Abstract
The intermediate steps in the biosynthesis of the ADP-l-glycero-d-manno-heptose precursor of inner core lipopolysaccharide (LPS) are not yet elucidated. We isolated a mini-Tn10 insertion that confers a heptoseless LPS phenotype in the chromosome of Escherichia coli K-12. The mutation was in a gene homologous to the previously reported rfaE gene from Haemophilus influenzae. The E. coli rfaE gene was cloned into an expression vector, and an in vitro transcription-translation experiment revealed a polypeptide of approximately 55 kDa in mass. Comparisons of the predicted amino acid sequence with other proteins in the database showed the presence of two clearly separate domains. Domain I (amino acids 1 to 318) shared structural features with members of the ribokinase family, while Domain II (amino acids 344 to 477) had conserved features of the cytidylyltransferase superfamily that includes the aut gene product of Ralstonia eutrophus. Each domain was expressed individually, demonstrating that only Domain I could complement the rfaE::Tn10 mutation in E. coli, as well as the rfaE543 mutation of Salmonella enterica SL1102. DNA sequencing of the rfaE543 gene revealed that Domain I had one amino acid substitution and a 12-bp in-frame deletion resulting in the loss of four amino acids, while Domain II remained intact. We also demonstrated that the aut::Tn5 mutation in R. eutrophus is associated with heptoseless LPS, and this phenotype was restored following the introduction of a plasmid expressing the E. coli Domain II. Thus, both domains of rfaE are functionally different and genetically separable confirming that the encoded protein is bifunctional. We propose that Domain I is involved in the synthesis of d-glycero-d-manno-heptose 1-phosphate, whereas Domain II catalyzes the ADP transfer to form ADP-d-glycero-d-manno-heptose.
Lipopolysaccharide (LPS) is a major nonprotein component of the outer membrane of enteric and nonenteric gram-negative bacteria (57). LPS is an amphipathic molecule consisting of lipid A and an oligosaccharide core domain. Some microorganisms also express a hydrophilic, surface-exposed O-specific polysaccharide that is found attached to the reducing end of the lipid A core (43, 57). In most cases, lipid A consists of five to seven saturated fatty acids attached to a glucosamine dimer and is responsible for the endotoxic activities of the LPS molecule (38). The core oligosaccharide can be further subdivided into inner and outer core domains. The outer core is generally made of hexoses, while the inner core oligosaccharide is composed of at least two residues of 3-deoxy-d-manno-octulosonic acid, and depending on the particular species of gram-negative bacteria, two or three residues of l-glycero-d-manno-heptose (LDHep) (19). The structure of the inner core has a high degree of conservation among enteric and nonenteric bacteria (19).
LPS plays an important role in maintaining the structural integrity of the bacterial outer membrane (35). Early studies by Tamaki et al. (47) have shown that Escherichia coli K-12 mutants lacking heptose in the LPS demonstrate hypersensitivity to hydrophobic antibiotics (such as novobiocin), detergents, and bile salts. Heptoseless E. coli K-12 mutants are also deficient in F plasmid conjugation (18) and transduction by the P1 bacteriophage (11). The collection of these phenotypes is referred to as the “deep rough” phenotype. The deep rough phenotype is related to a general defect in the assembly of outer membrane proteins, some of which are involved as receptors for conjugation and/or phage attachment (11, 18, 44), components of efflux systems (26), and F-pilus assembly (J. D. Klena, unpublished data). For other organisms, such as Haemophilus influenzae, a heptoseless mutant was found to be serum sensitive and displayed a reduced virulence in an animal model (20, 60).
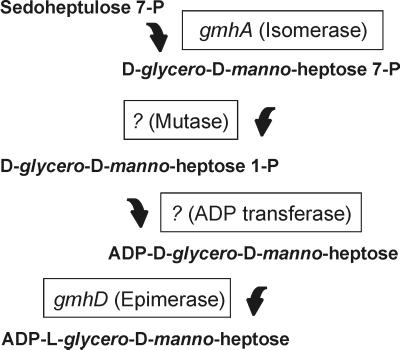
Because of the structural conservation of the inner core in gram-negative bacteria, we have hypothesized that the biosynthesis pathway for LDHep is also conserved. From studies using Salmonella enterica, Eidels and Osborn (15, 16) proposed that LDHep is synthesized from sedoheptulose 7-phosphate via four steps: (i) conversion of sedoheptulose 7-phosphate into d-glycero-d-manno-heptose 7-phosphate by a phosphoheptose isomerase, (ii) formation of d-glycero-d-manno-heptose 1-phosphate by a mutase reaction, (iii) transfer of a nucleotide via a phosphodiester linkage, and (iv) epimerization of d-glycero-d-manno-heptose 1-phosphate residue of the sugar nucleotide to LDHep (Fig. 1). Further investigations resulted in the identification of ADP as the nucleotide sugar residue attached to glycero-manno-heptose in Shigella sonnei and S. enterica (24, 25). Verification of this pathway requires the identification of genes and gene products and the biochemical demonstration of their functions. The epimerase gene, gmhD (previously rfaD, see reference 40 for a discussion on the current gene nomenclature), of several microorganisms was isolated and characterized (10, 14, 34, 46, 55). Also, Brooke and Valvano (7, 8) recently identified gmhA (formerly lpcA) in E. coli and H. influenzae as the first gene of this pathway, confirming that it encodes a phosphoheptose isomerase activity. We have also shown that gmhA is highly conserved in Neisseria gonorrhoeae, Neisseria meningitidis, Helicobacter pylori, Vibrio cholerae, and Pseudomonas aeruginosa (49).
FIG. 1.
Pathway for the synthesis of ADP-l-glycero-d-manno-heptose as proposed by Eidels and Osborn (15, 16). gmhA and gmhD are the only genes whose functions have been established biochemically (7, 10).
In this study, we report the identification and molecular characterization of an E. coli gene encoding a bifunctional protein consisting of two distinct domains, both of which are required for the intermediate steps in the synthesis of ADP-LDHep. We also show that these domains function independently, and in some microorganisms they are encoded by separate genes. The predicted functions of these domains suggest a novel pathway for the synthesis of ADP-glycero-manno-heptose.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Except when indicated, all chemicals used in this study were purchased from Sigma Chemical Co., St. Louis, Mo. Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were cultured in Luria broth (LB) (Sigma), supplemented when necessary with antibiotics at final concentrations of 50 μg/ml for novobiocin, 30 μg/ml for chloramphenicol, 100 μg/ml for ampicillin, and 20 μg/ml for tetracycline. S. enterica SL1102 was obtained by mutagenesis as described elsewhere (58). Gene libraries of E. coli VW187 (previously constructed in our laboratory by M. Handelsman) and VW194 were used to complement the rfaE mutants of E. coli K-12 KLC2926 and S. enterica SL1102. pMGC2 contained a 12.5-kb BamHI fragment from the E. coli VW194 chromosome cloned into the BamHI site of pACYC184. pTLS1 contained a 1.463-kb fragment from pMGC2, obtained by PCR with primers P54 and P56 (Table 2), cloned into the SmaI site of the expression vector pEX1 (37). pMAV47, containing Domain II from rfaE, was constructed by cloning a 0.5-kb fragment from pTLS1, obtained by PCR with primers PAB10542 and P56, into the SmaI site of pUC19. This strategy generates a protein fusion between Pro16 of β-galactosidase with Pro305 of RfaE. pMAV51, containing only Domain I, was generated by deleting a 0.5-kb HindIII fragment of pTLS1. pMAV44 contains the chloramphenicol acetyltransferase (cat) gene from pBR325 inserted into the KpnI site of pTLS1. The cat gene was obtained by PCR amplification with primers P39 and P40. Before ligation with amplified DNA, the KpnI site in pTLS1 was treated with the Klenow fragment of DNA polymerase. The cat gene in pMAV44 is transcribed in the opposite direction from the rfaE gene. pMB10 is a 1.7-kb BamHI fragment containing the plac promoter and the rfaE gene from pTLS1 cloned into the BamHI site of pME6000. pMAV52 was obtained by subcloning a 1.2-kb BamHI/HindIII fragment containing the plac promoter and Domain I from rfaE into the BamHI/HindIII sites in pME6000. To generate pMAV53, a 0.5-kb amplification product obtained with primers P147 and P148 (both present in vector sequences flanking the DNA insert) and pMAV47 as a template was cloned into the SpeI site in pME6000. pCM200 and pCM202 were generated by PCR amplification of a 1.4-kb fragment containing the wild-type and mutated rfaE genes from S. enterica strains SL1027 and SL1102, respectively. The primers used in the reaction were P181 and P182, and the amplified fragments were cloned into the SmaI site of pEX1. Plasmid pCM204 was generated by digesting pCM200 with BamHI and inserting the linearized DNA into the BamHI site in pME6000. pCM203 was constructed in a similar manner, except that digestion was carried out with HindIII. Primers P91 (ptac promoter) and P182 were used to confirm that the inserts in these plasmids contained the S. enterica rfaE gene. pME6000 is a low-copy-number, broad-host-range cloning vector kindly provided by S. Heeb, Laboratoire de Biologie Microbienne, Université de Lausanne. pME6000 was constructed by replacing the cat gene of pBBR1MCS (27) with the tetRA genes of pVK100 (23).
TABLE 1.
Bacterial strains, plasmids, and bacteriophages used in this study
| Strain or plasmid | Relevant genotype and/or characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| χ711 | F−leu-4 proAB118 arg-35 λr T6r Strr | 7 |
| BW19851 | RP4-2::Mu-1kan::Tn7/creB510 hsdR17 endA1 zbf-5 uidA(ΔMluI)::pir(wt) recA1 thi | 32 |
| KLC2926 | W1485, rfaE::Tn10, Tetr Novs | This study |
| VW187 | E. coli O7:K1, prototrophic | 50 |
| VW194 | E. coli O6:K?, prototrophic | 52 |
| W1485 | E. coli K-12, wild type | E. coli Genetic Stock Center, Yale University |
| S. enterica | ||
| SL1027 | metA22 trpC2 fla-66 rpsL120 xyl-404 metE551 | 58 |
| SL1102 | Same as for SL1027, rfaE543 | 58 |
| R. eutrophus | ||
| HB3 | H16, aut::Tn5, deficient in autotrophic growth, Kmr | |
| H16 | Prototrophic strain | |
| Plasmids | ||
| pACYC184 | Cloning vector, Cmr | 9 |
| pBR325 | Cloning vector, Cmr Ampr Tetr | 3 |
| pCM199 | 1.4-kb amplicon containing the rfaE::Tn10 junction cloned into pUC19, Ampr | This study |
| pCM200 | 1.4-kb amplicon containing rfaE+ from S. enterica SL1027 cloned into pEX1, Ampr | This study |
| pCM202 | 1.4-kb amplicon containing rfaE543 from S. enterica SL1102 cloned into pEX1, Ampr | This study |
| pCM204 | 1.7-kb BamHI fragment containing the plac promoter and rfaE+ from pCM200 cloned into pME6000, Tetr | This study |
| pCM205 | 1.7-kb BamHI fragment containing the plac promoter and rfaE543 from pCM202 cloned into pME6000, Tetr | This study |
| pEX1 | Expression vector, Ampr | 37 |
| pHI3 | 1.9-kb HindIII-BglII fragment carrying part of the rfaE gene of H. influenzae 2019 cloned into pUC19, Ampr | 28 |
| pJB2 | 3-kb EcoRI fragment containing gmhA cloned into pMAV3, Cmr | 7 |
| pJK2252 | pGEM4 carrying a 5.5-kb BglII fragment harboring waaFCLKZ from E. coli K-12, Ampr | 22 |
| pMAV3 | Cloning vector, Cmr | 30 |
| pMAV44 | cat cassette inserted into the KpnI site of pTLS1, Cmr Ampr | This study |
| pMAV47 | 541-bp amplicon from pTLS1 cloned into pUC19, Ampr | This study |
| pMAV51 | 500-bp HindIII deletion from pTLS1, Ampr | This study |
| pMAV52 | 1.2-kb BamHI-HindIII fragment from pMAV51 cloned into pME6000, Tetr | This study |
| pMAV53 | 550-bp amplicon from pMAV47 cloned into pME6000, Tetr | This study |
| pMB10 | 1.9-kb BamHI fragment from pTLS1 cloned into pME6000, Tetr | This study |
| pME6000 | Cloning vector, Tetr | S. Heeb |
| pMGC2 | 12.5-kb BamHI fragment from E. coli VW194 cloned into pACYC184, Cmr | |
| pTLS1 | 1.4-kb amplicon containing the rfaE gene from pMGC2 cloned in pEX1, Ampr | This study |
| pUC19 | Cloning vector, Ampr | 59 |
TABLE 2.
Primers used in this study
| Primer no. | Gene or plasmid and description (source) | Sequence |
|---|---|---|
| P39 | cat, N terminus (pACYC184) | 5′-GATCACTTCGCAGAAT-3′ |
| P40 | cat, C terminus (pACYC184) | 5′-AATTACGCCCCGCCCT-3′ |
| P54 | rfaE, N terminus (E. coli) | 5′-ATGAAAGTAACGCTGCC-3′ |
| P56 | rfaE, C terminus (E. coli) | 5′-GATCTGTGAACCGCTTTCC-3′ |
| P69 | tetA, near BamHI site (pACYC184) | 5′-GACTACGCGATCATGGC-3′ |
| P70 | tetA, near BamHI site (pACYC184) | 5′-CAGCAACCGCACCTGT-3′ |
| P91 | plac, forward (pEX1) | 5′-GCGGCAAGCTTCTGGCGT-3′ |
| P147 | pUC19, forward | 5′-GCAGACTAGTATACGCAAACCG-3′ |
| P148 | pUC19, reverse | 5′-GCAGACTAGTATACCGCACAGAT-3′ |
| P174 | tetA, N terminus outward (Tn10) | 5′-AGTGGTTAGCGATATCTTCC-3′ |
| P175 | tetA, C terminus outward (Tn10) | 5′-GATGGCTGGATTTGGATTA-3′ |
| P181 | rfaE, N terminus (S. enterica) | 5′-ATGAGAGTAAATCTGCCAGC-3′ |
| P182 | rfaE, C terminus (S. enterica) | 5′-ATAAGCTGTCCGACGCACCTTC-3′ |
| P192 | rfaE, internal (S. enterica) | 5′-GGAGGTCAATGTGAAGTGC-3′ |
| P193 | rfaE, internal (S. enterica) | 5′-GAGCTGCTGATTACGTGAT-3′ |
| P194 | rfaE, internal (S. enterica) | 5′-GGATTAACCGGACGGCTT-3′ |
| PAB10542 | rfaE, internal (E. coli) | 5′-CGATCGAGCTGGAAAATG-3′ |
| PAB10543 | rfaE, internal (E. coli) | 5′-ACCCAGTCGACCGCTT-3′ |
Mating in R. eutrophus.
Plasmids containing the different constructs in pME6000 were transferred into Ralstonia eutrophus HB3 by conjugation. The plasmids were first transformed by electroporation (13) into the E. coli K-12 strain BW19851. This strain contains an integrated RP4 plasmid carrying the necessary components to allow mobilization of recombinant plasmids containing the mob region present in vector pME6000. The donor strain BW19851, which carried the appropriate plasmids, and the recipient HB3 were mixed in equal proportions and washed once with prewarmed sterile LB. The bacterial mixture was spread on LB plates and incubated overnight at 30°C. The growth was resuspended and diluted in LB, and exconjugants were selected in LB plates containing kanamycin and tetracycline at final concentrations of 120 and 100 μg/ml, respectively.
Recombinant DNA methods.
Small- and large-scale plasmid DNA extractions and DNA gel electrophoresis were conducted as described (31). Chromosomal DNA was prepared by a clear lysis method (51). Transformations were done by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada) and 0.1-cm cuvettes, following the method described by Dower et al. (13). Restriction enzyme analysis and cloning were performed by following standard protocols (29). Restriction endonucleases were obtained from Roche Diagnostics, Dorval, Quebec, Canada, and Pharmacia Canada Inc., Baie d'Urfé, Quebec, Canada. Calf alkaline phosphatase, T4 DNA ligase, and polynucleotide kinase were from Roche Diagnostics. All enzymes were used as suggested by the manufacturers.
PCR.
PCR were carried out with a Hybaid Omnigene temperature cycler (Interscience, Markham, Ontario, Canada) and PwoI DNA polymerase (Roche Diagnostics). The primers used in this study are listed in Table 2. Prior to cloning, amplicons were treated with polynucleotide kinase and purified by gel electrophoresis followed by extraction of the DNA band with the QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.) according to the manufacturer's instructions.
LPS analysis.
LPS was isolated as described by Marolda et al. (31) and analyzed by Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Schagger and von Jagow (42). Commercially cast 16% polyacrylamide gels were purchased from Novex, San Diego, Calif., and LPS bands were visualized by silver staining (31).
Bacteriophage U3.
Aliquots of 400 μl of overnight E. coli cultures were added to 7 ml of warmed 0.8% LB agar containing the appropriate antibiotics. The mixture was applied as an overlay to LB media plates also containing 1.5% agar and antibiotics as necessary. Agar was allowed to harden, and a 20-μl drop of U3 bacteriophage lysate was carefully deposited on the center of the plate. When the drop was dry (after about 30 min at room temperature), plates were inverted and incubated at 37°C for 6 to 8 h. Lysis was evidenced by clear zones indicating growth inhibition.
Polypeptide analysis.
In vitro transcription-translation was performed with the prokaryotic DNA-directed translation kit from Amersham with [35S]methionine as recommended by the manufacturer. Polypeptides were separated by SDS-PAGE (31), followed by treatment with En3Hance (Dupont). Dried gels were exposed to Kodak X-Omat film at −80°C for 16 to 24 h.
DNA sequence analysis.
DNA sequencing of plasmid constructs was carried out with an automated sequencer at MOBIX, McMaster's University, Hamilton, Ontario, Canada, and at the DNA Sequencing Facility of the Robarts Research Institute, London, Ontario, Canada. DNA and protein sequence analysis was carried out with the University of Wisconsin Genetics Computer Group package, version 9 (12). Nucleotide similarity searches were performed with the program BLAST2 (1) via the National Center for Biotechnology Information. Protein sequence alignments were produced with the program Clustal X (48) and edited with GeneDoc (K. B. Nicholas and H. B. Nicholas, Jr., 1997, GeneDoc: analysis and visualization of genetic variation [http://www.cris.com/∼Ketchup/genedoc.shtml]).
Nucleotide sequence accession number.
The DNA sequences of the S. enterica SL1027 rfaE and SL1102 rfae543 genes have been deposited in GenBank with the accession no. AF163661 and AF163662, respectively.
RESULTS
Isolation and characterization of the heptoseless mutant E. coli strain KLC2926.
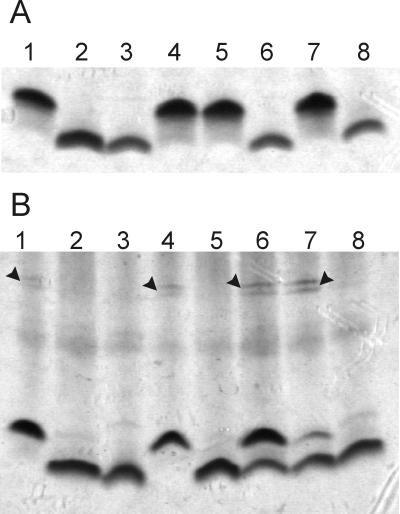
A transposon mutagenesis strategy using mini-Tn10 was utilized to isolate genes involved in the biosynthesis pathway of ADP-LDHep. Mutagenesis was conducted in the E. coli K-12 strain W1485. Since heptose-deficient mutants are usually nonviable in the presence of very low concentrations of novobiocin (49), 1,000 Tetr colonies were screened for sensitivity to novobiocin at a concentration of 200 μg/ml. Seven Tetr Novs colonies were identified. Only three of these colonies were also sensitive to novobiocin at a concentration of 50 μg/ml. One of them, KLC2926, had a mucoid appearance, a characteristic also found associated with the deep rough LPS phenotype (36). KLC2926 was also resistant to lysis by the core-specific bacteriophage U3, which recognizes a terminal galactose in the complete E. coli K-12 core (56). Therefore, mucoidity, phage U3 resistance, and high susceptibility to novobiocin suggested that KLC2926 makes a heptoseless LPS and consequently is unable to assemble the remaining sugar components of the outer core. This conclusion was supported by the electrophoretic analysis of KLC2926 LPS, which displayed a rapid migration in Tricine polyacrylamide gels comparable with that of the heptoseless LPS of strain χ711 (Fig. 2A, lanes 2 and 3). None of the above-mentioned phenotypes of strain KLC2926 were complemented by transformation with plasmids pJB2, which contains the gmhA gene (7), or pJK2252, which contains the heptosyltransferases encoded by waaC and waaF (22) (data not shown). Furthermore, hybridization experiments with a core gene-specific probe indicated that mini-Tn10 was not located in the LPS core oligosaccharide gene cluster waa (data not shown), also ruling out that the mutation in KLC2926 was in gmhD or in any of the three heptosyltransferase genes mapping within this cluster (19, 43). Therefore, we concluded that the Tn10 insertion in KLC2926 defined a novel gene or genes for the synthesis of ADP-heptose.
FIG. 2.
Analysis of LPS in E. coli and R. eutrophus. (A) LPS profiles of E. coli strains. Lanes: 1, W1485 (wild type); 2, KLC2926 (rfaE::Tn10); 3, χ711 (gmhA); 4, KLC2926(pTLS1); 5, KLC2926(pMAV51); 6, KLC2926(pMAV47); 7, KLC2926(pCM200); 8, KLC2926(pCM202). (B) LPS profiles of R. eutrophus strains. Lanes: 1, H16 (wild type); 2, HB3 (aut::Tn5); 3, E. coli χ711 (gmhA); 4, HB3(pMB10); 5, HB3(pMAV52); 6, HB3(pMAV53); 7, HB3(pCM204); 8, HB3(pCM205). Arrows indicate the bands corresponding to polymeric O antigen. In both cases, cell lysates were separated by Tricine-SDS-PAGE and stained with silver.
The Tn10 insertion in KLC2926 is located within the rfaE gene.
Preliminary attempts to clone the gene complementing the novobiocin-sensitive phenotype of KLC2926 from DNA fragments of strains W1485 and VW187 were unsuccessful. However, we were able to clone a 12.5-kb BamHI fragment from E. coli VW194, resulting in plasmid pMGC2 (Fig. 3A). KLC2926(pMGC2) was not only resistant to novobiocin but also became sensitive to bacteriophage U3 and displayed a complete lipid A core band in Tricine-SDS polyacrylamide gels, suggesting that the insert DNA carries gene or genes complementing the defect caused by the mini-Tn10 insertion in this strain (data not shown). pMGC2 in KLC2926 was unstable and yielded very little DNA upon plasmid purification. We speculated that the instability problems could be caused by recombination events involving sequences in the insert DNA near or within the locus defined by mini-Tn10. This appeared to be the case since pMGC2 was stable in the recA-defective strain DH5α.
FIG. 3.
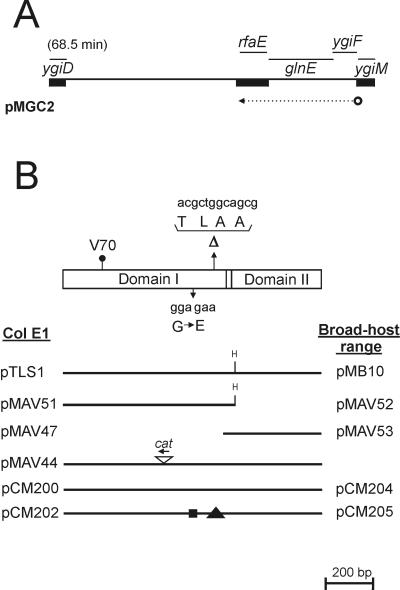
(A) Partial map of the chromosomal region cloned into pMGC2 that corresponds to the E. coli K-12 genetic map. Thick bars represent regions that were sequenced. The arrow indicates the direction of transcription of the putative ygiF-glnE-rfaE operon. (B) Map of the rfaE gene. Boxes correspond to the Domain I and Domain II regions. The positions of the various mutations are indicated: V70, the position of the Tn10 insertion in KLC2926 after the codon encoding valine 70 of the RfaE protein; ΔTLAA, the location of the 12-bp deletion in SL1102 rfaE and the corresponding amino acids; G→E, the glycine-to-glutamic acid substitution due to a base pair change (GGA to GAA). The partial maps show the various plasmids. The ColE1 vector was pEX1, and these plasmids were used in the experiments involving E. coli and S. enterica. The broad-host-range vector was pME6000, and these plasmids were used in the experiments involving R. eutrophus. The position and orientation (arrow) of the insertion of a cat cassette within the E. coli rfaE gene in pMAV44 is shown. pCM200 and pCM204 correspond to the cloned wild-type rfaE gene of S. enterica SL1027. pCM202 and pCM205 correspond to the cloned mutant rfaE543 gene of S. enterica SL1102. The box and triangle in pCM202/pCM205 denote the positions of the amino acid substitution and deletion, respectively.
Since the genomic sequence of E. coli has been recently determined (2), we sequenced the ends of the 12.5-kb BamHI fragment of pMGC2, using primers P69 and P70 to position the insert on the E. coli K-12 chromosomal map. One of the ends coincided with the termination codon of ygiD, encoding a hypothetical protein of unknown function, while the other end was at 234 bp into the sequence of ygiM, spanning a region of approximately 19 kb in length. This region contains several open reading frames with homologies to a fimbrial gene cluster and also an IS21 insertion element inserted downstream of the putative major fimbrial subunit gene. Thus, the discrepancy between 19 kb in E. coli K-12 and 12 kb in strain VW194 could be explained if some of the genes in this region are not present in the chromosome of strain VW194 or, alternatively, if part of these genes were deleted during the cloning procedure. An inspection of the E. coli K-12 genes located between ygiD and ygiM (Fig. 3A) revealed the presence of a gene homologous to the H. influenzae rfaE gene, which has been shown to complement the rfaE gene mutation in S. enterica (28). This mutation is characterized phenotypically by the production of heptoseless LPS (58). We cloned this gene in the expression vector pEX1, and the resulting plasmid, pTLS1 (Fig. 3B), was able to restore the heptoseless defect in KLC2926 (Fig. 2A, lane 4), confirming that this was the only gene from the 12.5-kb BamHI fragment necessary for complementation. The E. coli rfaE gene was located downstream of glnE, a gene encoding an adenylyltransferase involved in regulation of glutamine synthetase (53). In a previous work, van Heeswijk et al. (53) showed that ygiF(orfEX), of unknown function, and glnE are cotranscribed from a promoter located upstream of ygiF. There are 47 bp separating glnE and rfaE, with no indication of transcription termination sequences nor promoter features in the intervening sequence. Thus, the E. coli rfaE gene appears to be the last gene of a three-gene operon.
To precisely localize the Tn10 insertion, KLC2926 chromosomal DNA was isolated and used as a template in PCR amplifications with primer pair P54 and P174 and primer pair P56 and P175. Primers P54 and P56 each corresponded to an end of the rfaE coding region while P174 and P175 annealed to the tetA gene of Tn10 (Table 2). Amplification products were cloned into the SmaI site of pUC19. One of these plasmids, pCM199, was sequenced, and the endpoint of the Tn10 insertion was localized next to bp 210 of the rfaE coding region (Val70 in the protein sequence) (Fig. 3B).
The rfaE gene encodes a single protein with two distinct domains.
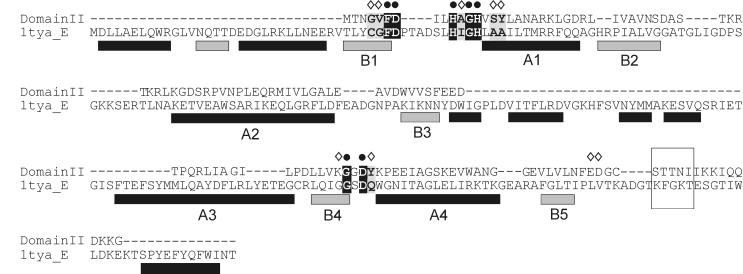
The predicted RfaE polypeptide was compared with the BLAST program (1) to amino acid protein sequences deposited in GenBank. It was strikingly apparent that the predicted protein contained two distinct regions. The region from amino acid 1 to 318 (Fig. 3B Domain I) showed conservation with members of the ribokinase family (Table 3) (5). The amino acid conservation was especially strong in critical regions for enzymatic activity, such as an aspartic acid located in an anion “hole” in the E. coli ribokinase that is essential for catalysis (Fig. 4) (45). Other regions of the enzyme containing amino acids that make contact with the sugar substrate are also conserved in Domain I (Fig. 4). The C-terminal region of the RfaE protein (Fig. 3B) from amino acid 344 to 477, designated Domain II, had features in common with the cytidylyltransferase superfamily (Table 3) (4). This family is a member of a large group of diverse enzymes hydrolyzing the alpha-beta pyrophosphate bond in nucleoside triphosphates, which have been known to show the characteristic HXGH motif of the class I aminoacyl-tRNA synthetases (Fig. 5) (4, 54).
TABLE 3.
Proteins containing Domains I and II within the same protein or in separate proteinsa
| Protein type | Description | Organism | No. of residuesb | GenBank accession no. |
|---|---|---|---|---|
| Domain I + II | Aut, autotrophic growth | Haemophilus influenzae | 476 | C64127 |
| WaaE, ADP-d-glycero-d-manno-heptose synthase | Helicobacter pylori J99 | 463 | AAD06368 | |
| RfaE, ADP-heptose synthase | Helicobacter pylori 29695 | 461 | AAD07904 | |
| SC2G5.08, bifunctional synthase/transferase | Streptomyces coelicolor | 463 | CAB36595 | |
| Domain I only | RfaE, ADP-heptose synthase | Neisseria meningitidis | 313 | AAD32179 |
| WaaE, ADP-heptose synthase | Burkholderia pseudomallei | 307 | AAD43345 | |
| RfaE, ADP-heptose synthase | Aquifex aeolicus | 315 | AC06797 | |
| TM0960, ribokinase | Thermotoga maritima | 299 | AAD36039 | |
| RbsK, ribokinase | Bacillus subtilis | 293 | P36945 | |
| F20D23.14, ribokinase | Arabidopsis thaliana | 378 | AAD50017 | |
| RbsK, ribokinase | Haemophilus influenzae | 306 | P44331 | |
| RbsK, ribokinase | Escherichia coli | 309 | P05054 | |
| YihV, sugar kinase | Escherichia coli | 298 | P32143 | |
| CscK, fructokinase | Synechocystis sp. | 307 | BAA17561 | |
| RbsK, ribokinase | Schizosaccharomyces pombe | 318 | O60116 | |
| RbsK, ribokinase | Pyrobaculum aerophilum | 305 | AAD00536 | |
| YdjE, sugar kinase | Bacillus subtilis | 320 | O34768 | |
| ScrK, fructokinase | Clostridium beijerinckii | 312 | AAC99323 | |
| RbsK, ribokinase | Lactobacillus sakei | 302 | AAD34338 | |
| K6P2, 6-phosphofructokinase isozyme 2 | Escherichia coli | 309 | P06999 | |
| Domain II only | TagD1, glycerol-3-phosphate cytidylyltransferase | Aquifex aeolicus | 157 | AAC06521 |
| Aut, autotrophic growth | Ralstonia eutropha | 164 | I39548 | |
| TagD, glycerol-3-phosphate cytidylyltransferase | Pyrococcus abyssi | 148 | CAB50222 | |
| PH0735, hypothetical protein | Pyrococcus horikoshii | 148 | BAA29826 | |
| TagD, glycerol-3-phosphate cytidylyltransferase | Archaeoglobus fulgidus | 137 | AAB89835 | |
| MJ1179, hypothetical protein | Methanococcus jannaschii | 149 | B64447 | |
| T6A23.13, phosphoethanolamine cytidylyltransferase | Arabidopsis thaliana | 139 (421) | AAC67351 | |
| TagD2, glycerol-3-phosphate cytidylyltransferase | Aquifex aeolicus | 168 | AAC07343 | |
| TagD, glycerol-3-phosphate cytidylyltransferase | Bacillus subtilis | 129 | P27623 | |
| C39D10.3, glycerol-3-phosphate cytidylyltransferase | Caenorhabditis elegans | 221 | AAA80419 | |
| TagD, glycerol-3-phosphate cytidylyltransferase | Staphylococcus aureus | 132 | AAB51063 | |
| MTH844, autotrophic growth | Methanobacterium thermoautotrophicum | 151 | AAB85342 | |
| CTPT, cholinephosphate cytidylyltransferase | Plasmodium falciparum | 179 (370) | P49587 |
Results from a BLAST search with the E. coli RfaE protein as a query. All listed proteins show strong identity with RfaE. Domain I corresponds to the ribokinase family. Domain II corresponds to the cytidylyltransferase superfamily.
The number indicates the length of the homologous region that coincides with the length of the protein(s) except for the eukaryotic homologues, for which the total length of the protein is indicated in parentheses.
FIG. 4.
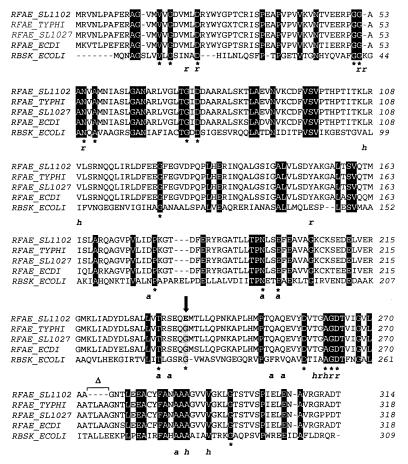
Amino acid sequencing alignment of Domain I of RfaE proteins from E. coli (RFAE_ECDI), S. enterica serovar Typhi (RFAE_TYPHI), S. enterica serovar Typhimurium strains SL11027 (RFAE_SL1027) and SL1102 (RFAE_SL1102), and E. coli ribokinase (RBSK_ECOLI). The alignment was produced with CLUSTAL W. Identical residues in all five proteins are boxed in black. ∗, residues that are also conserved in other members of the ribokinase family (45); r, a, and h, residues in the ribokinase that bind ribose (r) or ADP (a) via hydrogen bonds or that make van der Waals contacts with ribose and/or ADP (h) (for more details on the structure of ribokinase see reference 45). The arrow indicates the substitution of glycine (boxed in grey) by glutamic acid (bold) in the SL1102 RfaE mutant of SL1102. ▵, the deletion of four amino acids in the SL1102 RfaE mutant.
FIG. 5.
Comparison of the known structure of the tyrosyl tRNA synthetase TyrTS (1tya_E) with Domain II. Secondary structure elements in TyrTS (6) are shown below the sequence (black bar, helix; gray bar, strand). The numbers indicate the helices and strands placed around the substrate binding site (according to reference 4). ● and ◊, conserved residues that line the ATP binding pocket. The flexible loop KFGKT, also involved in catalysis, and its corresponding segment in Domain II, STTNI (conserved in all members of cytidylyltransferases), are also boxed.
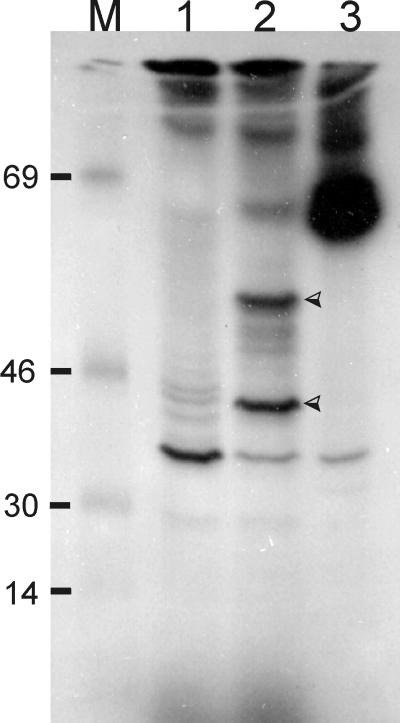
Because of this unusually clear-cut separation of domains, we suspected that the rfaE gene could consist of two genes. We designed primers close to the junction between the two domains (Table 2, PAB10542 and PAB10543) and sequenced both strands of the insert in pTLS1, confirming that there is a single open reading frame. More importantly, the insert in pTLS1 expressed only one polypeptide, as shown by an in vitro transcription-translation experiment with an isolated 1.7-kb BamHI fragment carrying a portion of the plac promoter and the rfaE gene (Fig. 6, lane 2). This polypeptide band had an apparent molecular mass of 52 kDa, consistent with the predicted size of the protein deduced from the amino acid sequence. A smaller polypeptide of about 30 kDa, the calculated mass of Domain I, was also detected. We interpreted this result as either premature termination of translation or posttranslational proteolytic processing of the 55-kDa polypeptide in the transcription-translation reaction. No unique polypeptide in the 14- to 18-kDa region was noticed, suggesting that if processing occurred and the 30-kDa band indeed corresponds to Domain I, the Domain II region of the protein was completely degraded. No similar bands were observed in the transcription-translation experiment with pMAV44 (Fig. 6, lane 1), which contains a cat gene cassette inserted in the KpnI site of rfaE (Fig. 3B). From all these experiments, we concluded that rfaE encodes a single protein with two different domains, suggesting the possibility of a bifunctional enzyme.
FIG. 6.
Autoradiograph showing rfaE expression by in vitro transcription-translation. Proteins were labeled with [14C]leucine as indicated by the supplier (Amersham) and separated by SDS-PAGE. M, [14C]methylated molecular weight markers (bovine serum albumin [69 kDa], ovalbumin [46 kDa], carbonic anhydrase [30 kDa], and lysozyme [14 kDa]). Lanes: 1, pMAV44; 2, pTLS1; 3, Kit's positive control. Arrowheads indicate unique polypeptides of ca. 55 and 38 kDa.
Expression of Domain I is sufficient to complement the Tn10 mutation in E. coli KLC2926.
Since RfaE has two domains, it was important to investigate whether one or both of these domains are involved in the synthesis of ADP-heptose. For this purpose, plasmid pMAV51, containing a deletion eliminating Domain II, and pMAV47, expressing only Domain II, were constructed (Fig. 3B). In the case of pMAV47, a protein fusion of Domain II was generated by fusing this domain with Pro16 from the LacZ protein, thus adding 16 N-terminal amino acids of LacZ and driving the expression of Domain II by the lac promoter. Both plasmids were transformed into KLC2926 and the LPS phenotypes examined before. Only pMAV51 could complement the mutant phenotype as indicated by restoration of a wild-type lipid A-core band (Fig. 2A, lane 5) and the sensitivity to bacteriophage U3. In contrast, pMAV47 could not complement the heptoseless phenotype of KLC2926 (Fig. 2A, lane 6). These results indicate that Domain I is sufficient to complement the LPS-deficient phenotype. Therefore, Domain II is either not involved in lipid A-core synthesis or, alternatively, the Tn10 insertion is nonpolar on the expression of this domain. This may be due to the presence of at least three intragenic initiation codons preceded by potential ribosomal binding sites in the sequence downstream from the site of the Tn10 insertion.
The S. enterica rfaE mutation has an in-frame deletion and an amino acid substitution in Domain I.
In a previous work, Lee et al. (28) reported that pHI3, a plasmid carrying the rfaE gene of H. influenzae, complements the rfaE543 mutation of S. enterica SL1102. The insert in pHI3 contains the entire Domain I homologue-encoding sequence but only a portion of that for Domain II. Following transformation of S. enterica SL1102 with pMAV51 and pMAV47, we determined that only pMAV51 could complement the deep rough LPS phenotype of this strain (data not shown). To establish the nature of the S. enterica rfaE mutation, we cloned and determined the DNA sequence of rfaE543. We took advantage of the recent genomic sequencing projects of S. enterica serovar Typhimurium and Serovar Typhi. A search of the available sequences released by the Sanger Centre resulted in the identification of the rfaE gene in S. enterica serovar Typhi, which we used to obtain primers P181 and P182 (Table 2). These primers amplified a 1,495-bp fragment from chromosomal DNA of S. enterica SL1102 and SL1027. Cloning of the SL1027 amplicon into pEX1 (pCM204) resulted in complementation of the heptoseless phenotype of E. coli KLC2926, while the corresponding plasmid containing the mutated rfaE543 allele from S. enterica SL1102 (pCM205) did not complement (Fig. 2A, lanes 7 and 8, respectively). Comparison of DNA sequences of both rfaE genes revealed an in-frame deletion of 12 bp within rfaE543, resulting in the loss of four amino acids, Thr-Leu-Ala-Ala, located after Ala272 and within Domain I of the mutated rfaE gene product. Additionally, there was one base pair change, which resulted in the replacement of Gly236 by glutamic acid. Leu274, absent in the mutated rfaE gene product, is a conserved residue present in α-helix 8 of the ribokinase (Fig. 4), next to one of the two regions critical for substrate binding (45). Also, the replaced Gly236 is near another region for substrate binding. The fact that both mutations are localized within Domain I and the reading frame is unaltered strongly suggests that Domain II is expressed. At the same time, Domain I is important for the synthesis of ADP-heptose. This was consistent with the inability of pCM202, which carries the mutated rfaE gene of S. enterica, to complement the phenotype of E. coli KLC2926 (Fig. 2A, lane 8).
Domain II functions independently of Domain I in synthesis of LPS.
Since the expression of Domain II was presumably not affected in the rfaE mutations studied, it was important to examine its role in LPS synthesis. For this purpose, we turned our attention to the Ralstonia (Alcaligenes) eutrophus strain HB3 containing a Tn5 insertion in the aut gene (17). This gene encodes a protein with strong amino acid conservation with Domain II. The aut::Tn5 mutation has been associated with pleiotropic effects, including autotrophic growth and changes in cell morphology and colony appearance (17). An examination of the available sequence flanking aut indicates no homologies with other known genes (data not shown). A comparison of the LPS electrophoretic profile of the aut::Tn5 mutant with that of the wild-type strain, H16 (Fig. 2B, lanes 1 and 2), revealed that the mutant has a rapid migrating lipid A-core band that comigrates with that of the E. coli K-12 heptoseless lipid A-core. H16 also exhibits slow migrating bands in the gel corresponding to polymeric O antigen which are not present in HB3. Furthermore, HB3 is novobiocin sensitive while H16 remains resistant to this antibiotic. Therefore, we concluded that the aut gene is involved in LPS synthesis. To confirm whether the E. coli rfaE gene can complement the function of aut in R. eutrophus, we subcloned the 1.7-kb BamHI fragment of pTLS1 into the vector pME6000 and mobilized this construct, pMB10, to strain HB3 by conjugation. The results indicate that this strain displayed a normal appearance, regained resistance to novobiocin, and expressed a complete LPS (Fig. 2B, lane 4). In contrast to the results with E. coli KLC2926, pMAV52 (encoding Domain I) failed to complement the aut mutation (Fig. 2B, lane 5), while pMAV53 (encoding Domain II) did complement, although a small proportion of LPS molecules remained heptoseless (Fig. 2B, lane 6). We concluded from these experiments that Domain II also functions in the synthesis of lipid A-core, and its function is independent from the activity mediated by Domain I.
pCM204 and pCM205, carrying the wild-type and mutated rfaE genes from Salmonella, respectively, were introduced in HB3. R. eutrophus HB3(pCM204) showed two species of lipid A-core bands corresponding to mutant and wild-type LPS, while no complementation was detected with pCM205 (Fig. 2B, lanes 7 and 8). This was not due to DNA rearrangements in R. eutrophus since pCM205 isolated from strain HB3 had the expected restriction endonuclease pattern and an intact DNA insert was recovered by PCR (data not shown). Since R. eutrophus and Salmonella have different G+C contents and codon usage, it is possible that the S. enterica Domain II polypeptide is not well expressed in the Ralstonia cell background.
DISCUSSION
In this study, we have identified a mini-Tn10 mutation in E. coli K-12 associated with heptoseless LPS that corresponds to an insertion in the rfaE gene. The predicted polypeptide consisted of two distinct domains clearly localized to two different regions. The N-terminal portion of RfaE showed features described for the ribokinase family, and it was designated Domain I. The RfaE C-terminal region had features in common with the cytidylyltransferase superfamily, and it was designated Domain II.
The presence of two distinct domains in the RfaE protein prompted us to investigate their role in the synthesis of heptose. For this purpose, each domain was cloned and expressed separately. Complementation experiments were conducted with E. coli KLC2926 (rfaE::Tn10) and S. enterica SL1102 (rfaE543). Our data showed that expression of Domain I was sufficient to complement the heptoseless phenotype of both mutant strains and demonstrated that the mutations were located in the sequence encoding this domain. These experiments did not elucidate the possible involvement of Domain II in LPS synthesis. We approached this question by conducting another set of complementations with the aut::Tn5 mutation in R. eutrophus. Freter and Bowien (17) previously described mutations in a R. eutrophus locus, designated aut, which conferred a pleiotropic phenotype characterized by slow heterotrophic growth on substrates catabolized via the glycolytic pathway and by an altered colony morphology. We noticed that the predicted amino acid sequence of the Aut protein was highly conserved as compared to the sequence of Domain II of RfaE. However, the aut gene does not have an N-terminal portion encoding a protein similar to Domain I. Since, in contrast to the transparent and round appearance of the wild-type colonies, aut mutant colonies are whitish-opaque and have rough edges, we hypothesized that aut could be involved in LPS synthesis. A comparison of the LPS profile of the R. eutrophus strain HB3 (aut::Tn5) with that of the wild-type strain suggested that HB3 produced a heptoseless LPS core and also lacked O antigen. Furthermore, only plasmids expressing the E. coli rfaE Domain II were capable of complementing the LPS phenotype of the HB3 mutant. Taken together, our data demonstrate that the R. eutrophus aut gene encodes a protein involved in core LPS synthesis, which is functionally equivalent to the RfaE Domain II. More importantly, these experiments provided a biological demonstration that E. coli rfaE encodes a bifunctional protein. Unequivocal proof of the bifunctional nature of this protein will require in vitro biochemical experiments clearly demonstrating that RfaE can actually catalyze two separate reactions. The purification of the complete RfaE protein as well as each of its separate domains is underway in our laboratory.
The rfaE genes in E. coli and S. enterica were found downstream of glnE, and the same DNA strand contains both genes. Because of the sequence conservation and gene organization in both microorganisms, rfaE may have been acquired before E. coli and Salmonella diverged. This is in agreement with the G+C content of the rfaE DNA sequence, which is very similar to the average G+C content of E. coli and S. enterica. In contrast, the rfaE homologues in other nonenteric bacteria are not located in the vicinity of glnE. Also, in these cases the G+C content of rfaE is similar to that of the corresponding genus and species. Therefore, although it is difficult to trace the evolutionary path of rfaE, the available evidence suggests that this gene is evolutionarily older than the genes for the synthesis of outer core oligosaccharide components and O antigens (39, 43), in agreement with the conserved nature of the LPS inner core structure. A search for rfaE homologues in the available genomic databases of gram-negative bacteria showed that in Campylobacter jejuni, H. influenzae, H. pylori, P. aeruginosa, Streptomyces coelicolor, V. cholerae, and Yersinia pestis, the gene encodes Domain I and II. Separate genes encoding each domain may be found in N. gonorrhoeae, N. meningitidis, Bordetella pertussis, and Burkholderia pseudomallei, but the reasons for this separation are not obvious.
The intermediate steps of the biosynthesis pathway for ADP-LDHep are not completely elucidated. Eidels and Osborn (15, 16) postulated a pathway involving the isomerization of sedoheptulose 7-phosphate into glycero-manno-heptose-7 phosphate followed by the activity of a phosphomutase responsible for the transfer of the phosphate residue from carbon 7 to carbon 1, resulting in glycero-manno-heptose 1-phosphate (Fig. 1). Transfer of this sugar to ADP and a subsequent epimerization step complete this pathway. Since the genes involved in the isomerization and epimerization reactions have been previously identified and characterized (7, 8, 10) and the heptosyltransferase genes are also known (19), we conclude that rfaE must be involved in the intermediate steps of ADP-LDHep biosynthesis. However, the fact that Domain I is a putative kinase from the ribokinase family is difficult to reconcile with its role as a phosphomutase. The ribokinase family includes a large number of proteins whose function is the phosphorylation of sugars at positions 1 or 6 in the case of hexoses and at positions 1 or 5 in the case of riboses. None of the enzymes currently classified within the ribokinase family have phosphomutase activity. Recently, the crystal structure of the E. coli ribokinase in complex with ribose and dinucleotide has been determined (45), resulting in the identification of the critical amino acids for kinase activity. The majority of these residues are all conserved in Domain I, especially an aspartic acid located in an anion hole that is essential for catalysis (45). If Domain I is a sugar kinase, its role in the synthesis of ADP-LDHep as well as in the pathway of synthesis are not clear. One possibility is that the pathway lacks a phosphomutase reaction and a sugar kinase phosphorylates C1. This reaction, however, would result in glycero-manno-heptose-1,7-diphosphate, and there is no evidence of the existence of this sugar or its derivative ADP-glycero-manno-heptose 7-phosphate. Although some of the heptoses in the core are phosphorylated on carbon 7 (38), the phosphorylation appears to be due to the activity of enzyme(s) encoded by the waa cluster and probably occurs after the heptose residue has been incorporated onto the LPS core (19). Alternatively, Domain I may have phosphomutase activity, and the structural similarity to ribokinase may only be due to similar features involved in the recognition of the sugar phosphate. However, this would not explain the strong conservation of the kinase catalytic site in RfaE. If a sugar kinase step exists for the synthesis of glycero-manno heptose 1-phosphate, we have to postulate yet another step involving dephosphorylation of the 7-phosphate precursor. Thus, it is possible that instead of a phosphomutase, an unidentified sugar phosphate phosphatase acts to remove the phosphate in position 7 from either glycero-manno-heptose 7-phosphate or from a putative ADP-glycero-manno-heptose-1,7-diphosphate (49). We are currently exploring these possibilities by investigating a number of uncharacterized genes in E. coli K-12 with homologies to sugar phosphate phosphatases.
Domain II has strong homologies with the cytidylyltransferase superfamily. This family includes enzymes like glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis and the eukaryotic choline phosphate cytidylyltransferase, which are involved in teichoic acid and phospholipid biosynthesis, respectively (4). Also, three members of this family, pantoate β-alanine ligase, acetate:SH-citrate lyase, and phosphopantetheine adenylyltransferase, are ADP transferases (4, 21). Bork et al. (4) have shown that this family has structural conservation with the class I tRNA synthetases, all of which also use ATP. In addition, there are several bifunctional enzymes, such as FAD synthetase, PAPS synthase (forming 3′-phosphoadenosine 5′-phosphosulfate), and the transcriptional regulator NadR, that contain this domain (33). The crystal structure of one of the ADP transferases of this family, phosphopantetheine adenylyltransferase, has been recently established (21). Almost identical residues were found in the fold corresponding to the nucleotide-binding site (Fig. 5), which has the characteristics of a dinucleotide-binding fold (41). Therefore, it is possible that Domain II functions as a nucleotide sugar transferase. ADP-glycero-manno-heptose can be isolated from yeast and bacteria (24, 25), and in a previous study, Coleman has shown that the gmhD (formerly rfaD) gene codes for ADP-LDHep-6-epimerase (10), the last step prior to the transfer of LDHep to the core LPS. Therefore, it is likely that ADP is the nucleotide added in vivo to the phosphorylated d-glycero-d-manno-heptose. Since the sugar precursors for the biosynthesis of ADP-heptose are not readily available, it is difficult to determine the function of these two domains with certainty.
According to the current nomenclature for LPS biosynthesis genes (40), rfaE should be renamed with the prefix gmh (for glycero-manno-heptose synthesis). However, the lack of a clear functional assignment, as well as the presence of separate genes encoding each domain in some microorganisms, makes it difficult to reassign a meaningful new gene name to rfaE. To avoid more confusion, we prefer not to introduce a new gene name until further biochemical investigations clarify the intermediate steps of the biosynthesis pathway for ADP-heptose.
ACKNOWLEDGMENTS
We thank the colleagues mentioned or referenced in Table 1 for the gifts of strains and plasmids used in this study.
This study was supported by grants from the Natural Sciences and Engineering Research Council and the Medical Research Council of Canada to M.A.V.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 4.Bork P, Holm L, Koonin E V, Sander C. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins. 1995;22:259–266. doi: 10.1002/prot.340220306. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brick P, Bhat T N, Blow D M. Structure of the tyrosyl tRNA synthetase refined at 2.3 Ångströms resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- 7.Brooke J S, Valvano M A. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J Biol Chem. 1996;271:3608–3614. doi: 10.1074/jbc.271.7.3608. [DOI] [PubMed] [Google Scholar]
- 8.Brooke J S, Valvano M A. Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. J Bacteriol. 1996;178:3339–3341. doi: 10.1128/jb.178.11.3339-3341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable DNA cloning vectors derived from P15A cryptic plasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman W G., Jr The rfaD gene codes for ADP-l-glycero-d-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J Biol Chem. 1983;258:1985–1990. [PubMed] [Google Scholar]
- 11.Curtiss R, III, Charamella J, Stallions D R, Mays J A. Parental functions during conjugation. Bacteriol Rev. 1968;32:320–348. [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drazek E S, Stein D C, Deal C D. A mutation in the Neisseria gonorrhoeae rfaD homolog results in altered lipooligosaccharide expression. J Bacteriol. 1995;177:2321–2327. doi: 10.1128/jb.177.9.2321-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eidels L, Osborn M J. Lipopolysaccharide and aldoheptose biosynthesis in transketolase mutants of Salmonella typhimurium. J Biol Chem. 1971;68:1673–1677. doi: 10.1073/pnas.68.8.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eidels L, Osborn M J. Phosphoheptose isomerase, first enzyme in the biosynthesis of aldoheptose in Salmonella typhimurium. J Biol Chem. 1974;249:5642–5648. [PubMed] [Google Scholar]
- 17.Freter A, Bowien O. Identification of a novel gene, aut, involved in autotrophic growth of Alcaligenes eutrophus. J Bacteriol. 1994;176:5401–5408. doi: 10.1128/jb.176.17.5401-5408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havekes L M, Lugtenberg B J J, Hoekstra W P M. Conjugation deficient E. coli K-12 F-mutants with heptose-less lipopolysaccharide. Mol Gen Genet. 1976;146:43–50. doi: 10.1007/BF00267981. [DOI] [PubMed] [Google Scholar]
- 19.Heinrichs D E, Valvano M A, Whitfield C. Biosynthesis and genetics of lipopolysaccharide core. In: Brade H, Morrison D C, Vogel S, Opal S, editors. Endotoxin in health and disease. New York, N.Y: Marcel Dekker, Inc.; 1999. pp. 305–330. [Google Scholar]
- 20.Helander I M, Lindner B, Brade H, Altmann K, Lindberg A A, Rietschel E T, Zahringer U. Chemical structure of the lipopolysaccharide of Haemophilus influenzae strain I-69 Rd−/B+: description of a novel deep-rough chemotype. Eur J Biochem. 1988;177:483–492. doi: 10.1111/j.1432-1033.1988.tb14398.x. [DOI] [PubMed] [Google Scholar]
- 21.Izard T, Geerlof A. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 1999;18:2021–2030. doi: 10.1093/emboj/18.8.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klena J D, Ashford R S, Schnaitman C A. Role of the Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J Bacteriol. 1992;174:7297–7307. doi: 10.1128/jb.174.22.7297-7307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 24.Kocsis B, Kontrohr T. Isolation of adenosine 5′-diphosphate-l-glycero-d-mannoheptose, the assumed substrate of heptose transferase(s), from Salmonella minnesota R595 and Shigella sonnei Re mutants. J Biol Chem. 1984;259:11858–11860. [PubMed] [Google Scholar]
- 25.Kontrohr T, Kocsis B. Isolation of adenosine 5′-diphosphate-d-glycero-d-mannoheptose. An intermediate in lipopolysaccharide biosynthesis of Shigella sonnei. J Biol Chem. 1981;256:7715–7718. [PubMed] [Google Scholar]
- 26.Koronakis V, Koronakis E, Li J, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 27.Kovach M E, Phillips R W, Elzer P H, Roop R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 28.Lee N-G, Sunshine M G, Apicella M A. Molecular cloning and characterization of the nontypeable Haemophilus influenzae 2019 rfaE gene required for lipopolysaccharide biosynthesis. Infect Immun. 1995;63:818–824. doi: 10.1128/iai.63.3.818-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 30.Marolda C L, Valvano M A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1) J Bacteriol. 1993;175:148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marolda C L, Welsh J, Dafoe L, Valvano M A. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J Bacteriol. 1990;172:3590–3599. doi: 10.1128/jb.172.7.3590-3599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 33.Mushegian A. The purloined letter: bacterial orthologs of archeal NMN adenylyltransferase are domains within multifunctional transcription regulator NadR. J Mol Microbiol Biotechnol. 1999;1:127–128. [PubMed] [Google Scholar]
- 34.Nichols W A, Gibson B W, Melaugh W, Lee N-G, Sunshine M, Apicella M A. Identification of the ADP-l-glycero-d-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect Immun. 1997;65:1377–1386. doi: 10.1128/iai.65.4.1377-1386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Passador L, Linn T. Autogenous regulation of the RNA polymerase β subunit of Escherichia coli occurs at the translational level in vivo. J Bacteriol. 1989;171:6234–6242. doi: 10.1128/jb.171.11.6234-6242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive molecules. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 39.Reeves P R. Variation in O-antigens, niche-specific selection and bacterial populations. FEMS Microbiol Lett. 1992;79:509–516. doi: 10.1111/j.1574-6968.1992.tb14085.x. [DOI] [PubMed] [Google Scholar]
- 40.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 41.Rossmann M G, Liljas A, Branden C I, Banaszak L J. Evolutionary and structural relationships among dehydrogenases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 11. New York, N.Y: Academic Press; 1975. pp. 62–102. [Google Scholar]
- 42.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 43.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherburne C, Taylor D E. Effect of lipopolysaccharide mutations on recipient ability of Salmonella typhimurium for incompatibility group H plasmids. J Bacteriol. 1997;179:952–955. doi: 10.1128/jb.179.3.952-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigrell J A, Cameron A D, Jones T A, Mowbray S L. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 Å resolution: insights into a new family of kinase structures. Structure. 1998;6:183–193. doi: 10.1016/s0969-2126(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 46.Sirisena D M, MacLachlan P R, Liu S L, Hessel A, Sanderson K E. Molecular analysis of the rfaD gene, for heptose synthesis, and the rfaF gene, for heptose transfer, in lipopolysaccharide synthesis in Salmonella typhimurium. J Bacteriol. 1994;176:2379–2385. doi: 10.1128/jb.176.8.2379-2385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamaki S, Sato T, Matsuhashi M. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J Bacteriol. 1971;105:968–975. doi: 10.1128/jb.105.3.968-975.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valvano M A. Biosynthesis and genetics of ADP-heptose. J Endotoxin Res. 1999;5:90–95. [Google Scholar]
- 50.Valvano M A, Crosa J H. Aerobactin iron transport genes commonly encoded by certain ColV plasmids occur in the chromosome of a human invasive strain of Escherichia coli K1. Infect Immun. 1984;46:159–167. doi: 10.1128/iai.46.1.159-167.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valvano M A, Crosa J H. Molecular cloning, expression, and regulation in Escherichia coli K-12 of a chromosome-mediated aerobactin iron transport system from a human invasive isolate of E. coli K1. J Bacteriol. 1988;170:5529–5538. doi: 10.1128/jb.170.12.5529-5538.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valvano M A, Silver R P, Crosa J H. Occurrence of chromosome- or plasmid-mediated aerobactin iron transport systems and hemolysin production among clonal groups of human invasive strains of Escherichia coli K1. Infect Immun. 1986;52:192–199. doi: 10.1128/iai.52.1.192-199.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Heeswijk W C, Rabenberg M, Westerhoff H V, Kahn D. The genes of the glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol Microbiol. 1993;9:443–457. doi: 10.1111/j.1365-2958.1993.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 54.Venkatachalam K V, Fuda H, Koonin E V, Strott C A. Site-selected mutagenesis of a conserved nucleotide binding HXGH motif located in ATP sulfurylase domain of a human bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase. J Biol Chem. 1999;274:2601–2604. doi: 10.1074/jbc.274.5.2601. [DOI] [PubMed] [Google Scholar]
- 55.Vimont S, Dumontier S, Escuyer V, Berche P. The rfaD locus: a region of rearrangement in Vibrio cholerae O139. Gene. 1997;185:43–47. doi: 10.1016/s0378-1119(96)00625-7. [DOI] [PubMed] [Google Scholar]
- 56.Watson G, Paigen K. Isolation and characterization of an Escherichia coli bacteriophage requiring cell wall galactose. J Virol. 1971;8:669–674. doi: 10.1128/jvi.8.5.669-674.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitfield C, Valvano M A. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson R G, Gemski P, Stocker B A D. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972;70:527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 60.Zwahlen A, Rubin L G, Connelly C J, Inzana T J, Moxon E R. Alteration of the cell wall in Haemophilus influenzae type b by transformation with cloned DNA: association with attenuated virulence. J Infect Dis. 1985;152:485–492. doi: 10.1093/infdis/152.3.485. [DOI] [PubMed] [Google Scholar]