Abstract
The correct site for translation initiation for Escherichia coli WecA (Rfe), presumably involved in catalyzing the transfer of N-acetylglucosamine 1-phosphate to undecaprenylphosphate, was determined by using its FLAG-tagged derivatives. The N-terminal region containing three predicted transmembrane helices was found to be necessary for function but not for membrane localization of this protein.
Lipopolysaccharide (LPS), a complex glycolipid composed of lipid A, core oligosaccharide, and O antigen, is found in the outer membrane of gram-negative bacteria (15, 35). Previous work by Alexander and Valvano (1) showed that WecA (formerly rfe [27]) is required for the first step in O7 LPS synthesis, which involves the transfer of N-acetylglucosamine (GlcNAc) to an undecaprenylphosphate lipid carrier. Several studies have shown that WecA is also required for the biosynthesis of many O antigens containing GlcNAc in strains of Escherichia coli, Klebsiella pneumoniae O1, Shigella dysenteriae, S. flexneri, Salmonella enterica, and Yersinia enterocolitica (1, 5, 16, 17, 19, 28, 30, 37, 38), suggesting a general role for this protein in the biosynthesis of O-specific polysaccharides.
wecA is the first gene of the wec cluster, which governs the synthesis of enterobacterial common antigen (18), a GlcNAc-containing surface glycolipid shared by enteric bacteria (4). Genetic and biochemical data strongly support the conclusion that wecA is the structural gene for a tunicamycin-sensitive UDP-GlcNAc:undecaprenylphosphate GlcNAc-1-phosphate transferase (22, 23, 28). Interestingly, amino acid sequence similarities to the eukaryotic UDP-GlcNAc:dolicholphosphate GlcNAc-1-phosphate transferase (GPT) have been found in discrete regions of WecA (8, 39). Preliminary experiments in our laboratory (3) indicated that WecA is poorly expressed, making it difficult to obtain a protein preparation of sufficient purity and quantity for raising specific antibodies and for detailed structure-function studies. Furthermore, there are contradicting claims in the literature regarding the location of the initiation codon of wecA, since this site has not been determined experimentally (29), and the analysis of the published wecA sequence reveals three possible initiation codons. Thus, we designed a vector system to generate C-terminal protein fusions to the FLAG epitope, which we used to find conditions to monitor WecA by immunoblot analysis with anti-FLAG antibodies, to determine the correct site for initiation of translation, and to investigate the localization of hybrid proteins in the cytoplasmic membrane.
Construction of a WecA protein with a C-terminal FLAG epitope tag.
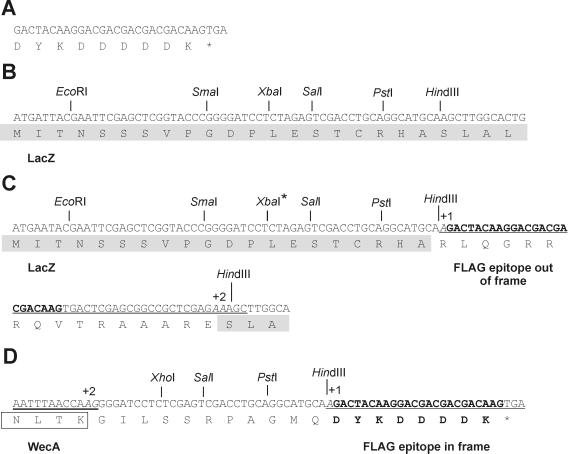
The properties of the strains and plasmids used in this work are indicated in Table 1. The FLAG C-terminal fusion vector pAA8 was constructed by modifying pUC18 (36). A 41-bp fragment encoding the FLAG epitope tag (14), followed by a UGA termination codon (Fig. 1A), was introduced into the HindIII site of pUC18 (Fig. 1B). For this purpose, pUC18 was digested with HindIII and the linearized DNA was used as a template in a PCR. The primers 5′-CCGCTCGAGAAAGCTTGGCACTGGCCGTCGTTTTAC-3′ and 5′-CCG CTCGAGTCACTTGTCGTCGTCGTCCTTGTAGTCTTGC ATGCCTG (FLAG oligonucleotide sequence underlined) were designed in such a manner that the final product after ligation would have one additional base at the 5′ end of the FLAG sequence and two extra bases 3′ from the FLAG's terminal UGA. Extra bases were necessary to restore the reading frame of the lacZ gene and at the same time cause a shift in the reading frame of the FLAG DNA sequence (Fig. 1C). The amplification product was gel purified, treated with T4 DNA kinase, ligated, and transformed into DH5α. Blue colonies expressing β-galactosidase activity on Luria broth plates supplemented with 0.2% (wt/vol) 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Roche Diagnostics, Dorval, Quebec, Canada) were isolated, and the recombinant plasmids were examined by restriction endonuclease analysis. The correct construct was verified by DNA sequencing to confirm the incorporation of the FLAG-encoding oligonucleotide within the coding region of the β-galactosidase gene (Fig. 1C). Thus, pAA8 could be used for the cloning of any protein gene sequence containing two extra bases at the C-terminal end to restore the reading frame of the FLAG sequence, resulting in the expression of a tagged fusion protein. At the same time, the UGA codon located downstream from the FLAG epitope sequence (Fig. 1A) would preclude the expression of β-galactosidase, facilitating the identification of fusion-positive clones as white colonies on X-Gal plates.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | endA hsdR supE thi-1 recA relA? gyrA φ80lacZΔM15 Δ(argF-lacZYA)U169 | Laboratory collection |
| JM109DE3 | endA recA gyrA thi hsdR17(rK− mK+) relA supE Δ(lac-proAB) [F′ traD36 proAB lacIq ZΔM15] | K. E. Sanderson |
| MV501 | VW187; wecA::Tn10 Tetr | 1 |
| VW187 | O7:K1; clinical isolate | 32 |
| Plasmids | ||
| pEX1 | Ampr ptac lacIqrrnBt | 26 |
| pKS | Bluescript II KS P, cloning vector; Ampr | 2 |
| pRL100 | 1.46-kb ClaI-XmaIII fragment containing wecA cloned in pBR322; Ampr | 22 |
| pUC18 | Cloning vector; Ampr | 36 |
| pAA8 | 41-bp fragment containing FLAG sequence in pUC18 HindIII site; Ampr | This work |
| pAA11 | 1.4-kb PCR fragment containing wecA cloned in pAA8; Ampr | This work |
| pAA12 | 1.4-kb fragment containing tagged wecA cloned in Bluescript II KS P; wecA+ Ampr | This work |
| pAA14 | 1.2-kb fragment containing part of wecA gene cloned in pEX1; wecA+ Ampr | This work |
| pAA15 | 0.98-kb fragment containing part of wecA gene cloned in pEX1; Ampr | This work |
| pAA16 | 0.76-kb fragment containing part of wecA gene cloned in pEX1; Ampr | This work |
| pAA19 | 1.5-kb fragment containing galF gene cloned in pAA8; Ampr | This work |
FIG. 1.
Construction of pAA8 (FLAG fusion cloning vector) and pAA11 (carrying the WecA-FLAG fusion). (A) DNA sequence of the oligonucleotide encoding the FLAG epitope tag. (B) DNA sequence of the multiple cloning site of pUC18 as described in the published sequence. The first 24 amino acids of the LacZ protein are indicated (shaded box). (C) Modifications of the pUC18 multiple cloning site giving rise to pAA8. A 41-bp fragment (underlined) encoding the FLAG epitope sequence (bold) followed by a stop codon was added into the HindIII site of pUC18. The FLAG sequence was shifted by 1 base (+1, base is italic) at the 5′ end. The reading frame of the lacZ gene was corrected by addition of 2 bases (+2, bases are italic) following the FLAG-encoding sequence. (D) Construction of WecA-FLAG fusion plasmid pAA11. The sequence of the C-terminal end of WecA (double underlining) and the last four amino acids of WecA (open box) are shown. Two base pairs (+2, italic) were added to correct the reading frame such that the FLAG epitope (bold) is fused in frame to the C terminus of WecA. The XbaI site in the multiple cloning site of pUC18 (B, asterisk) was changed to XhoI (asterisk), and this strategy also added 11 intervening amino acids between the end of WecA and the FLAG epitope tag.
The WecA-FLAG hybrid was constructed by PCR amplification of a 1.4-kb fragment from pRL100. This fragment was obtained by using a 5′ primer incorporating an EcoRI site (5′-TCGATGCAATGGAAT-3′) and a 3′ primer (5′-TTGGTTAAATTGGGGCT-3′) containing two extra bases at its 5′-terminal end to allow the epitope tag to be expressed in frame with WecA. The resulting 1.4-kb amplicon was digested with EcoRI, ligated to the EcoRI and SmaI sites of pAA8 (Fig. 1D), and transformed into DH5α. The correct fusion in one of the recombinant plasmids, designated pAA11, was verified by DNA sequencing (Fig. 1D). As a positive control, we constructed a GalF-FLAG (pAA19) fusion by using a similar strategy (data not shown). GalF is a cytoplasmic protein involved in the regulation of UDP-glucose pyrophosphorylase and has been shown to be expressed under different conditions (20).
Detection of WecA-FLAG and GalF-FLAG protein fusions by immunoblotting and functional complementation of a wecA::Tn10 mutant.
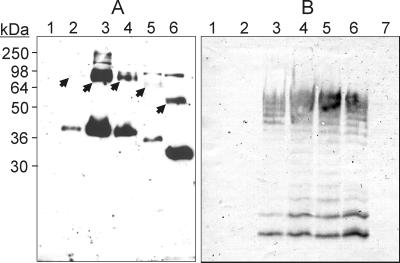
E. coli JM109DE3 containing either pAA11 or pAA19 was lysed following induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. The lysis buffer consisted of 0.01 M sodium phosphate, 1% β-mercaptoethanol, 1% sodium dodecyl sulfate (SDS), and 6 M urea. Lysates were mixed with equal volumes of loading sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.1% bromophenol blue) and incubated at 45°C for 30 min before loading onto an SDS–10% polyacrylamide gel. The denaturing temperature of 45°C was crucial, since incubation at higher temperatures resulted in failure to detect WecA-FLAG. Similar difficulties with boiling have been previously encountered with other integral membrane proteins (10, 31). In contrast, the GalF-FLAG fusion protein was readily detected under all of the conditions examined (data not shown). Transfer of protein to nitrocellulose membranes was performed according to standard procedures, and membranes were incubated with the FLAG M2 monoclonal antibody (MAb) (Sigma Chemical Company, St. Louis, Mo.) and then with horseradish peroxidase-linked sheep anti-mouse immunoglobulin G (Amersham Pharmacia Biotechnology, Piscataway, N.J.). Detection by chemiluminescence assay was performed by using the BM Chemiluminescence Blotting Substrate (Roche Diagnostics) as recommended by the manufacturer. A polypeptide band with a molecular mass of approximately 38 kDa (see lane 2 in Fig. 3A) did not appear in the control lysate prepared from JM109DE3 transformed with pAA8 alone (see Fig. 3A, lane 1). The 38-kDa polypeptide was about 2 kDa lighter than the predicted mass of WecA as deduced from its translated DNA sequence (40.9 kDa). A similar aberrant migration has been observed in the case of the WecA eukaryotic homologue GPT (9) and may be due to the hydrophobic nature of integral membrane proteins (13).
FIG. 3.
(A) Detection of WecA derivatives tagged with the FLAG epitope by immunoblotting with MAb M2. JM109DE3 cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes, and processed as described in the text. Lanes: 1, pAA8; 2, pAA11; 3, pAA12; 4, pAA14; 5, pAA15; 6, pAA16. Arrows indicate polypeptides with higher molecular masses (barely visible in lanes 2 and 5) that may represent oligomers of WecA. The positions of the following molecular mass standards are shown: myosin (250 kDa), bovine serum albumin (98 kDa), glutamic acid dehydrogenase (64 kDa), alcohol dehydrogenase (50 kDa), carbonic anhydrase (36 kDa), and myoglobin (30 kDa). (B) Immunoblot of LPS samples prepared from MV501 (lanes 1 to 5 and 7) and VW187 (lane 6), which were reacted with anti-O7 antibodies. MV501 was transformed with the various wecA derivatives as follows: lane 1, pAA16; lane 2, pAA15; lane 3, pAA14; lane 4, pAA12; lane 5, pAA11.
To increase the expression of the WecA-FLAG, we cloned a 1.4-kb EcoRI-PvuII fragment of pAA11 containing the gene fusion into the EcoRI-SmaI sites of pBluescript KS+, resulting in pAA12. Extracts from cells containing this plasmid had large amounts of WecA-FLAG (see Fig. 3A, lane 3), suggesting that the presence of the strong T7 promoter increased the level of gene expression. A larger protein band of approximately 75 kDa with a strong reaction with the M2 MAb was also detected in cells with pAA12 and pAA11 (see Fig. 3A, arrows). The mild denaturing conditions used for the preparation of cell lysates may not be sufficient to disperse protein aggregates and could explain the presence of this band, suggesting the possibility that WecA is able to oligomerize. Similar observations have been made with GPT (9). In addition, we also observed another band of approximately 83 kDa that appeared often in all samples, including those prepared from cells containing either vector DNA or no plasmid. This band was not due to cross-reaction with the horseradish peroxidase-labeled secondary antibody and probably represents the existence of an M2 cross-reactive epitope in cellular proteins of E. coli. The nature of this band was not studied further.
To confirm that the unique 38-kDa polypeptide band expressed by pAA11 and pAA12 was indeed WecA, we carried out a functional complementation experiment by using E. coli MV501. This strain is a derivative of VW187 containing a wecA::Tn10 insertion and is thus unable to express O7-specific LPS (see Fig. 3B, lane 7) (1). LPS was extracted and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by immunoblotting using anti-O7 antibodies (21). The formation of O7 LPS in MV501 was restored to wild-type levels following transformation with pAA11 and pAA12 (see Fig. 3B, lanes 4 and 5). This experiment demonstrated that addition of 19 amino acids, including the 8-amino-acid FLAG epitope tag, to the C terminus of WecA did not affect its function.
Use of WecA-FLAG to determine the precise translation initiation site of the wecA coding sequence.
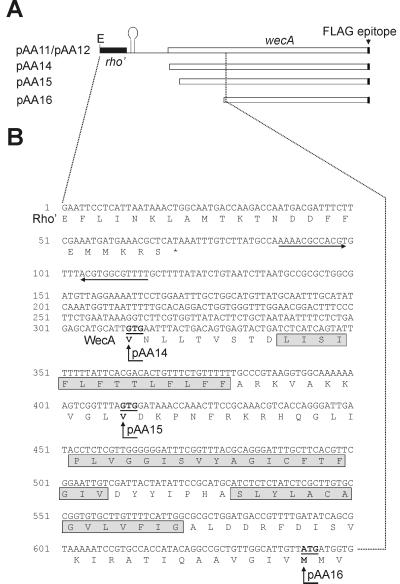
An examination of the E. coli wecA DNA sequence reported in the literature shows several potential starts for translation. Ohta et al. (24) and Meier-Dieter et al. (23) predicted a putative initiation codon located 570 nucleotides downstream from the termination codon of rho (Fig. 2B). However, the coding region can be extended upstream from this site until nucleotide 312, and this is the start annotated in the GenBank entry for this gene by the E. coli Genome Project (11). To experimentally identify the initiation of translation of the wecA gene, several fragments were amplified by PCR from pAA11 and cloned into pEX1 (26). This plasmid is a derivative of the expression vector pKK223-3 with the lacIq gene inserted into the PvuII site. pEX1 contains the ptac promoter and a ribosomal binding site that can be utilized to express inserts cloned in the EcoRI and SmaI sites, provided they have an initiation codon. We investigated three potential start sites of translation for the wecA gene. The first start site to be examined was ATG642 (Fig. 2A and B, pAA16). A PCR fragment encoding the WecA-FLAG fusion starting at ATG642 with an EcoRI site at the 5′ end was obtained. The 0.76-kb fragment was digested with EcoRI and ligated into the EcoRI-SmaI site of pEX1. Two additional plasmids with the other potential translation start sites were constructed in a similar manner by cloning PCR fragments of 0.98 and 1.2 kb encoding wecA starting at GTG411 and GTG312, respectively (Fig. 2A and B, pAA15 and pAA16). The correct junctions in all of these plasmids were verified by DNA sequencing. pAA14, pAA15, and pAA16 were tested for WecA function by complementation analysis in strain MV501. pAA14 was the only plasmid that restored the formation of O7 LPS as determined by slide agglutination and anti-O7 immunoblotting (Fig. 3B, lane 3). pAA14 expressed a polypeptide detectable with MAb M2 that migrated with a gel mobility identical to that of the polypeptide expressed by pAA11 and pAA12 (Fig. 3A, lane 4), while pAA15 and pAA16 expressed MAb M2-reacting polypeptides with lower molecular masses (Fig. 3A, lanes 5 and 6). The reduction in the masses of these polypeptides was consistent with the predicted sizes of the deletion-containing proteins. Since pAA14 was the only construct expressing a functional protein with a mass comparable to that of wild-type WecA, we concluded that GTG312 is the initiation codon of the wecA gene. Interestingly, a larger protein band was also found in all cases. This band had a decrease in mass that was proportional to the decrease found in the monomeric proteins. The molecular mass of this band, together with its migration behavior in the various constructs, provides further evidence consistent with a dimeric form of WecA. Results also suggest that if dimerization indeed occurs, it may depend on sequences located outside of the missing N-terminal regions.
FIG. 2.
(A) Partial maps of the plasmids containing wecA. pAA11 and pAA12 contain the 3′ end of rho. E, EcoRI site. The putative hairpin structure involved in transcription termination of rho is indicated. pAA14, pAA15, and pAA16 do not include sequences upstream of the putative start codons of wecA. (B) Nucleotide sequence of the rho-wecA region of E. coli K-12 and deduced amino acid sequences (GenBank accession no. M76129) (22). Horizontal arrows indicate the putative transcriptional terminator of rho. Putative starting sites for translation of wecA (underlined) are GTG312, GTG411, and ATG642. The 5′ ends of cloned wecA derivatives in pAA14, pAA15, and pAA16 are also indicated (vertical arrows). Amino acids involved in predicted transmembrane domains I, II, and III are indicated (shaded boxes).
N-terminal amino acid sequences are not required for membrane insertion of WecA.
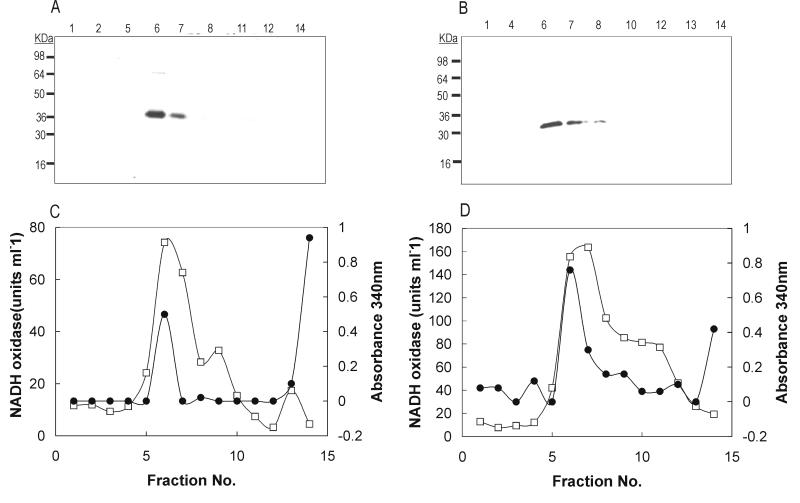
Analysis of WecA with the dense alignment surface-transmembrane segment prediction algorithm that can predict the presence of α helices in bacterial membrane proteins (6) showed that WecA has 11 predicted transmembrane domains (data not shown). Transmembrane domain I was not present in WecA334 (pAA15), while WecA257 (pAA16) lacked transmembrane domains I, II, and III (Fig. 2B). Since these two derivatives of WecA were nonfunctional, it is possible that the missing region of the N terminus is required for WecA function. Alternatively, the missing region may contain information necessary for the proper insertion of WecA within the cytoplasmic membrane. To distinguish between these two possibilities, we analyzed membrane extracts of cells expressing WecA367 (wild type) and WecA257 (lacking transmembrane domains I to III), which were fractionated by sucrose density gradient centrifugation as previously described (12). Identification of the isolated fractions was confirmed by the distribution of NADH oxidase, which was estimated by measuring the rate of decrease in A340 at 22°C (25). The presence of outer membrane porins in the fractions was examined by SDS-PAGE, followed by silver staining. Immunoblot analysis using MAb M2 showed that both WecA367 (Fig. 4A, lanes 6 and 7) and WecA257 (Fig. 4B, lanes 6 and 7) are present in fractions containing cytoplasmic membrane components, as identified by high NADH oxidase activity (Fig. 4C and D) and lack of outer membrane porins (data not shown). No detectable WecA protein was found in the pellet of the sucrose density gradients, ruling out the presence of inclusion bodies (12). These results confirm that WecA367 (wild-type WecA) is a membrane protein. Moreover, since WecA257 was also found in the cytoplasmic membrane fraction, we concluded that the first 110 N-terminal amino acids of WecA are not required for membrane insertion. Polytopic cytoplasmic membrane proteins are not processed by signal peptidase, and their export process has not been completely elucidated (7). A model has been proposed involving the spontaneous insertion into the membrane of pairs of hydrophobic segments that are eventually resolved so that each hydrophobic segment is flanked on its cytoplasmic side by positively charged amino acids (33, 34). This mechanism of membrane insertion is independent of the common pathway for secretion of proteins. Given that the N-terminal region of WecA appears not to be required for membrane insertion, it is possible that this protein is inserted in the membrane by a sec-independent mechanism involving amino acids in the middle and/or the C-terminal part of the protein. This conclusion is further supported by preliminary results indicating that the last transmembrane helix is essential for WecA protein expression (3).
FIG. 4.
Cell fractionation by sucrose density gradients of JM109DE3 cells containing pAA14 (A and C) and pAA16 (B and D), expressing WecA367 and WecA257, respectively. (A and B) Immunoblot analysis with MAb M2 of fractions collected from sucrose gradients. (C and D) Protein concentration (●) and NADH oxidase activity (□) profiles of the various fractions, as determined by measurement of A340 (right vertical axis) and units of enzyme activity per milliliter (left vertical axis), respectively, are shown.
Further experiments concerning the confirmation of the predicted topological model and identification of functional domains are under way, facilitated by our ability to detect the WecA protein. The pAA8 vector can also be used for epitope tagging of other proteins participating in LPS biosynthesis. This strategy may prove to be useful for the examination of protein-protein interactions among the various components that are presumed to be implicated in the processing of O antigen.
Acknowledgments
A.O.A. was supported in part by an Ontario Graduate Scholarship. This work was supported by grant MT10206 to M.A.V. from the Medical Research Council of Canada.
We thank the members of our laboratory for helpful discussions and suggestions.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alting-Mees M A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amer, A., and M. A. Valvano. Unpublished results.
- 4.Barr K, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. J Biol Chem. 1987;262:7142–7150. [PubMed] [Google Scholar]
- 5.Clarke B R, Bronner D, Keenleyside W J, Severn W B, Richard J C, Whitfield C. Role of Rfe and RfbF in the initiation of biosynthesis of d-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J Bacteriol. 1995;177:5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 7.Dalbey R D, Kuhn A, von Heijne G. Directionality in protein translocation across membranes: the N-tail phenomenon. Trends Cell Biol. 1995;5:380–383. doi: 10.1016/s0962-8924(00)89079-0. [DOI] [PubMed] [Google Scholar]
- 8.Dal Nogare A R, Lehrman M A. Conserved sequences in enzymes of the UDP-GlcNAc/MurNAc family are essential in hamster UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase. Glycobiology. 1988;8:625–632. doi: 10.1093/glycob/8.6.625. [DOI] [PubMed] [Google Scholar]
- 9.Dan N, Lehrman M A. Oligomerization of hamster UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase, an enzyme with multiple transmembrane spans. J Biol Chem. 1997;272:14214–14219. doi: 10.1074/jbc.272.22.14214. [DOI] [PubMed] [Google Scholar]
- 10.Dan N, Middleton R B, Lehrman M A. Hamster UDP-N-acetylglucosamine:dolichol-P N-acetylglucosamine-1-P transferase has multiple transmembrane spans and a critical cytosolic loop. J Biol Chem. 1996;271:30717–30724. doi: 10.1074/jbc.271.48.30717. [DOI] [PubMed] [Google Scholar]
- 11.Daniels D L, Plunket III G, Burland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 12.Fiermonte G, Walker J E, Palmieri F. Abundant bacterial expression and reconstitution of an intrinsic membrane-transport protein from bovine mitochondria. Biochem J. 1993;294:293–299. doi: 10.1042/bj2940293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grisshammer R, Tate C G. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- 14.Howard M, Du Vall D M, Devor D C, Dong J, Henze K, Frizzell R A. Epitope tagging permits cell surface detection of functional CFTR. Am J Physiol. 1995;269:C1565–C1576. doi: 10.1152/ajpcell.1995.269.6.C1565. [DOI] [PubMed] [Google Scholar]
- 15.Jann K, Jann B. Structure and biosynthesis of O-antigens. In: Rietschel E T, editor. Handbook of endotoxin. Chemistry of endotoxin. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 138–186. [Google Scholar]
- 16.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 17.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn H-M, Meier U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 19.Mäkelä P H, Jahkola M, Lüderitz O. A new gene cluster rfe concerned with the biosynthesis of Salmonella lipopolysaccharide. J Gen Microbiol. 1970;60:91–106. doi: 10.1099/00221287-60-1-91. [DOI] [PubMed] [Google Scholar]
- 20.Marolda C L, Valvano M A. The GalF protein of Escherichia coli is not a UDP-glucose pyrophosphorylase but interacts with GalU protein possibly to regulate cellular levels of UDP-glucose. Mol Microbiol. 1996;22:827–840. doi: 10.1046/j.1365-2958.1996.01531.x. [DOI] [PubMed] [Google Scholar]
- 21.Marolda C L, Welsh J, Dafoe L, Valvano M A. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7-K1 strain VW187. J Bacteriol. 1990;172:3590–3599. doi: 10.1128/jb.172.7.3590-3599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 23.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 24.Ohta M, Ina K, Kusuzaki K, Kido N, Arakawa Y, Kato N. Cloning and expression of the rfe-rff gene cluster of Escherichia coli. Mol Microbiol. 1991;5:1853–1862. doi: 10.1111/j.1365-2958.1991.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 25.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 26.Passador L, Linn T. Autogenous regulation of the RNA polymerase β subunit of Escherichia coli occurs at the translational level in vivo. J Bacteriol. 1989;171:6234–6242. doi: 10.1128/jb.171.11.6234-6242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 28.Rick P D, Hubbard G L, Barr K. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rick P D, Silver R P. Enterobacterial common antigen and capsular polysaccharides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 104–122. [Google Scholar]
- 30.Schmidt G, Mayer H, Makela P H. Presence on the rfe genes in Escherichia coli: their participation in biosynthesis of O antigen and enterobacterial common antigen. J Bacteriol. 1976;127:755–762. doi: 10.1128/jb.127.2.755-762.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seol W, Shatkin A J. Membrane topology model of Escherichia coli α-ketoglutarate permease by PhoA fusion analysis. J Bacteriol. 1993;175:565–567. doi: 10.1128/jb.175.2.565-567.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valvano M A, Crosa J H. Aerobactin iron transport genes commonly encoded by certain ColV plasmids occur in the chromosome of a human invasive strain of Escherichia coli K1. Infect Immun. 1984;46:159–167. doi: 10.1128/iai.46.1.159-167.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Heijne G. Getting greasy: how transmembrane polypeptide segments integrate into the lipid bilayer. Mol Microbiol. 1997;24:249–253. doi: 10.1046/j.1365-2958.1997.3351702.x. [DOI] [PubMed] [Google Scholar]
- 34.von Heijne G. Membrane protein assembly: rules of the game. Bioessays. 1995;17:25–30. doi: 10.1002/bies.950170107. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 37.Yao Z, Valvano M A. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in virulence of Yersinia enterocolitica serotype O:8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X, Lehrman M A. Cloning, sequence, and expression of a cDNA encoding hamster UDP-GlcNAc:dolichol phosphate-N-acetylglucosamine-1-phosphate transferase. J Biol Chem. 1990;265:14250–14255. [PubMed] [Google Scholar]