulses; and (4) insensitivity to B1 inhomogeneity, especially when using surface coils. There are no region-specific requirements for the most frequently studied regions: cortex, striatum, corpus callosum, hippocampus. However, some specific regions (e.g. cerebellum, olfactory bulb) are more difficult to shim due to intrinsic properties of the tissue or are outside the sensitive volume of RF coils and need careful B1 and B0 shim adjustments.
Basic pulse sequences for localized spectroscopy were designed a long time ago and are still used in most preclinical studies. The most popular localization methods are based either on a stimulated echo (e.g. stimulated echo acquisition mode spectroscopy, STEAM92,93) or on a double spin echo (e.g. point-resolved spectroscopy, PRESS94).
The STEAM sequence uses three slice-selective 90 pulses to form a stimulated echo; however, half of the magnetization available in the VOI is lost with this pulse sequence. STEAM is suitable for short or even ultra-short-echo-time measurements (TE = 1 ms; Reference 93). Because of the use of 90 °pulses for localization, this pulse sequence has very small CSD error. The flatness of the sine function around an angle of 90° leads to a reduced sensitivity of STEAM to B1 variation compared with pulse sequences employing amplitude modulated refocusing pulses.
The PRESS sequence preserves all the magnetization available in a selected VOI. On the other hand, it is quite difficult to suppress all undesired echoes created by pairs of slice-selective pulses in the double spin echo sequence. Thus, this sequence is mainly used at a longer echo time (≥10 ms). In addition, its conventional 180° pulse cannot achieve a bandwidth as broad as adiabatic pulses, which limits the localization performance of PRESS and increases CSD error in two spatial directions.
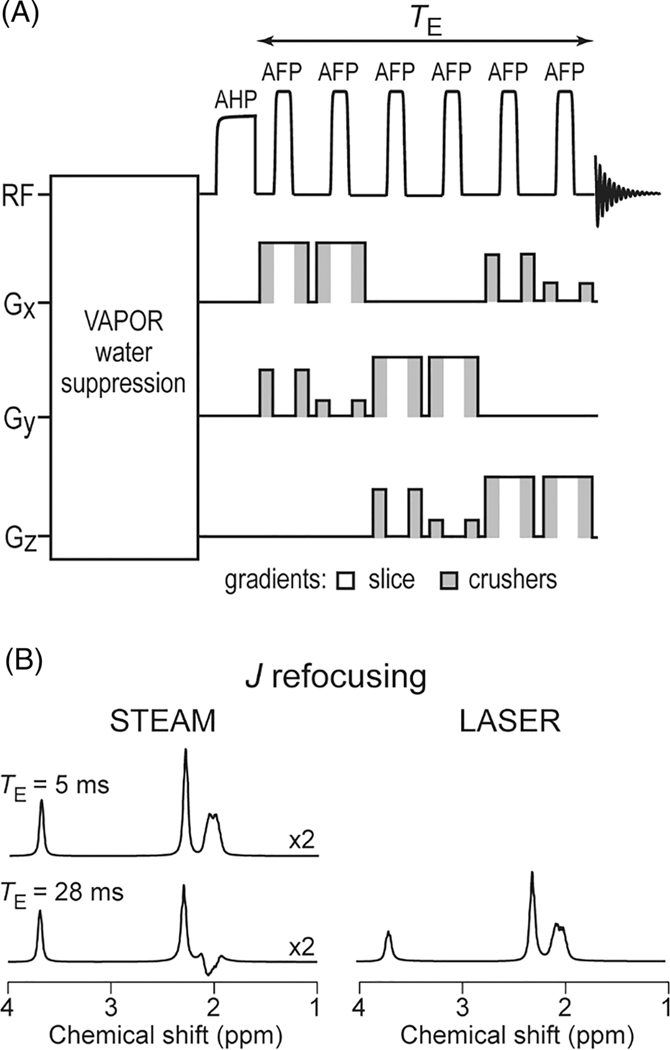
In recent years, novel methods of localized spectroscopy suitable for preclinical studies have appeared using adiabatic selective refocusing RF pulses. The first sequence performing an accurate volume localization with seven adiabatic pulses (SADLOVE95) evolved into the full LASER (localization by adiabatic selective refocusing) pulse sequence,96 which is a fully adiabatic single-shot 3D localization sequence, and does not require outer volume suppression (OVS). It consists of a non-selective adiabatic half-passage (AHP) pulse followed by three pairs of slice-selective adiabatic full-passage (AFP) pulses (Figure 2A). On preclinical scanners, an optimized LASER sequence can result in TE values ranging from 15 to 28 ms, similar to PRESS.52,53,97–99 Due to the properties of the AFP pulses, clean profiles with sharp transitions are obtained and the CSD error is minimal due to the large bandwidth of the AFP pulses, which are typically higher than 10 kHz. At 9.4 T on a preclinical scanner, the resulting CSD error is typically 2.4%/ppm. The successive application of multiple AFP pulses in LASER suppresses J evolution in coupled spin systems and prolongs apparent T2,99–101 resulting in much smaller signal loss for LASER than observed for other sequences at similar TE. For TE values between 15 and 28 ms at 9.4 T, the loss due to J evolution and T2 is minimal99 (Figure 2B).
FIGURE 2.
LASER sequence. A, LASER sequence with RF and gradient pulses shown schematically. Volume selection with LASER is performed with AFP pulses. Pulsed field gradients are used for suppressing outer-volume signals (gray shading) and for slice selection (white). B, Simulated scalar coupling evolution of glutamate at 9.4 T for STEAM sequence at TE = 5 ms and 28 ms, and LASER sequence at TE = 28 ms. The successive application of multiple AFP pulses in the LASER sequence suppresses J evolution in coupled spin systems. The vertical scale for the STEAM sequence has been multiplied by 2
One-dimensional image-selected in vivo spectroscopy (ISIS)102 and a slice-selective single spin echo have been combined in a technique with the acronym SPECIAL (spin echo, full intensity acquired localized spectroscopy).103 Standard and semi-adiabatic versions of this sequence as well as the advantages and disadvantages of this technique are described in detail in another paper of this issue.104 The method has been successfully used for short-echo-time localized spectroscopy (TE = 2.8 ms) in mice and rats.105–107 Localization efficiency of all these sequences can be improved by saturation of the magnetization outside the VOI using a series of slice-selective saturation pulses.
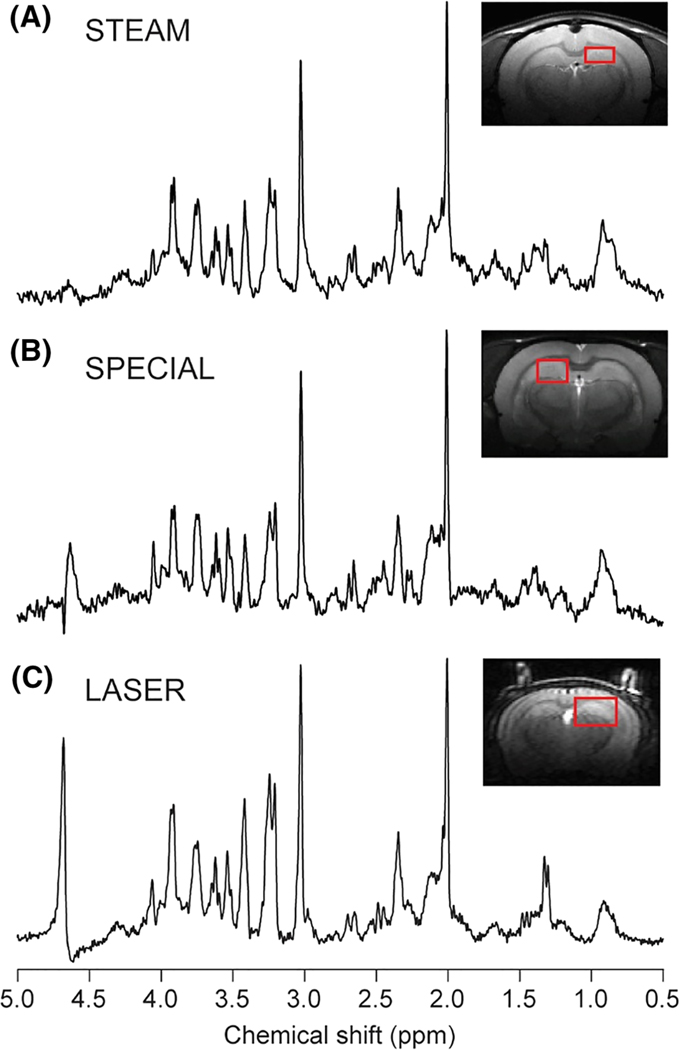
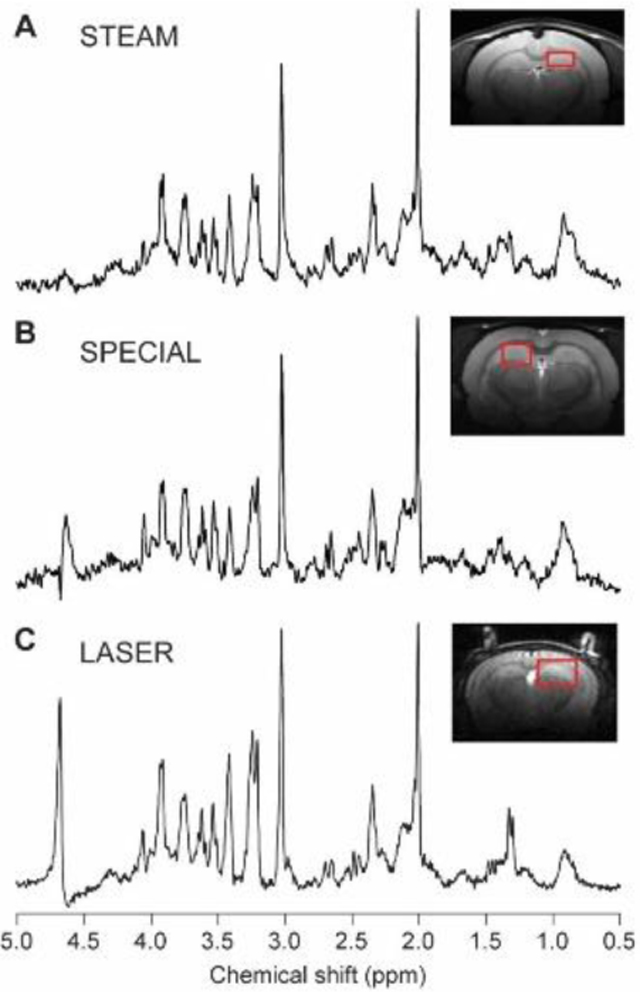
In all pulse sequences, efficient water suppression is important to eliminate the strong water signal, which can overlap with metabolite signals and cause baseline distortion.108 Total elimination of the residual water signal is possible with different methods, e.g. variable pulse power and optimized relaxation delays (VAPOR) water suppression.93 Typical 1H MRS acquisitions with STEAM, SPECIAL and LASER in rodents are presented in Figure 3, while a comparison of the features of those MRS pulse sequences used in preclinical studies is given in Table 3.
FIGURE 3.
Example 1H MR spectra obtained in rodent brains at 9.4 T with STEAM (A), SPECIAL (B) and LASER (C) sequences. A, STEAM spectrum: rat brain, 2.3 × 1.3 × 2.5 mm3 voxel placed in the hippocampus, TR = 5 s, TE = 2 ms, TM = 20 ms, number of averages = 448. Spectrum is shown with Gaussian factor = 0.15. B, SPECIAL spectrum: rat brain, 2 × 2.8 × 2 mm3 voxel placed in the hippocampus, TR = 4, TE = 2.8 ms, number of averages = 160. C, LASER spectrum: mouse brain, 1.7 × 2.25 × 2.25 mm3 voxel placed in hippocampus, TR = 4 s, TE = 27 ms, number of averages = 384. The STEAM spectrum was provided by Ivan Tkáč
TABLE 3.
Comparison of features of 1H MRS localization pulse sequences used in preclinical studies.
| Sequence characteristics | STEAM | SPECIAL | LASER |
|---|---|---|---|
| Fraction of available signal (%) | 50 | 100 | 100 |
| Single-shot method | yes | no | yes |
| Localization performance | ++ | ++ | +++ |
| Sensitivity to B1 inhomogeneity | −− | −− | − |
| Sensitivity to motion | − | −−− | − |
| TE (ms) | 2 | 2.8 | 15–28 |
| CSDE/ppm in 3 directions at 9.4 T | (9, 9, 9%)a | (4, 12, 4%)b | (2.4, 2.4, 2.4%)c |
| Flexibility for spectral editing | +++ | +++ | ++ |
| Requirement of T2 or T1ρ decay knowledge for quantification | no | no | yes |
For this table, the original form of SPECIAL is considered rather than semi-adiabatic form of SPECIAL. STEAM refers to the in-house implementation of the typical vendor provided STEAM sequence with improved features, such as shorter TE, better localization and OVS performance.
The evaluation of the localization performance considers the sequences as currently implemented, including OVS modules for STEAM and SPECIAL. The requirement for B1 max is not very different between sequences because to achieve such short TE for STEAM and SPECIAL very short localization pulses (which require high B1) are used.
Large numbers of + signs indicate positive attributes, e.g. enhanced localization performance.
Large numbers of − signs indicate negative attributes, e.g. increased motion sensitivity.
0.5 ms 90° asymmetric sinc pulses for three directions.
0.5 ms 90° and 180° asymmetric sinc pulses for excitation and refocusing; 2 ms AFP for inversion in the 1D ISIS.
4 ms AHP (non-selective) pulse for excitation and six 1.5 ms AFP pulses for refocusing.
The calibration of the B1 field for the VOI is a prerequisite for achieving excellent performance of MRS sequences with OVS and water suppression, especially at high magnetic fields and using surface coils. Various methods can be used, e.g. adjusting amplitudes of the localization RF pulses for the maximal signal, or B1 mapping methods based on double-angle,109 stimulated echo110 or Bloch-Siegert shift.111 When the VAPOR water suppression scheme is optimized, the amplitudes of the water suppression pulses as well as the last inter-pulse delay can be finely adjusted to minimize the residual water signal.
Similar to human neurochemical profile data, the acquired (i.e. raw) pre-clinical data are handled as follows: (1) data are preprocessed, a procedure sometimes just called “processing”, (e.g. combination of signals from different RF coils, removal of motion corrupted scans, frequency and phase drift correction, combining averages, eddy current correction and, if needed, water peak removal); (2) the intensity of the metabolite signal(s) of interest is often estimated by linear combination model fitting; and (3) the dimensionless signal intensity units are converted to scaled concentration estimates, a process called quantification. For pre-clinical data the quantification is slightly simpler due to the fact that the rodent brain contains mainly gray matter and thus no brain segmentation is performed provided that the MRS voxel is localized in a specific brain region with no cerebrospinal fluid contamination. Moreover, pre-clinical data are often acquired under almost fully relaxed conditions (ultra-short TE and long TR) and thus relaxation corrections are not required. Finally, water or total creatine is usually used as an internal reference. For more details on state-of-the-art processing, analysis and quantification, the reader is referred to the experts’ recommendation article on this topic in this special issue.112 For the analysis of already preprocessed 1H MRS data we recommend the use of a linear combination model fitting, e.g. using a software that allows the decomposition of the spectrum into individual spectra of particular metabolites, using a metabolite basis set such as LCModel,113,114 jMRUI/QUEST115 or others.116 In addition to metabolites, the basis set used should include the experimentally acquired macromolecular spectrum. It has been reported that the macromolecular content and spectral pattern are not different in healthy rodents between the hippocampus, cortex and striatum,117,118 mainly due to the fact that the rodent brain contains mostly grey matter. Thus, assuming a uniform spectral pattern for the macromolecule (MM) spectra is a practical approach when fitting metabolite concentrations. Additionally, the total macromolecular content was shown to change during development,119 with no change in the macromolecular pattern in normal brain. Note that one recent study reported variation in macromolecular patterns during astrocyte reactivity in mice,120 suggesting that group-specific macromolecular spectra might be necessary in some disease applications. More information on the spectrum of MMs can be found in the next consensus paper of this special issue.121
1H MRSI is an approach that is becoming more popular in clinical scans. In rodents, MRSI is not widely applied essentially because of the difficulties related to the small rodent brain, the shimming of large volumes with many tissue interfaces and the limited SNR linked to a large partial volume of muscle, skin and fat in the field of view of the RF coil. 1H MRSI is therefore still challenging to implement in preclinical studies in terms of shim, water suppression artefacts and lipid contamination, particularly long measurement times,122 the quality assessment of a huge number of spectra, absolute quantification, precision and reliability of derived metabolite maps.
5 |. SEQUENCES AND ACQUISITION PROTOCOLS: 31P MRS
31P is the 100% naturally abundant, NMR visible isotope of phosphorus. 31P MRS allows non-invasive measurement of the concentration of phosphorylated metabolites such as adenosine triphosphate (ATP), inorganic phosphate (Pi) or PCr, which are involved in energy metabolism. Furthermore, 31P MRS combined with magnetization/saturation transfer can be used to quantify reaction rates of key metabolic enzymes, such as creatine kinase or ATP synthase.123 In addition, pH can be determined from the chemical shift difference between Pi and PCr, while [Mg2+] can be determined from the chemical shift difference between the α- and β-ATP resonances.124 Hence, 31P MRS has an enormous potential to probe metabolic features that cannot be assessed with other non-invasive techniques and is a complementary technique to 1H MRS and 13C MRS. However, despite the abundance of 31P nuclei in vivo, 31P MRS entails three major hurdles compared with 1H MRS.
The gyromagnetic ratio is low (~2.5 times lower than 1H), thus resulting in intrinsically low sensitivity. Going to higher magnetic fields as generally available for preclinical studies is beneficial to increase 31P MRS sensitivity,125 especially considering the fact that metabolite T1 decreases with the field, possibly due to an increased contribution of chemical shift anisotropy to the relaxation.126
T2 relaxation time constants are short for some important metabolites such as Pi (<80 ms) and ATP (<40 ms),127,128 so signal loss during the echo time can be significant when using conventional single-shot localization sequences (e.g. STEAM, PRESS or LASER).
The frequency range spanned by metabolites is large (~25 ppm, i.e. ~4000 Hz at 9.4 T), thus requiring broadband pulses, in particular to avoid CSD error.
In a clinical context, the last two points are usually circumvented by using MRSI, or no spectroscopic localization at all, e.g. just exploiting the sensitivity profile of a surface coil. These approaches are possible in humans because the contribution from skin, muscle and fat surrounding the brain is small due to their small volume fraction, and because the large brain size allows MRSI with sufficient SNR. However, this can hardly be translated to a preclinical context, because animal brains are smaller, which makes MRSI quite inefficient, and in general surrounded by a significant amount of muscle in rodents and even more in primates. Hence for 31P MRS in a preclinical context it is recommended to use one of the two following localization approaches.
ISIS combines several advantages: it is basically a zero echo time sequence although someT2 relaxation occurs during the RF pulses, thus reducing signal loss; localization can be achieved solely by adiabatic inversion pulses, which ensures efficient inversion throughout the volume even when surface coils are used, provided sufficiently high transmit B1 fields can be reached, thus avoiding signal loss due to incorrect flip angles; and the large bandwidth that can be achieved with adiabatic pulses (with less constraint on maximal transmit B1 than with conventional pulses) reduces CSD error. For excitation, AHP pulses can be used, also alleviating the need for accurate B1 calibration. If conventional pulses are used, B1 calibration should be performed. This can be done for each experiment on the strongest in vivo peak (PCr) provided that the signal is strong enough, or during a separate preliminary experiment on a 31P-phantom (e.g. 100 mM tripolyphosphate in saline). Because it relies on the combination of signals collected over eight-scan cycles, ISIS is unfortunately less robust to motion and drifts than single-shot localization techniques. In addition, optimal acquisition scheme should be used to avoid signal contamination due toT1 smearing.129
In OVS-based localization, the magnetization surrounding the VOI is destroyed by trains of RF pulses and crusher gradients. Because the magnetization within the VOI is (ideally) not perturbed, such localization limits signal loss due to relaxation. Also, as RF pulses used for OVS are not meant to perform large flip angles, their bandwidth is less constrained by transmit B1 than that of 180° pulses, thus reducing CSD. If surface coils are used, OVS can be made largely insensitive to B1 inhomogeneity using BISTRO-type OVS trains.130
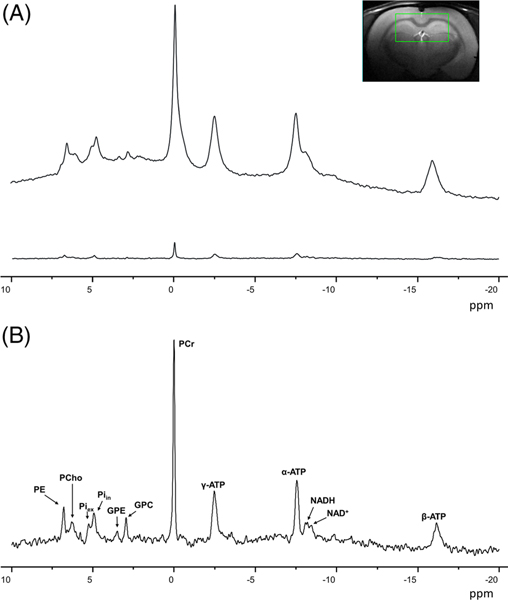
Note that OVS and ISIS can be combined to further improve localization.131,132 In the end, when localization is performed, adequate shimming within the VOI will allow observation of subtle spectral features, such as resolving two Pi resonances at 4.9 and 5.3 ppm,133 presumably corresponding to intracellular and extracellular Pi (see Figure 4). Distinguishing the two Pi resonances in the brain should be considered as a signature of excellent spectral quality (in terms of both shim and SNR). 31P spectra can be processed with fitting algorithms that include prior knowledge, such as AMARES134 or LCModel.135
FIGURE 4.
A, Effect of localization on 31P spectra in the rat brain at 9.4 T. The upper spectrum was acquired in a pulse-acquire experiment (no localization), while the lower spectrum was acquired using an ISIS localization sequence (voxel size 5 × 9 × 9 mm3). Spectra were acquired with TR = 8 s and 512 averages in both cases. A strong signal reduction and baseline flattening are observed. B, Magnification of localized spectrum, with the corresponding metabolites
6 |. SEQUENCES AND ACQUISITION PROTOCOLS: 13C MRS
In contrast to 1H MRS, which is mostly used to measure concentrations of metabolites, 13C MRS allows convenient measurement of metabolic rates in vivo. The low natural abundance of 13C (1.1%) makes it possible to use it as a non-radioactive tracer and to follow incorporation of 13C label into downstream metabolites after injection of a 13C-labeled precursor. The injected 13C-labelled molecules are called tracers for their ability to trace biochemical pathways. Due to the limited sensitivity of in vivo 13C MRS and in order to increase significantly the substrate enrichment in blood, they are typically not injected in trace amounts.
Detection of 13C label can be achieved either using direct 13C detection at 13C frequency, or indirect detection via protons attached to 13C via heteronuclear editing (proton-observed carbon-edited, POCE) (see References 136–139 for reviews). Factors to consider for successful experiments are detailed below.
6.1 |. Pulse sequences
Direct detection yields more biochemical information than indirect detection but requires larger detection volumes due to the lower sensitivity of 13 C detection. Indirect detection is more suitable for small volumes (higher sensitivity), but greater spectral overlap makes detection of certain resonances difficult (e.g. resolved detection of glutamate and glutamine C2, or of glutamate and glutamine C3; see Figure 5).
FIGURE 5.
In vivo 13C spectra in the rat brain after infusion of 70%-enriched [1,6-13C2]glucose and mouse brain after infusion of 70%-enriched [U-13C6]glucose. A-C, Rat. A, RF coil, viewed from the top, consisting of a 1H quadrature surface coil (two loops of 14 mm diameter) and an inner 13C linearly polarized surface coil (12 mm diameter). B, The MRS voxel, shown on axial and sagittal T2 images, was 9 × 5 × 9 mm3 (400 mL). C, Spectrum acquired using a semi-adiabatic DEPT sequence.142 Data were acquired for 1.8 h (2560 averages, TR 2.5 s) starting 1.8 h after the beginning of glucose infusion. D-F, Mouse. D, RF coil, viewed from the top, consisting of a 1H quadrature surface coil (two loops of 13 mm diameter) and an inner 13C linearly polarized surface coil (10 mm diameter). E, Representative coronal and sagittal fast spinecho images of the mouse brain with the VOI for 13C MRS measurement. F, Averaged edited 1H-[13C] MR spectra acquired in the mouse brain during the first hour of [U-13C6] glucose infusion (VOI = 60 μL, 960 averages, TR = 4.0 s)140
For direct detection, most recent preclinical studies have used 1H localization (e.g. ISIS) followed by polarization transfer,82,141–143 which provides better sensitivity and excellent localization with a smaller CSD error than 13C localization and excitation + nuclear Overhauser effect. Proton localization is most often done using 3D-ISIS, because adiabatic inversion pulses provide large bandwidth (small CSD) and B1 insensitivity, and multi-shot localization is not an issue in anesthetized animals (minimal motion). Although most studies have used 3D-ISIS, in principle any 1H localization can be used prior to polarization transfer to 13C.
Polarization transfer, however, cannot be used for carbons with no directly attached protons (e.g. carbonyl/carboxyl carbons). In that case, direct 13C localization must be used (e.g. 3D-ISIS). Polarization transfer also cannot be used to detect metabolites with very short T2 (e.g. glycogen).144,145 For glycogen, localization with well optimized OVS and a short TR is recommended.144,145
For indirect detection, virtually every 1H MRS sequence can be modified for heteronuclear editing by adding a 13C inversion pulse to every other scan. Examples are ACED-STEAM,81 POCE with ISIS,146 POCE-PRESS,74,147 POCE-LASER148 or BISEP-SPECIAL.149 Adiabatic pulses mitigate the effects of inhomogeneous B1 with surface coils. Semi-selective pulses can be used in the 13C channel to separate overlapping resonances.146,150
B1 calibration for RF pulses in the 13C channel is not straightforward, as the low 13C signal in vivo is not sufficient for general routines such as those used for 1H or 31P. Therefore, a pre-calibration experiment for B1 (in the 13C channel) is generally performed with a phantom containing abundant 13C signal. In addition, a sphere containing 99% 13C formic acid is typically placed in the center of the 13C coil to correct for coil B1 efficiency differences between phantom and in vivo conditions due to different sample loading.
6.2 |. Heteronuclear decoupling
Excellent coil design and additional RF filters are necessary to achieve sufficient electrical isolation between 1H and 13C channels and avoid injection of unwanted noise during decoupling. WALTZ-16 is the most commonly used decoupling scheme in preclinical 13C studies. Adiabatic decoupling can be used to further improve performance.151 Unlike the case in humans, power deposition is generally not an issue in preclinical studies.
6.3 |. Data processing
Both direct 13C MRS and indirect 1H-13C MRS spectra should be processed with fitting algorithms that include prior knowledge, such as LCModel or jMRUI/AMARES.152,153
6.4 |. 13C labeled substrates
Most 13C studies in rodents have been performed in the brain and have used [1-13C]glucose or [1,6-13C2]glucose as infused substrate. These substrates generate [3-13C]pyruvate, which is then metabolized in the TCA cycle (primarily in neurons, with a smaller fraction metabolized in astrocytes). Labeling time courses are measured for downstream metabolites such as glutamate C2, C3, C4, glutamine C2, C3, C4 and (if there is sufficient SNR) GABA C2, C3, C4. These time courses are then fitted with metabolic models (see below).
Other commonly used substrates are the following.
[2-13C]acetate or [1,2-13C2]acetate149,154–157: to study glial metabolism (acetate is a glial-specific substrate).
[U-13C6]glucose140: advantageous because it doubles enrichment in downstream metabolites compared with [1-13C]glucose, and is much cheaper than [1,6-13C2]glucose. When using indirect detection (with 13C decoupling), [U-13C6]glucose gives identical results to [1,6-13C2]glucose. When using direct detection, spectra are more complex with [U-13C6]glucose than with [1-13C] or [1,6-13C2]glucose due to labeling of additional carbons (13C-13C couplings).142
[2-13C]glucose: to measure metabolism through pyruvate carboxylase.158
6.5 |. Infusion protocols
13C infusion protocols aim to raise the blood fractional enrichment rapidly (within minutes) from natural abundance (1.1%) to a high enrichment (60% or higher) and keep it elevated for the duration of the measurement. Blood samples are taken at regular intervals to determine the actual fractional enrichment in each animal, which is then used as “input function” in the metabolic model.
6.6 |. Metabolic modeling
Brain metabolic models can be divided into so-called one-compartment models and two (or more)-compartment models (see136,138,139,159–161 for reviews). One-compartment models comprise one (primarily neuronal) TCA cycle rate. More complex two- and three-compartment models allow determination of neuronal-glial metabolic rates such as glial TCA cycle rate, pyruvate carboxylase or glutamate-glutamine cycle.
More recently, models have also been developed to take into account the additional information from 13C-13C isotopomers.162–164
6.7 |. Hyperpolarized 13C
The above section focused on conventional (non-hyperpolarized) 13C MRS. In vivo hyperpolarized 13C MRS is a relatively new technique that dramatically increases the SNR of the starting 13C magnetization, but only for a few minutes until magnetization returns to thermal equilibrium with T1 relaxation. Hyperpolarized 13C allows fast measurement of the initial steps of substrate metabolism. With hyperpolarized 13C, labeled substrates are chosen for their long T1, with 13C label on carbons with no protons attached (e.g. [1-13C]pyruvate). Most conventional localization sequences cannot be used because they destroy the hyperpolarized magnetization after one shot. Most studies thus use fast MRSI sequences. Hyperpolarized 13C is outside the scope of this paper, and we refer the reader to recent reviews.165–167
7 |. GENERAL CONSENSUS AND RECOMMENDATIONS
The anesthesia protocol should be carefully chosen and optimized considering the biological question to be addressed.
It is essential to monitor and record physiological parameters (at least body temperature and respiratory rate) under anesthesia.
Transmit/receive surface coils are recommended to maximize transmit B1 (and thus maximize RF pulse bandwidth) and increase SNR for MRS.
B1 calibration should always be performed in preclinical scanners. For 13C MRS, a precalibration with a phantom is recommended.
1H MRS sequences enabling robust and efficient localization (low CSD, no extracerebral lipid contamination) are recommended. Currently recommended MRS sequences are adiabatic full-intensity sequences (e.g. LASER) for their robustness towards B1 inhomogeneity; when ultra-short echo times are required, SPECIAL or STEAM are recommended. OVS has to be included for advanced ultra-short TE STEAM or SPECIAL sequences.
In relation to the previous point: we recommend CSD not to exceed 10% over the range of metabolites of interest (e.g., for a spectral region from ~1.3 to ~4.3 ppm in the case of 1H MRS, the pulse bandwidth should be ~1000 Hz/T or more).
Efficient water suppression (e.g. using a VAPOR module) should be used for 1H MRS.
Minimal quality standard should be met on 1H spectra: symmetric line shape; linewidth smaller than ~0.05 ppm for singlets (ideally ~0.03 ppm); water residual not much higher than the highest metabolite peak (typically NAA); no lipid contamination from the scalp, no baseline distortions.
For 31P MRS, OVS or ISIS + OVS are recommended.
For 13C MRS, ISIS + adiabatic DEPT is recommended in order to maximize the available biochemical information, if detection efficiency is sufficient (depending on voxel size and depth, final enrichment achieved with the chosen labelled substrate); in the case of low final 13C labelling of the molecules of interest or a small/deep acquired voxel, POCE-LASER or POCE-SPECIAL are recommended for 1H{13C} MRS.
Preclinical MRS data should be quantified using a fitting algorithm that allows for a robust decomposition of the spectrum into a combination of individual metabolite spectra, after careful visual inspection of the acquired spectra with regard to good water suppression, outer-volume signal contamination, SNR and linewidth.
In 1H MRS, MMs should be included as components in the analysis model and should be based on an in vivo acquired MM spectrum with careful inspection and elimination of metabolite residuals.
Supplementary Material
ACKNOWLEDGEMENTS
An initial group of authors had discussed the outline of this paper and most of the authors wrote draft sections. All of the coauthors edited and contributed to the final form of the article. A further group of MRS experts – recruited by personal invitation with expertise in the field of MRS in the rodent brain was collected to support the recommendations as the “Experts’ Working Group on MRS in the rodent brain”. The members of the group are listed in Appendix A. The authors are grateful to Dr Ivan Tkáč for providing part of Figure 3 and for insightful comments and input on Table 3, and to Dr Veronika Rackayová for providing the 31P MR spectra plotted in Figure 4.
The preparation of this manuscript was in part supported by the National Institutes of Health (NIH grants AG062677, NS100620, NS080816, AG063911, KK, BTRC P41 EB027061, P30 NS076408 MM) and by the Swiss National Science Foundation (SNF grant 310030_173222, CC).
Funding information
National Institutes of Health (NIH), Grant/Award Numbers: AG062677 NS100620, NS080816, AG063911, KK BTRC P41 EB027061 P30 NS076408 MM; Swiss National Science Foundation (SNF), Grant/Award Number: 310030_173222, CC
Abbreviations:
- AFP
adiabatic full passage
- AHP
adiabatic half passage
- ATP
adenosine triphosphate
- CNS
central nervous system
- CSD
chemical shift displacement
- GABA
gamma-aminobutyric acid
- ISIS
image-selected in vivo spectroscopy
- LASER
localization by adiabatic selective refocusing
- MM
macromolecule
- MRSI
magnetic resonance spectroscopic imaging
- NAA
N-acetylaspartate
- OVS
outer volume suppression
- PCr
phosphocreatine
- Pi
inorganic phosphate
- POCE
proton-observed carbon-edited
- PRESS
point-resolved spectroscopy
- SNR
signal-tonoise ratio
- SPECIAL
spin echo, full intensity acquired localized spectroscopy
- STEAM
stimulated echo acquisition mode spectroscopy
- TE
echo time
- TM
mixing time
- VAPOR
variable pulse power and optimized relaxation delays water suppression
- VOI
volume of interest
APPENDIX A
In addition to the co-authors of this article, the following researchers, who constitute the Experts’ Working Group on magnetic resonance spectroscopy in the rodent brain, support the consensus paper and the recommendations therein:
Kevin Behar, Department of Psychiatry and Magnetic Resonance Research Center, Yale University School of Medicine, New Haven, USA;
Fawzi Boumezbeur, Commissariat à l’Energie Atomique et aux Energies Alternatives, NeuroSpin, Gif-sur-Yvette, France; Centre National de la Recherche Scientifique, Université Paris-Sud, Université Paris-Saclay, BAOBAB, Gif-sur-Yvette, France;
Dinesh Kumar Deelchand, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, USA;
Wolfgang Dreher, Department of Chemistry, In vivo-MR Group, University Bremen, Bremen, Germany;
Brenda A. Klaunberg, National Institutes of Health Mouse Imaging Facility, Bethesda, MD, USA;
Clemence Ligneul, Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom;
Diana M. Lindquist, Cincinnati Children’s Hospital Medical Center, Imaging Research Center, University of Cincinnati Dept. of Radiology, Cincinnati, OH, USA;
Jamie Near, Douglas Mental Health University Institute and Department of Psychiatry, McGill University, Montreal, QC, Canada;
Gülin Öz, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, USA;
IvanTkáč, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, USA;
Steve R. Williams, Centre for Imaging Science and Manchester Academic Health Sciences Centre, University of Manchester, Manchester, UK
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Cudalbu C. In vivo studies of brain metabolism in animal models of hepatic encephalopathy using 1H magnetic resonance spectroscopy. Metab Brain Dis. 2013; 28:167–174. 10.1007/s11011-012-9368-9 [DOI] [PubMed] [Google Scholar]
- 2.Oz G, Alger JR, Barker PB, et al. Clinical proton MR spectroscopy in central nervous system disorders. Radiology. 2014;270(3):658–679. 10.1148/radiol.13130531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rackayova V, Cudalbu C, Pouwels PJW, Braissant O. Creatine in the central nervous system: from magnetic resonance spectroscopy to creatine deficiencies. Anal Biochem. 2017;529:144–157. 10.1016/j.ab.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 4.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. [DOI] [PubMed] [Google Scholar]
- 5.Duarte JM, Lei H, Mlynarik V, Gruetter R. The neurochemical profile quantified by in vivo 1H NMR spectroscopy. NeuroImage. 2012;61:342–362. 10.1016/j.neuroimage.2011.12.038 [DOI] [PubMed] [Google Scholar]
- 6.Braissant O, Rackayová V, Pierzchala K, Grosse J, McLin VA, Cudalbu C. Longitudinal neurometabolic changes in the hippocampus of a rat model of chronic hepatic encephalopathy. J Hepatol. 2019;71(3):505–515. 10.1016/j.jhep.2019.05.022 [DOI] [PubMed] [Google Scholar]
- 7.Lanz B, Rackayova V, Braissant O, Cudalbu C. MRS studies of neuroenergetics and glutamate/glutamine exchange in rats: extensions to hyper-ammonemic models. Anal Biochem. 2017;529:245–269. 10.1016/j.ab.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 8.Lai M, Gruetter R, Lanz B. Progress towards in vivo brain 13C-MRS in mice: metabolic flux analysis in small tissue volumes. Anal Biochem. 2017;529: 229–244. 10.1016/j.ab.2017.01.019 [DOI] [PubMed] [Google Scholar]
- 9.Valette J, Tiret B, Boumezbeur F. Experimental strategies for in vivo C-13 NMR spectroscopy. Anal Biochem. 2017;529:216–228. 10.1016/j.ab.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 10.Kjell J, Olson L. Rat models of spinal cord injury: from pathology to potential therapies. Dis Model Mech. 2016;9:1125–1137. 10.1242/dmm.025833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. 10.1016/0092-8674(87)90646-5 [DOI] [PubMed] [Google Scholar]
- 12.Zan Y, Haag JD, Chen KS, et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21(6): 645–651. 10.1038/nbt830 [DOI] [PubMed] [Google Scholar]
- 13.Geurts AM, Cost GJ, Rémy S, et al. Generation of gene-specific mutated rats using zinc-finger nucleases. Methods Mol Biol. 2010;597:211–225. 10.1007/978-1-60327-389-3_15 [DOI] [PubMed] [Google Scholar]
- 14.Tong C, Huang G, Ashton C, Li P, Ying QL. Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc. 2011; 6:827–844. 10.1038/nprot.2011.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao Y, Guan Y, Wang L, et al. CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc. 2014;9(10):2493–2512. 10.1038/nprot.2014.171 [DOI] [PubMed] [Google Scholar]
- 16.Francis C, Natarajan S, Lee MT, et al. Divergence of RNA localization between rat and mouse neurons reveals the potential for rapid brain evolution. BMC Genomics. 2014;15(1):883. 10.1186/1471-2164-15-883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellenbroek B, Youn J. Rodent models in neuroscience research: is it a rat race? Dis Model Mech. 2016;9:1079–1087. 10.1242/dmm.026120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfensohn S, Lloyd M. Handbook of Laboratory Animal Management and Welfare. 3rd ed. Chichester, UK: Wiley; 2003;1–18. [Google Scholar]
- 19.Tremoleda JL, Kerton A, Gsell W. Anaesthesia and physiological monitoring during in vivo imaging of laboratory rodents: considerations on experimental outcomes and animal welfare. EJNMMI Res. 2012;2:44-44. 10.1186/2191-219X-2-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukasik VM, Gillies RJ. Animal anaesthesia for in vivo magnetic resonance. NMR Biomed. 2003;16:459–467. 10.1002/nbm.836 [DOI] [PubMed] [Google Scholar]
- 21.Makaryus R, Lee H, Yu M, et al. The metabolomic profile during isoflurane anesthesia differs from propofol anesthesia in the live rodent brain. J Cereb Blood Flow Metab. 2011;31(6):1432–1442. 10.1038/jcbfm.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gargiulo S, Greco A, Gramanzini M, et al. Mice anesthesia, analgesia, and care, part I: anesthetic considerations in preclinical research. ILAR J. 2012; 53(1):E55–E69. 10.1093/ilar.53.1.55 [DOI] [PubMed] [Google Scholar]
- 23.Boretius S, Tammer R, Michaelis T, Brockmöller J, Frahm J. Halogenated volatile anesthetics alter brain metabolism as revealed by proton magnetic resonance spectroscopy of mice in vivo. NeuroImage. 2013;69:244–255. 10.1016/j.neuroimage.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 24.Lei H, Duarte JMN, Mlynarik V, Python A, Gruetter R. Deep thiopental anesthesia alters steady-state glucose homeostasis but not the neurochemical profile of rat cortex. J Neurosci Res. 2010;88:413–419. 10.1002/jnr.22212 [DOI] [PubMed] [Google Scholar]
- 25.Du F, Zhang Y, Iltis I, et al. In vivo proton MRS to quantify anesthetic effects of pentobarbital on cerebral metabolism and brain activity in rat. Magn Reson Med. 2009;62(6):1385–1393. 10.1002/mrm.22146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi IY, Lei H, Gruetter R. Effect of deep pentobarbital anesthesia on neurotransmitter metabolism in vivo: on the correlation of total glucose consumption with glutamatergic action. J Cereb Blood Flow Metab. 2002;22:1343–1351. 10.1097/01.WCB.0000040945.89393.46 [DOI] [PubMed] [Google Scholar]
- 27.Gaertner D, Hallman TM, Hankenson FC, Batchelder MA. Anesthesia and analgesia for laboratory rodents. In: Fish R, Danneman PJ, Brown M, Karas A, eds. Anesthesia and Analgesia in Laboratory Animals. London, UK: Academic Press; 2008:239–297. [Google Scholar]
- 28.Chowdhury GMI, Behar KL, Cho W, Thomas MA, Rothman DL, Sanacora G. 1H-[13C]-nuclear magnetic resonance spectroscopy measures of ketamine’s effect on amino acid neurotransmitter metabolism. Biol Psychiatry. 2012;71(11):1022–1025. 10.1016/j.biopsych.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Pelt LF. Ketamine and xylazine for surgical anesthesia in rats. J am Vet Med Assoc. 1977;171:842–844. [PubMed] [Google Scholar]
- 30.Bannova AV, Akulov AE, Menshanov PN, Dygalo NN. Estimation of an area between the baseline and the effect curve parameter for lactate levels in the hippocampi of neonatal rats during anesthesia. J Pharmaceut Biomed. 2018;150:327–332. 10.1016/j.jpba.2017.12.021 [DOI] [PubMed] [Google Scholar]
- 31.Gonsenhauser I, Wilson CG, Han F, Strohl KP, Dick TE. Strain differences in murine ventilatory behavior persist after urethane anesthesia. J Appl Physiol. 2004;97:888–894. 10.1152/japplphysiol.01346.2003 [DOI] [PubMed] [Google Scholar]
- 32.Gaertner DH, Hallman TM, Hankenson FC, Batchelder MA. Anesthesia and analgesia for laboratory rodents. In: Fish R, Danneman PJ, Brown M, Karas A, eds. Anesthesia and Analgesia in Laboratory Animals. London, UK: Academic Press; 2008:239–297. [Google Scholar]
- 33.Hildebrandt IJ, Su H, Weber WA. Anesthesia and other considerations for in vivo imaging of small animals. ILAR J. 2008;49:17–26. [DOI] [PubMed] [Google Scholar]
- 34.Bresnen A, Duong TQ. Brain high-energy phosphates and creatine kinase synthesis rate under graded isoflurane anesthesia: an in vivo 31P magnetization transfer study at 11.7 tesla. Magn Reson Med. 2015;73:726–730. 10.1002/mrm.25136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obernier JA, Baldwin RL. Establishing an appropriate period of acclimatization following transportation of laboratory animals. ILAR J. 2006;47:364–369. 10.1093/ilar.47.4.364 [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Abdallah CG, Chen Y, et al. Behavioral deficits, abnormal corticosterone, and reduced prefrontal metabolites of adolescent rats subject to early life stress. Neurosci Lett. 2013;545:132–137. 10.1016/j.neulet.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magalh~aes R, Novais A, Barrière DA, et al. A resting-state functional MR imaging and spectroscopy study of the dorsal hippocampus in the chronic unpredictable stress rat model. J Neurosci. 2019;39(19):3640–3650. 10.1523/jneurosci.2192-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovel DP. Variation in pentobarbitone sleeping time in mice 1. Strain and sex differences. Lab Anim. 1986;20:85–90. 10.1258/002367786780865142 [DOI] [PubMed] [Google Scholar]
- 39.Lovel DP. Variation in pentobarbitone sleeping time in mice 2. Variables affecting test results. Lab Anim. 1986;20:91–96. 10.1258/002367786780865089 [DOI] [PubMed] [Google Scholar]
- 40.Bogaert MJVV, Groenink L, Oosting RS, Westphal KGC, van der Gugten J, Olivier B. Mouse strain differences in autonomic responses to stress. Genes Brain Behav. 2006;5(2):139–149. 10.1111/j.1601-183X.2005.00143.x [DOI] [PubMed] [Google Scholar]
- 41.Schwarcz A, Natt O, Watanabe T, Boretius S, Frahm J, Michaelis T. Localized proton MRS of cerebral metabolite profiles in different mouse strains. Magn Reson Med. 2003;49(5):822–827. 10.1002/mrm.10445 [DOI] [PubMed] [Google Scholar]
- 42.Roy U, Stute L, Höfling C, et al. Sex- and age-specific modulation of brain GABA levels in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2018;62:168–179. 10.1016/j.neurobiolaging.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 43.van Duijn S, Nabuurs RJA, van Duinen SG, Natté R, van Buchem MA, Alia A. Longitudinal monitoring of sex-related in vivo metabolic changes in the brain of Alzheimer’s disease transgenic mouse using magnetic resonance spectroscopy. J Alzheimer’s Dis. 2013;34(4):1051–1059. 10.3233/JAD-122188 [DOI] [PubMed] [Google Scholar]
- 44.Hjelmervik H, Hausmann M, Craven AR, Hirnstein M, Hugdahl K, Specht K. Sex- and sex hormone-related variations in energy-metabolic frontal brain asymmetries: a magnetic resonance spectroscopy study. NeuroImage. 172, 2018;817–825. 10.1016/j.neuroimage.2018.01.043 [DOI] [PubMed] [Google Scholar]
- 45.Zambricki EA, D’Alecy LG. Rat sex differences in anesthesia. Comp Med. 2004;54:49–53. [PubMed] [Google Scholar]
- 46.Waterman AE, Livingston A. Effects of age and sex on ketamine anaesthesia in the rat. Br J Anaesth. 1978;50:885–889. 10.1093/bja/50.9.885 [DOI] [PubMed] [Google Scholar]
- 47.O’Connor CA, Cernak I, Vink R. Interaction between anesthesia, gender, and functional outcome task following diffuse traumatic brain injury in rats. J Neurotrauma. 2003;20:533–541. 10.1089/089771503767168465 [DOI] [PubMed] [Google Scholar]
- 48.Czerniak R. Gender-based differences in pharmacokinetics in laboratory animal models. Int J Toxicol. 2001;20:161–163. 10.1080/109158101317097746 [DOI] [PubMed] [Google Scholar]
- 49.Aschoff J. Circadian timing. Ann N Y Acad Sci. 1984;423:442–468. [DOI] [PubMed] [Google Scholar]
- 50.Liachenko S, Ramu J. 1H-MRS changes in the rat brain due to circadian cycle. Poster presented at: ISMRM 20th Annual Meeting & Exhibition; May 8, 2012; Melbourne, Australia; 1826. [Google Scholar]
- 51.Duarte JM, Do KQ, Gruetter R. Longitudinal neurochemical modifications in the aging mouse brain measured in vivo by 1H magnetic resonance spectroscopy. Neurobiol Aging. 2014;35:1660–1668. 10.1016/j.neurobiolaging.2014.01.135 [DOI] [PubMed] [Google Scholar]
- 52.Marjanska M, Curran GL, Wengenack TM, et al. Monitoring disease progression in transgenic mouse models of Alzheimer’s disease with proton magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 2005;102(33):11906–11910. 10.1073/pnas.0505513102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oz G, Nelson CD, Koski DM, et al. Noninvasive detection of presymptomatic and progressive neurodegeneration in a mouse model of spinocerebellar ataxia type 1. J Neurosci. 2010;30(10):3831–3838. 10.1523/JNEUROSCI.5612-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albrecht M, Henke J, Tacke S, Markert M, Guth B. Influence of repeated anaesthesia on physiological parameters in male Wistar rats: a telemetric study about isoflurane, ketamine-xylazine and a combination of medetomidine, midazolam and fentanyl. BMC Vet Res. 2014;10:310–310. 10.1186/s12917-014-0310-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furtado KS, Andrade FO. Comparison of the beneficial and adverse effects of inhaled and injectable anaesthetics: a mini-review. OA Anaesthetics. 2013;1(2):20. [Google Scholar]
- 56.Horn T, Klein J. Lactate levels in the brain are elevated upon exposure to volatile anesthetics: a microdialysis study. Neurochem Int. 2010;57:940–947. 10.1016/j.neuint.2010.09.014 [DOI] [PubMed] [Google Scholar]
- 57.Ewald AJ, Werb Z, Egeblad M. Monitoring of vital signs for long-term survival of mice under anesthesia. Cold Spring Harb Protoc. 2011;2011:174–178. 10.1101/pdb.prot5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tremoleda JL, Macholl S, Sosabowski JK. Anesthesia and monitoring of animals during MRI Studies. Methods Mol Biol. 2018;1718:423–439. 10.1007/978-1-4939-7531-0_25 [DOI] [PubMed] [Google Scholar]
- 59.Council NR. Guide for the Care and Use of Laboratory Animals. 8th ed. National Academies Press; 2011. [PubMed] [Google Scholar]
- 60.Xu S, Ji Y, Chen X, Yang Y, Gullapalli RP, Masri R. In vivo high-resolution localized 1H MR spectroscopy in the awake rat brain at 7 T. Magn Reson Med. 2013;69(4):937–943. 10.1002/mrm.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Low LA, Bauer LC, Pitcher MH, Bushnell MC. Restraint training for awake functional brain scanning of rodents can cause long-lasting changes in pain and stress responses. Pain. 2016;157:1761–1772. 10.1097/j.pain.0000000000000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stenroos P, Paasonen J, Salo RA, et al. Awake rat brain functional magnetic resonance imaging using standard radio frequency coils and a 3D printed restraint kit. Front Neurosci. 2018;12:1–14. 10.3389/fnins.2018.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madularu D, Mathieu AP, Kumaragamage C, et al. A non-invasive restraining system for awake mouse imaging. J Neurosci Methods. 2017;287:53–57. 10.1016/j.jneumeth.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang P-C, Procissi D, Bao Q, Centeno MV, Baria A, Apkarian AV. Novel method for functional brain imaging in awake minimally restrained rats. J Neurophysiol. 2016;116(1):61–80. 10.1152/jn.01078.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becerra L, Chang PC, Bishop J, Borsook D. CNS activation maps in awake rats exposed to thermal stimuli to the dorsum of the hindpaw. NeuroImage. 2011;54:1355–1366. 10.1016/j.neuroimage.2010.08.056 [DOI] [PubMed] [Google Scholar]
- 66.Tkac I, Henry P-G, Andersen P, Keene CD, Low WC, Gruetter R. Highly resolved in vivo 1H NMR spectroscopy of the mouse brain at 9.4 T. Magn Reson Med. 2004;52(3):478–484. 10.1002/mrm.20184 [DOI] [PubMed] [Google Scholar]
- 67.Lei H, Poitry-Yamate C, Preitner F, Thorens B, Gruetter R. Neurochemical profile of the mouse hypothalamus using in vivo 1H MRS at 14.1T. NMR Biomed. 2010;23:578–583. 10.1002/nbm.1498 [DOI] [PubMed] [Google Scholar]
- 68.Juchem C. B0 shimming techniques: experts’ consensus recommendations. NMR Biomed. 2019. [DOI] [PubMed]
- 69.Automatic Gruetter R., localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993;29:804–811. [DOI] [PubMed] [Google Scholar]
- 70.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43:319–323. [DOI] [PubMed] [Google Scholar]
- 71.Miyasaka N, Takahashi K, Hetherington HP. Fully automated shim mapping method for spectroscopic imaging of the mouse brain at 9.4 T. Magn Reson Med. 2006;55:198–202. 10.1002/mrm.20731 [DOI] [PubMed] [Google Scholar]
- 72.Kreis R, Boer V, Choi I–Y, et al. Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: background and experts’ consensus recommendations. NMR Biomed. 2019. [DOI] [PMC free article] [PubMed]
- 73.Lim H, Thind K, Martinez-Santiesteban FM, Scholl TJ. Construction and evaluation of a switch-tuned 13C-1H birdcage radiofrequency coil for imaging the metabolism of hyperpolarized 13C-enriched compounds. J Magn Reson Imaging. 2014;40:1082–1090. 10.1002/jmri.24458 [DOI] [PubMed] [Google Scholar]
- 74.Kumaragamage C, Madularu D, Mathieu AP, de Feyter H, Rajah MN, Near J. In vivo proton observed carbon edited (POCE) 13C magnetic resonance spectroscopy of the rat brain using a volumetric transmitter and receive-only surface coil on the proton channel. Magn Reson Med. 2018;79(2):628635. 10.1002/mrm.26751 [DOI] [PubMed] [Google Scholar]
- 75.Rizzo F, Abaei A, Nespoli E, et al. Aripiprazole and Riluzole treatment alters behavior and neurometabolites in young ADHD rats: a longitudinal 1H-NMR spectroscopy study at 11.7T. Transl Psychiatry. 2017;7(8):1–8. e1189. 10.1038/tp.2017.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rizzo F, Nespoli E, Abaei A, et al. Aripiprazole selectively reduces motor tics in a young animal model for Tourette’s syndrome and comorbid attention deficit and hyperactivity disorder. Front Neurol. 2018;9:1–11. 10.3389/fneur.2018.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adriany G, Gruetter R. A half-volume coil for efficient proton decoupling in humans at 4 tesla. J Magn Reson. 1997;125:178–184. 10.1006/jmre.1997.1113 [DOI] [PubMed] [Google Scholar]
- 78.Chen W, Adriany G, Zhu XH, Gruetter R, Ugurbil K. Detecting natural abundance carbon signal of NAA metabolite within 12-cm3 localized volume of human brain using 1H-{13C} NMR spectroscopy. Magn Reson Med. 1998;40:180–184. [DOI] [PubMed] [Google Scholar]
- 79.de Graaf RA, Mason GF, Patel AB, Behar KL, Rothman DL. In vivo 1H-[13C]-NMR spectroscopy of cerebral metabolism. NMR Biomed. 2003;16:339–357. 10.1002/nbm.847 [DOI] [PubMed] [Google Scholar]
- 80.Lizarbe B, Lei H, Duarte JMN, Lanz B, Cherix A, Gruetter R. Feasibility of in vivo measurement of glucose metabolism in the mouse hypothalamus by 1H-[13C] MRS at 14.1T. Magn Reson Med. 2018;80(3):874–884. 10.1002/mrm.27129 [DOI] [PubMed] [Google Scholar]
- 81.Pfeuffer J, Tkác I, Choi IY, et al. Localized in vivo 1H NMR detection of neurotransmitter labeling in rat brain during infusion of [1-13C] D-glucose. Magn Reson Med. 1999;41(6):1077–1083. [DOI] [PubMed] [Google Scholar]
- 82.Lai M, Lanz B, Poitry-Yamate C, et al. In vivo 13C MRS in the mouse brain at 14.1 Tesla and metabolic flux quantification under infusion of [1,6-(3C2] glucose. J Cereb Blood Flow Metab. 2018;38(10):1701–1714. 10.1177/0271678X17734101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seres Roig E, Magill AW, Donati G, et al. A double-quadrature radiofrequency coil design for proton-decoupled carbon-13 magnetic resonance spectroscopy in humans at 7T. Magn Reson Med. 2015;73(2):894–900. 10.1002/mrm.25171 [DOI] [PubMed] [Google Scholar]
- 84.Klomp DW, Renema WKJ, van der Graaf M, de Galan BE, Kentgens APM, Heerschap A. Sensitivity-enhanced 13C MR spectroscopy of the human brain at 3 Tesla. Magn Reson Med. 2006;55(2):271–278. 10.1002/mrm.20745 [DOI] [PubMed] [Google Scholar]
- 85.Kovacs H, Moskau D, Spraul M. Cryogenically cooled probes—a leap in NMR technology. Prog Nucl Magn Reson Spectrosc. 2005;46:131–155. 10.1016/j.pnmrs.2005.03.001 [DOI] [Google Scholar]
- 86.Baltes C, Radzwill N, Bosshard S, Marek D, Rudin M. Micro MRI of the mouse brain using a novel 400 MHz cryogenic quadrature RF probe. NMR Biomed. 2009;22:834–842. 10.1002/nbm.1396 [DOI] [PubMed] [Google Scholar]
- 87.Mispelter J, Lupu M, Briguet A. NMR Probeheads for Biophysical and Biomedical Experiments: Theoretical Principles & Practical Guidelines. London, UK: Imperial College Press; 2006. [Google Scholar]
- 88.Junge S. Cryogenic and superconducting coils for MRI. eMagRes. 2012;2007:505–514. 10.1002/9780470034590.emrstm1162 [DOI] [Google Scholar]
- 89.Harris AD, Saleh MG, Edden RA. Edited 1H magnetic resonance spectroscopy in vivo: methods and metabolites. Magn Reson Med. 2017;77:1377–1389. 10.1002/mrm.26619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi IY et al. Spectral editing in 1H magnetic resonance spectroscopy: experts’ consensus recommendations. NMR Biomed. 2020. [DOI] [PMC free article] [PubMed]
- 91.Cudalbu C, Mlynarik V, Xin L, Gruetter R. Comparison of T1 relaxation times of the neurochemical profile in rat brain at 9.4 tesla and 14.1 tesla. Magn Reson Med. 2009;62:862–867. 10.1002/mrm.22022 [DOI] [PubMed] [Google Scholar]
- 92.Frahm J, Merboldt KD, Hanicke W. Localized proton spectroscopy using stimulated echoes. J Magn Reson. 1987;72:502–508. 10.1016/0022-2364(87)90154-5 [DOI] [PubMed] [Google Scholar]
- 93.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41:649–656. [DOI] [PubMed] [Google Scholar]
- 94.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. [DOI] [PubMed] [Google Scholar]
- 95.Slotboom J, Mehlkopf AF, Bovée WMMJ. A single-shot localization pulse sequence suited for coils with inhomogeneous RF fields using adiabatic slice-selective RF pulses. J Magn Reson. 1991;95:396–404. 10.1016/0022-2364(91)90229-M [DOI] [Google Scholar]
- 96.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153:155–177. 10.1006/jmre.2001.2340 [DOI] [PubMed] [Google Scholar]
- 97.Santin MD, Valabrègue R, Rivals I, et al. In vivo 1H MRS study in microlitre voxels in the hippocampus of a mouse model of Down syndrome at 11.7 T. NMR Biomed. 2014;27(10):1143–1150. 10.1002/nbm.3155 [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Kolkovsky AL, Meriaux S, Boumezbeur F. Metabolite and macromolecule T1 and T2 relaxation times in the rat brain in vivo at 17.2T. Magn Reson Med. 2016;75:503–514. 10.1002/mrm.25602 [DOI] [PubMed] [Google Scholar]
- 99.Deelchand DK, Henry PG, Marjanska M. Effect of Carr-Purcell refocusing pulse trains on transverse relaxation times of metabolites in rat brain at 9.4 Tesla. Magn Reson Med. 2015;73:13–20. 10.1002/mrm.25088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Michaeli S, Garwood M, Zhu XH, et al. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and CarrP-urcell spin echoes at 4T and 7T. Magn Reson Med. 2002;47(4):629–633. 10.1002/mrm.10135 [DOI] [PubMed] [Google Scholar]
- 101.Deelchand DK, Auerbach EJ, Marjanska M. Properties of localization by adiabatic refocusing (LASER) sequence. Paper presented at: ISMRM 25th Annual Meeting & Exhibition; Honululu, HI; 2017;5585. [Google Scholar]
- 102.Ordidge RJ, Connelly A, Lohman JAB. Image-selected in vivo spectroscopy (ISIS). A new technique for spatially selective NMR spectroscopy. J Magn Reson. 1986;66:283–294. 10.1016/0022-2364(86)90031-4 [DOI] [Google Scholar]
- 103.Mlynarik V, Gambarota G, Frenkel H, Gruetter R. Localized short-echo-time proton MR spectroscopy with full signal-intensity acquisition. Magn Reson Med. 2006;56:965–970. 10.1002/mrm.21043 [DOI] [PubMed] [Google Scholar]
- 104.Oz G, Deelchand DK, Wijnen JP, et al. Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: experts’ consensus recommendations. NMR Biomed. 2020;e4236. 10.1002/nbm.4236 [DOI] [PMC free article] [PubMed]
- 105.Dhamala E, Abdelkefi I, Nguyen M, Hennessy TJ, Nadeau H, Near J. Validation of in vivo MRS measures of metabolite concentrations in the human brain. NMR Biomed. 2019;32(3):1–15, e4058. 10.1002/nbm.4058 [DOI] [PubMed] [Google Scholar]
- 106.Heo H, Kim S, Lee HH, et al. On the utility of short echo time (TE) single voxel 1H-MRS in non-invasive detection of 2-hydroxyglutarate (2HG); challenges and potential improvement illustrated with animal models using MRUI and LCModel. PloS ONE. 2016;11(1):1–18, e0147794. 10.1371/journal.pone.0147794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duarte JMN, Skoug C, Silva HB, Carvalho RA, Gruetter R, Cunha RA. Impact of caffeine consumption on type 2 diabetes-induced spatial memory impairment and neurochemical alterations in the hippocampus. Front Neurosci. 2018;12(1015):1–15. 10.3389/fnins.2018.01015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tkac I et al. Water and lipid suppression techniques for advanced 1H MRS and MRSI: experts’ consensus recommendations. NMR Biomed. 2020. [DOI] [PMC free article] [PubMed]
- 109.Insko EK, Bolinger L. Mapping of the radiofrequency field. J Magn Reson A. 1993;103:82–85. 10.1006/jmra.1993.1133 [DOI] [Google Scholar]
- 110.Helms G, Finsterbusch J, Weiskopf N, Dechent P. Rapid radiofrequency field mapping in vivo using single-shot STEAM MRI. Magn Reson Med. 2008; 60:739–743. 10.1002/mrm.21676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sacolick LI, Wiesinger F, Hancu I, Vogel MW. B1 mapping by Bloch-Siegert shift. Magn Reson Med. 2010;63:1315–1322. 10.1002/mrm.22357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts’ consensus recommendations. NMR Biomed. 2020;e4257. 10.1002/nbm.4257 [DOI] [PMC free article] [PubMed]
- 113.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. 10.1002/mrm.1910300604 [DOI] [PubMed] [Google Scholar]
- 114.Provencher SW. Automatic quantitation of localized in vivo H-1 spectra with LCModel. NMR Biomed. 2001;14:260–264. 10.1002/Nbm.698 [DOI] [PubMed] [Google Scholar]
- 115.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31:269–286. 10.1016/S0010-4825(01)00006-3 [DOI] [PubMed] [Google Scholar]
- 116.Bottomley PA, Griffiths JR. Handbook of Magnetic Resonance Spectroscopy In Vivo: MRS Theory, Practice and Applications. Chichester, UK:Wiley; 2016. [Google Scholar]
- 117.Craveiro M, Clement-Schatlo V, Marino D, Gruetter R, Cudalbu C. In vivo brain macromolecule signals in healthy and glioblastoma mouse models: 1H magnetic resonance spectroscopy, post-processing and metabolite quantification at 14.1 T. J Neurochem. 2014;129:806–815. 10.1111/jnc.12673 [DOI] [PubMed] [Google Scholar]
- 118.Xin L, Mlynarik V, Lei H, Gruetter R. Influence of regional macromolecule baseline on the quantification of neurochemical profile in rat brain. Proc Int Soc Magn Reson Med. 2010;18:321. [Google Scholar]
- 119.Tkac I, Rao R, Georgieff MK, Gruetter R. Developmental and regional changes in the neurochemical profile of the rat brain determined by in vivo H-1 NMR spectroscopy. Magn Reson Med. 2003;50:24–32. 10.1002/mrm.10497 [DOI] [PubMed] [Google Scholar]
- 120.Ligneul C, Palombo M, Hernández-Garzón E, et al. Diffusion-weighted magnetic resonance spectroscopy enables cell-specific monitoring of astrocyte reactivity in vivo. NeuroImage. 2019;191:457–469. 10.1016/j.neuroimage.2019.02.046 [DOI] [PubMed] [Google Scholar]
- 121.Cudalbu C. Contribution of macromolecules to magnetic resonance spectra: experts’ consensus recommendations. NMR Biomed. 2019. [DOI] [PMC free article] [PubMed]
- 122.Mlynarik V, Kohler I, Gambarota G, Vaslin A, Clarke PGH, Gruetter R. Quantitative proton spectroscopic imaging of the neurochemical profile in rat brain with microliter resolution at ultra-short echo times. Magn Reson Med. 2008;59(1):52–58. 10.1002/mrm.21447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Du F, Zhu XH, Qiao HY, Zhang XL, Chen W. Efficient in vivo P-31 magnetization transfer approach for noninvasively determining multiple kinetic parameters and metabolic fluxes of ATP metabolism in the human brain. Magn Reson Med. 2007;57:103–114. 10.1002/mrm.21107 [DOI] [PubMed] [Google Scholar]
- 124.De Graaf RA. In Vivo NMR Spectroscopy: Principles and Techniques. 2nd ed. Chichester, UK: Wiley; 2007. [Google Scholar]
- 125.Lu M, Chen W, Zhu XH. Field dependence study of in vivo brain P-31 MRS up to 16.4 T. NMR Biomed. 2014;27:1135–1141. 10.1002/nbm.3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Evelhoch JL, Ewy CS, Siegfried BA, Ackerman JJH, Rice DW, Briggs RW. 31P spin-lattice relaxation times and resonance linewidths of rat tissue in vivo: dependence upon the static magnetic field strength. Magn Reson Med. 1985;2(4):410–417. [DOI] [PubMed] [Google Scholar]
- 127.Remy C, Albrand JP, Benabid AL, et al. In vivo 31P nuclear magnetic resonance studies of T1 and T2 relaxation times in rat brain and in rat brain tumors implanted to nude mice. Magn Reson Med. 1987;4(2):144–152. [DOI] [PubMed] [Google Scholar]
- 128.van der Kemp WJM, Klomp DWJ, Wijnen JP. 31P T2s of phosphomonoesters, phosphodiesters, and inorganic phosphate in the human brain at 7T. Magn Reson Med. 2018;80:29–35. 10.1002/mrm.27026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Burger C, Buchli R, Mckinnon G, Meier D, Boesiger P. The impact of the ISIS experiment order on spatial contamination. Magn Reson Med. 1992;26: 218–230. 10.1002/mrm.1910260204 [DOI] [PubMed] [Google Scholar]
- 130.de Graaf RA, Luo Y, Garwood M, Nicolay K. B1-insensitive, single-shot localization and water suppression. J Magn Reson B. 1996;113:35–45. [DOI] [PubMed] [Google Scholar]
- 131.Connelly A, Counsell C, Lohman JAB, Ordidge RJ. Outer volume suppressed image related in vivo spectroscopy (OSIRIS), a high-sensitivity localization technique. J Magn Reson. 1988;78:519–525. 10.1016/0022-2364(88)90136-9 [DOI] [Google Scholar]
- 132.Rackayova V, Braissant O, McLin VA, Berset C, Lanz B, Cudalbu C. 1H and 31P magnetic resonance spectroscopy in a rat model of chronic hepatic encephalopathy: in vivo longitudinal measurements of brain energy metabolism. Metab Brain Dis. 2016;31(6):1303–1314. 10.1007/s11011-015-9715-8 [DOI] [PubMed] [Google Scholar]
- 133.Tiret B, Brouillet E, Valette J. Evidence for a “metabolically inactive” inorganic phosphate pool in adenosine triphosphate synthase reaction using localized 31P saturation transfer magnetic resonance spectroscopy in the rat brain at 11.7 T. J Cereb Blood Flow Metab. 2016;36:1513–1518. 10.1177/0271678X16657095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. 10.1006/jmre.1997.1244 [DOI] [PubMed] [Google Scholar]
- 135.Deelchand DK, Nguyen TM, Zhu XH, Mochel F, Henry PG. Quantification of in vivo P-31 NMR brain spectra using LCModel. NMR Biomed. 2015;28: 633–641. 10.1002/nbm.3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gruetter R, Adriany G, Choi IY, Henry PG, Lei H, Öz G. Localized in vivo 13C NMR spectroscopy of the brain. NMR Biomed. 2003;16(67):313–338. 10.1002/nbm.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lizarbe B, Cherix A, Gruetter R. In vivo heteronuclear magnetic resonance spectroscopy. Methods Mol Biol. 2018;1718:169–187. 10.1007/978-1-4939-7531-0_11 [DOI] [PubMed] [Google Scholar]
- 138.de Graaf RA, Rothman DL, Behar KL. State of the art direct 13C and indirect 1H-[13C] NMR spectroscopy in vivo. A practical guide. NMR Biomed. 2011;24:958–972. 10.1002/nbm.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Henry PG, Adriany G, Deelchand D, et al. In vivo 13C NMR spectroscopy and metabolic modeling in the brain: a practical perspective. Magn Reson Imaging. 2006;24(4):527–539. 10.1016/j.mri.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 140.Xin LJ, Lanz B, Lei HX, Gruetter R. Assessment of metabolic fluxes in the mouse brain in vivo using H-1-[C-13] NMR spectroscopy at 14.1 Tesla. J Cereb Blood Flow Metab. 2015;35:759–765. 10.1038/jcbfm.2014.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li S, Chen Z, Zhang Y, et al. In vivo single-shot, proton-localized 13C MRS of rhesus monkey brain. NMR Biomed. 2005;18(8):560–569. 10.1002/nbm.993 [DOI] [PubMed] [Google Scholar]
- 142.Henry PG, Tkac I, Gruetter R. 1H-localized broadband 13C NMR spectroscopy of the rat brain in vivo at 9.4 T. Magn Reson Med. 2003;50:684–692. 10.1002/mrm.10601 [DOI] [PubMed] [Google Scholar]
- 143.Duarte JM, Gruetter R. Glutamatergic and GABAergic energy metabolism measured in the rat brain by 13C NMR spectroscopy at 14.1 T. J Neurochem. 2013;126:579–590. 10.1111/jnc.12333 [DOI] [PubMed] [Google Scholar]
- 144.Oz G, Seaquist ER, Kumar A. Human brain glycogen content and metabolism: implications on its role in brain energy metabolism. Am J Physiol Endocrinol Metab. 2007;292(3):E946–E951. 10.1152/ajpendo.00424.2006 [DOI] [PubMed] [Google Scholar]
- 145.Choi IY, Tkac I, Ugurbil K, Gruetter R. Noninvasive measurements of [1-13C]glycogen concentrations and metabolism in rat brain in vivo. Neurochem. 1999;73:1300–1308. [DOI] [PubMed] [Google Scholar]
- 146.Henry PG, Roussel R, Vaufrey F, Dautry C, Bloch G. Semiselective POCE NMR spectroscopy. Magn Reson Med. 2000;44:395–400. [DOI] [PubMed] [Google Scholar]
- 147.Chen X, Boesiger P, Henning A. J-refocused 1H PRESS DEPT for localized 13C MR spectroscopy. NMR Biomed. 2013;26:1113–1124. 10.1002/nbm.2925 [DOI] [PubMed] [Google Scholar]
- 148.Marjanska M, Henry PG, Gruetter R, Garwood M, Ugurbil K. A new method for proton detected carbon edited spectroscopy using LASER. Proc Int Soc Magn Reson Med. 2004;11:679. [Google Scholar]
- 149.Xin L, Mlynarik V, Lanz B, Frenkel H, Gruetter R. 1H-[13C] NMR spectroscopy of the rat brain during infusion of [2-13C] acetate at 14.1 T. Magn Reson Med. 2010;64:334–340. 10.1002/mrm.22359 [DOI] [PubMed] [Google Scholar]
- 150.de Feyter HM, Herzog RI, Steensma BR, et al. Selective proton-observed, carbon-edited (selPOCE) MRS method for measurement of glutamate and glutamine 13C-labeling in the human frontal cortex. Magn Reson Med. 2018;80(1):11–20. 10.1002/mrm.27003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.de Graaf RA. Theoretical and experimental evaluation of broadband decoupling techniques for in vivo nuclear magnetic resonance spectroscopy. Magn Reson Med. 2005;53:1297–1306. 10.1002/mrm.20507 [DOI] [PubMed] [Google Scholar]
- 152.Lanz B, Duarte JMN, Kunz N, Mlynárik V, Gruetter R, Cudalbu C. Which prior knowledge? Quantification of in vivo brain 13C MR spectra following 13 C glucose infusion using AMARES. Magn Reson Med. 2013;69(6):1512–1522. 10.1002/mrm.24406 [DOI] [PubMed] [Google Scholar]
- 153.Henry PG, Oz G, Provencher S, Gruetter R. Toward dynamic isotopomer analysis in the rat brain in vivo: automatic quantitation of C-13 NMR spectra using LCModel. NMR Biomed. 2003;16:400–412. 10.1002/nbm.840 [DOI] [PubMed] [Google Scholar]
- 154.Deelchand DK, Nelson C, Shestov AA, Ugurbil K, Henry PG. Simultaneous measurement of neuronal and glial metabolism in rat brain in vivo using co-infusion of [1,6-13C2]glucose and [1,2-13C2]acetate. J Magn Reson. 2009;196:157–163. 10.1016/j.jmr.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Patel AB, de Graaf RA, Rothman DL, Behar KL, Mason GF. Evaluation of cerebral acetate transport and metabolic rates in the rat brain in vivo using 1H-[13C]-NMR. J Cereb Blood Flow Metab. 2010;30:1200–1213. 10.1038/jcbfm.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang J, Li SS, Bacher J, Shen J. Quantification of cortical GABA-glutamine cycling rate using in vivo magnetic resonance signal of [2-13C]GABA derived from glia-specific substrate [2-13C]acetate. Neurochem Int. 2007;50:371–378. 10.1016/j.neuint.2006.09.011 [DOI] [PubMed] [Google Scholar]
- 157.Lanz B, Xin L, Millet P, Gruetter R. In vivo quantification of neuro-glial metabolism and glial glutamate concentration using H-[C] MRS at 14.1T. J Neurochem. 2014;128(1):125–139. [DOI] [PubMed] [Google Scholar]
- 158.Sibson NR, Mason GF, Shen J, et al. In vivo 13C NMR measurement of neurotransmitter glutamate cycling, anaplerosis and TCA cycle flux in rat brain during [2-13C]glucose infusion. J Neurochem. 2001;76(4):975–989. [DOI] [PubMed] [Google Scholar]
- 159.Sonnay S, Gruetter R, Duarte JMN. How energy metabolism supports cerebral function: insights from 13C magnetic resonance studies in vivo. Front Neurosci. 2017;11(288):1–20. 10.3389/fnins.2017.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rothman DL, De Feyter HM, de Graaf RA, Mason GF, Behar KL. 13C MRS studies of neuroenergetics and neurotransmitter cycling in humans. NMR Biomed. 2011;24:943–957. 10.1002/nbm.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lanz B, Gruetter R, Duarte JM. Metabolic flux and compartmentation analysis in the brain. Front Endocrinol. 2013;4(156):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shestov AA, Valette J, Deelchand DK, Ugurbil K, Henry PG. Metabolic modeling of dynamic brain 13C NMR multiplet data: concepts and simulations with a two-compartment neuronal-glial model. Neurochem Res. 2012;37:2388–2401. 10.1007/s11064-012-0782-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tiret B, Shestov AA, Valette J, Henry PG. Metabolic modeling of dynamic 13C NMR isotopomer data in the brain in vivo: fast screening of metabolic models using automated generation of differential equations. Neurochem Res. 2015;40:2482–2492. 10.1007/s11064-015-1748-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dehghani MM, Lanz B, Duarte JM, Kunz N, Gruetter R. Refined analysis of brain energy metabolism using in vivo dynamic enrichment of 13C multiplets. ASN Neuro. 2016;8:1–8. 10.1177/1759091416632342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang ZJ, Ohliger MA, Larson PEZ, et al. Hyperpolarized 13C MRI: state of the art and future directions. Radiology. 2019;291(2):273–284. 10.1148/radiol.2019182391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stewart NJ, Matsumoto S. Biomedical applications of the dynamic nuclear polarization and parahydrogen induced polarization techniques for hyperpolarized 13C MR imaging. Magn Reson Med Sci. 2019;1–17. 10.2463/mrms.rev.2019-0094 [DOI] [PMC free article] [PubMed]
- 167.Singh J, Suh EH, Sharma G, Khemtong C, Sherry AD, Kovacs Z. Probing carbohydrate metabolism using hyperpolarized 13C-labeled molecules. NMR Biomed. 2019;32(10):1–23, e4018. 10.1002/nbm.4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.