ABSTRACT
Chronic obstructive pulmonary disease (COPD) is one of the underlying conditions in adults of any age that place them at risk for developing severe illnesses associated with COVID-19. To determine whether SARS-CoV-2’s cellular tropism plays a critical role in severe pathophysiology in the lung, we investigated its host cell entry receptor distribution in the bronchial airway epithelium of healthy adults and high-risk adults (those with COPD). We found that SARS-CoV-2 preferentially infects goblet cells in the bronchial airway epithelium, as mostly goblet cells harbor the entry receptor angiotensin-converting enzyme 2 (ACE2) and its cofactor transmembrane serine protease 2 (TMPRSS2). We also found that SARS-CoV-2 replication was substantially increased in the COPD bronchial airway epithelium, likely due to COPD-associated goblet cell hyperplasia. Likewise, SARS-CoV and Middle East respiratory syndrome (MERS-CoV) infection increased disease pathophysiology (e.g., syncytium formation) in the COPD bronchial airway epithelium. Our results reveal that goblet cells play a critical role in SARS-CoV-2-induced pathophysiology in the lung.
IMPORTANCE SARS-CoV-2 or COVID-19’s first case was discovered in December 2019 in Wuhan, China, and by March 2020 it was declared a pandemic by the WHO. It has been shown that various underlying conditions can increase the chance of having severe COVID-19. COPD, which is the third leading cause of death worldwide, is one of the conditions listed by the CDC which can increase the chance of severe COVID-19. The present study uses a healthy and COPD-derived bronchial airway epithelial model to study the COVID-19 and host factors which could explain the reason for COPD patients developing severe infection due to COVID-19.
KEYWORDS: SARS-CoV-2, COVID-19, goblet cells, ciliated cells, COPD, airway epithelium, air-liquid interface, syncytium, cell sloughing, goblet cell hyperplasia, MUC5AC, MUC5B
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, a causative agent of coronavirus disease 2019, COVID-19) emerged in December 2019 in Wuhan, China. Since then, this pathogen has caused havoc in health care systems worldwide and consequentially ravaged the economy of countries with COVID-19 outbreaks. SARS-CoV-2 is a nonsegmented, positive-sense, single-strand RNA virus that causes both upper and lower respiratory tract infections. Coronavirus infection begins with the host cell entry, particularly by receptor-mediated endocytosis (1, 2). The tissue expression and distribution of the SARS-CoV-2 entry receptor angiotensin-converting enzyme 2 (ACE2) and its cofactor transmembrane serine protease 2 (TMPRSS2) determine the tropism of virus infection (3, 4). The ACE2 expression on cells dictates entry of both SARS-CoV and SARS-CoV-2 in the human airway epithelium (5, 6). For successful entry into cells, SARS-CoV-2 uses the serine protease TMPRSS2 for S protein priming (4). ACE2 is highly expressed in the small intestine, testes, kidneys, heart, thyroid, and adipose tissue, is expressed at moderate expression levels in the lung, colon, liver, bladder, and adrenal gland and at the lowest levels in the blood, spleen, bone marrow, brain, blood vessels, and muscle (5, 7). Previous studies reported that ACE2 expression in the lungs is predominantly observed in alveolar type 2 (AT2) cells (8–11), but ciliated cells also express ACE2 in the respiratory epithelium (12). Recent transcriptome sequencing (RNA-seq)-based studies have suggested that ACE2 is highly expressed on goblet cells in the nasal airways and on secretory cells in subsegmental bronchial branches of the lung (9, 11, 13). Although ACE2 and TMPRSS2 expressions are higher in goblet cells than in ciliated cells (9, 11, 13, 14), it appears that goblet cells are underappreciated in the SARS-CoV-2 infection studies. There are four major modes of SARS-CoV-2 transmissions: direct contact, indirect contact, droplet, and aerosol (15, 16). However, the dynamics of SARS-CoV-2 transmission are complex and have not been fully characterized (17–21). Although aerosol transmission was not considered a mode of transmission at the beginning of the SARS-CoV-2 outbreak, a number of studies have shown that it is a major contributor to the outbreak (22–24); however, both respiratory droplet and direct contact are continually contributing to person-person transmission during the outbreak (17, 21, 25–28). Hypersecretion of mucus has been reported in SARS-CoV-2-infected patients (29, 30). The possibility that SARS-CoV-2 infects goblet cells could explain the presence of viral RNA in sputum (31) and might explain the efficient transmission of the virus from person to person (32, 33).
SARS-CoV-2 infection induced COVID-19 clinical symptoms including fever, cough, and myalgias often similar to influenza virus infection (34). However, a subset of COVID-19 patients advances to severe acute respiratory distress syndrome (ARDS) (35, 36). Chest computed tomography (CT) scans of COVID-19 cases revealed bilateral pulmonary parenchymal ground-glass opacity with pulmonary consolidation in bilateral diffuse distribution and, less often, with a rounded morphology in a peripheral lung distribution (37). The histopathological findings of the fatal COVID-19 cases appeared to be consistent with the SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) infections (38, 39) and demonstrated damage of pneumocytes with hyaline membrane formation, interstitial lymphocyte infiltration, and multinucleated syncytial cells (40, 41). Importantly, respiratory failure and airway obstruction in COVID-19 patients are mostly caused by the formation of mucus plugs. One study reported that severe mucoid tracheitis was identified in 33% of COVID-19 autopsies (30). Pathological analysis of COVID-19 patients showed that bronchioles were plugged by mucus, accompanied by goblet cell hyperplasia in the airway epithelium (42). Goblet cells in the surface epithelium produce mucus. Importantly, goblet cell hyperplasia is a characteristic pathological feature of chronic obstructive pulmonary disease (COPD) patients, who are vulnerable to severe disease associated with COVID-19 (43–45). In fact, COPD is one of the high-risk factors for severe illness associated with COVID-19 (46–48). Therefore, it is prudent to determine to what extent SARS-CoV-2 infects goblet cells in the lung.
Here, we used multicellular pseudostratified bronchial airway epithelium models (in vitro) for identification of the SARS-CoV-2 entry receptor and its cofactor distributions. We found that SARS-CoV-2 primarily infects goblet cells due to the high expression of both ACE2 and TMPRSS2 in these cells. Goblet cell hyperplasia increases SARS-CoV-2 infection in the COPD airway epithelium. Thus, SARS-CoV-2 replication in goblet cells may explain the development of more severe COVID-19 in COPD patients.
RESULTS
The airway epithelium model recapitulates the chronic bronchial characteristics of COPD.
We established in vitro airway epithelium models by differentiating normal human bronchial epithelial (NHBE) cells from either healthy adults or high-risk adults (those with COPD) at the air-liquid interface (ALI) using an optimized protocol (49). We found that 4 weeks of differentiation provides a fully differentiated pseudostratified mucociliary airway epithelium for both conditions (Fig. 1A and B), which allows the use of this model for side-by-side comparisons. Primary NHBE cells from healthy adults differentiated primarily into a pseudostratified columnar epithelium, whereas the NHBE cells of COPD donors differentiated into a mix of pseudostratified and stratified columnar epithelium that contained all three main cell types of the respiratory epithelium (50–52): ciliated cells, goblet cells, and basal cells (Fig. 1B). We found that the apical site of the epithelium mainly consists of ciliated and goblet cells (Fig. 1C and D). Studies have shown that mucin 5AC (MUC5AC) is predominately expressed by airway goblet cells and that mucin 5B (MUC5B) is expressed by goblet cells of submucosal glands (53). COPD patients have higher expression of MUC5B in the bronchiolar lumen and MUC5AC in the bronchial epithelium (54). MUC5AC and MUC5B play a crucial role in goblet cell hyperplasia in COPD patients (55, 56). Club cells are the progenitors of goblet cells, which might express both MUC5AC and MUC5B (57, 58). As expected, we found heterogenicity in the cell population with differential expression of common goblet cell markers (Fig. 1E) (11). Basal cells are known for their self-renewal property and give rise to multiple types of differentiated airway epithelial cells (59). For the detection of basal cells, we sectioned the epithelium and stained the sections for the basal cell marker P63 (60, 61). We found that basal cells reside at the basal membrane of the epithelium and that the COPD epithelium has more basal cells than the healthy epithelium, which is known from the respiratory epithelium of COPD patients (Fig. 1F) (62, 63).
FIG 1.
The airway epithelium model recapitulates the bronchial characteristics of COPD. (A) Airway epithelium derived from differentiated NHBE cells obtained from a healthy adult or COPD patient were fixed, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). The nuclei are stained dark purple, and the cytoplasmic components are pink. Bar = 50 μm. (B) The sectioned epithelia were stained with alcian blue. Mucosubstances are stained blue, whereas the cytoplasmic components are pale pink, and the nuclei are pink/red. Bar = 50 μm. (C) The apical sites of the airway epithelia were fixed, permeabilized, and stained for the goblet cell marker MUC5AC (anti-MUC5AC, green), the ciliated cell marker acetyl-alpha tubulin (anti-acetyl-alpha tubulin, cyan), and F-actin (rhodamine phalloidin, red); the nuclei were also stained with DAPI (blue). Bar = 10 μm. (D) The sectioned epithelia were stained for the ciliated cell marker acetyl-alpha tubulin (anti-acetyl-alpha tubulin, cyan), the goblet cell marker MUC5AC (anti-MUC5AC, green), and F-actin (rhodamine phalloidin, red); the nuclei were also stained with DAPI (blue). Bar = 20 μm. (E) The apical sites of the airway epithelia were fixed and stained for two goblet cell markers, MUC5AC and MUC5B (anti-MUC5AC [green] and anti-MUC5B [cyan], respectively) and F-actin (rhodamine phalloidin, red); the nuclei were also stained with DAPI (blue). Bar = 10 μm. (F) The sectioned epithelia were stained for the basal cell marker P63 (anti-P63, green), the goblet cell marker MUC5B (anti-MUC5B, cyan), and F-actin (rhodamine phalloidin, red); the nuclei were also stained with DAPI (blue). Bar = 20 μm. (G) The MUC5B+ goblet cells were counted from tiled images of the airway epithelium stained for MUC5B (anti-MUC5B, cyan) (described above) obtained with a Leica DMi8 epifluorescence microscope. A total of approximately 3,000 cells were counted using ImageJ software from each epithelium to determine the ratio. The error bar represents the SEMs. The statistical significance was determined by unpaired two-tailed t tests. The results from one independent experiment are shown. (H) The MUC5AC+ goblet cells were counted using ImageJ software from random fields in confocal images of the airway epithelium stained for MUC5AC (anti-MUCAC, cyan) (described above). A total of approximately 2,000 cells were counted from each epithelium to determine the ratio. The error bar represents the SEMs. The statistical significance was determined by unpaired two-tailed t tests. The results from two independent experiments are shown. (I) The sectioned epithelia were stained for MUC5B (anti-MUC5B, cyan); the nuclei were stained with DAPI (blue). Bar = 20 μm. (J) Transepithelial electrical measurements (TEERs) of the airway epithelia were obtained. The data were obtained by combining three independent Transwell reads, and each Transwell read was an average of three independent reads. The error bars represent the SEMs. (K) The ciliary beat frequency (CBF) was measured on the airway epithelia. The data were obtained by combining three independent Transwell reads, and each Transwell read was an average of six random point reads. The error bars represent the SEMs. The statistical significance was determined by unpaired two-tailed t tests. The results from one independent experiment are shown.
COPD is associated with abnormal airway and alveolar responses during exposure to noxious stimuli (64). Because our COPD airway epithelium model was differentiated from NHBE cells, it should recapitulate the bronchial airway phenotypes instead of the alveolar phenotype as more commonly associated with emphysema (65). To determine whether our in vitro COPD airway epithelium model recapitulates some of the bronchial pathophysiological characteristics of COPD, we focused on two important phenotypes: goblet cell hyperplasia and squamous metaplasia. First, we compared the number of MUC5AC+ or MUC5B+ goblet cells between healthy and COPD epithelium. With respect to airway mucins, MUC5AC is predominantly expressed in asthma patients (66, 67); in contrast, MUC5B is known to be highly expressed in COPD patients (67, 68). Indeed, we found that MUC5B+ cells were significantly higher in COPD airway epithelium than in healthy epithelium (Fig. 1G). Although MUC5AC+ cells were also higher in COPD epithelium, the difference is not statistically significant compared to healthy samples (Fig. 1H). Thus, we found that MUC5B- or MUC5AC-positive goblet cells or both were higher in COPD than healthy epithelium, which is in line previously published COPD clinical features: the presence of abnormally high MUC5B and MUC5AC in the sputum of COPD patients and MUC5B is the predominant mucin in COPD (67, 69). The higher number of goblet cells in the COPD epithelium suggests a persistent goblet cell differentiation, which results in goblet cell hyperplasia (70–72). Indeed, we found a patch of a higher number of goblet cells with extensive mucus secretion in the COPD epithelium (Fig. 1I, center). We also found an apparent loss of pseudostratified epithelium accompanied by squamous metaplasia in the COPD epithelium, which is a common pathological phenotype in COPD (Fig. 1I, right) (52).
To determine the biophysical properties of respiratory epithelium, we assessed the tissue membrane integrity (transepithelial electrical resistance, TEER) and ciliary function (ciliary beat frequency, CBF), and we found no significant difference in these biophysical properties between healthy and COPD epithelium (Fig. 1J and K). These results indicated that NHBE cells from patients with COPD produced a mix of pseudostratified and stratified highly differentiated mucociliary epithelium with appropriate biophysical properties.
Goblet cells harbor SARS-CoV-2 entry receptor ACE2 and its cofactor TMPRSS2.
SARS-CoV-2 infects the human airway epithelium, and virus entry depends on the host cell receptors ACE2 and its cofactor TMPRSS2 (4). We quantified the ACE2 transcript levels in a lung epithelial cell line (A549 cells) and primary NHBE cells in a monolayer or in the differentiated airway epithelium by real-time PCR. We did not detect ACE2 transcripts in the A549 cells (see Fig. S1A in the supplemental material), which might indicate low or no expression of ACE2, confirming previous data (4, 73–75). However, we detected ACE2 transcripts in primary NHBE cells in both the monolayer and differentiated airway epithelium (Fig. S1A). Despite their similar biophysical properties (tissue barrier integrity and ciliary function [Fig. 1J and K, respectively]), ACE2 expression was higher in the COPD epithelium compared to those derived from a healthy donor (Fig. S1A to C). We then visualized the expression of ACE2 in the airway epithelium by immunofluorescence imaging. We observed ACE2 expression in both the healthy and COPD airway epithelium and found that ACE2 expression hardly overlapped with cilia on the apical site of the epithelium (Fig. 2A and Fig. S1D). Nevertheless, low levels of ACE2 expression were observed on ciliated cells in our model (Fig. S1E). These results suggest that the SARS-CoV-2 entry receptor ACE2 is mainly expressed on nonciliated cells in the respiratory epithelium.
FIG 2.
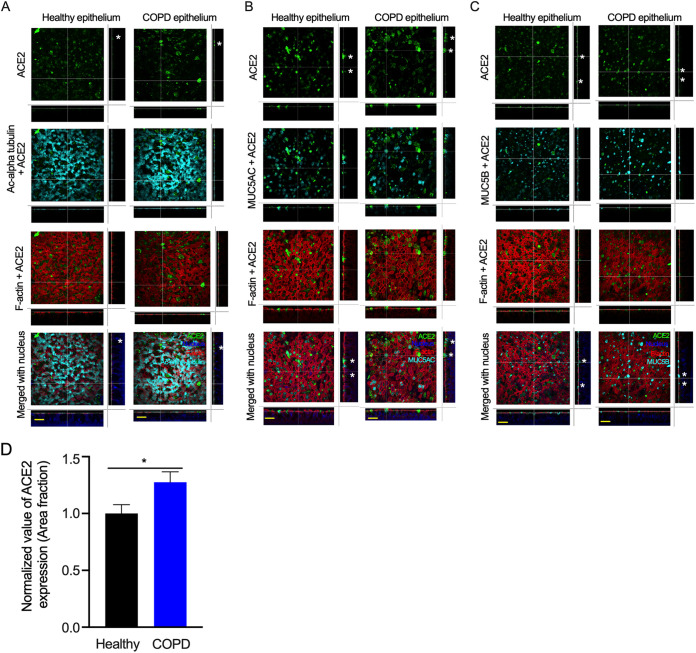
ACE2 is expressed at higher levels in goblet cells. The airway epithelia were generated and fixed as described in Fig. 1 (A) Cells were stained for ACE2 (anti-ACE2, green), cilia (anti-acetyl-alpha tubulin, cyan) F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). Bar = 30 μm. (B) Cells were stained for ACE2 (anti-ACE2, green), MUC5AC+ goblet cells (anti-MUC5AC, cyan), F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). Bar = 30 μm. (C) Cells were stained for ACE2 (anti-ACE2, green), MUC5B+ goblet cells (anti-MUC5B, cyan), F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). Bar =30 μm. The asterisks (*) indicate ACE2 on the epithelial surface. (D) The ACE2-stained area fraction of healthy and COPD samples was measured using ImageJ software. The error bars represent the SEMs. The statistical significance was determined by unpaired two-tailed t tests. The results from three independent experiments are shown.
Because ACE2 staining hardly overlapped with acetyl-alpha-tubulin (Ac-alpha tubulin), we tested the expression of ACE2 along with that of the goblet and club cell markers MUC5AC and MUC5B (11). ACE2 overlapped with MUC5AC (Fig. 2B) and MUC5B (Fig. 2C and Fig. S1F). We also compared ACE2 expression with the expression of E-cadherin and zonula occludens-1 (ZO-1), markers for adherens junction and tight junction proteins, respectively. The results showed that ACE2 expression did not overlap apical tight junctions or adherens junctions (Fig. S2), which suggests that ACE2 expression is primarily located within the cellular boundary and does not impact the tissue barrier integrity of the respiratory epithelium. In addition, ACE2-expressing goblet cells, including ACE2+MUC5AC+ cells and ACE2+MUC5B+ cells were higher in the COPD epithelium than in the healthy epithelium (Fig. S1G and H). Overall, ACE2 expression was higher in the COPD than in the healthy epithelium (Fig. 2D and Fig. S1 and S2), which is likely due to the presence of goblet cell hyperplasia in the COPD epithelium and thus a higher number of goblet cells.
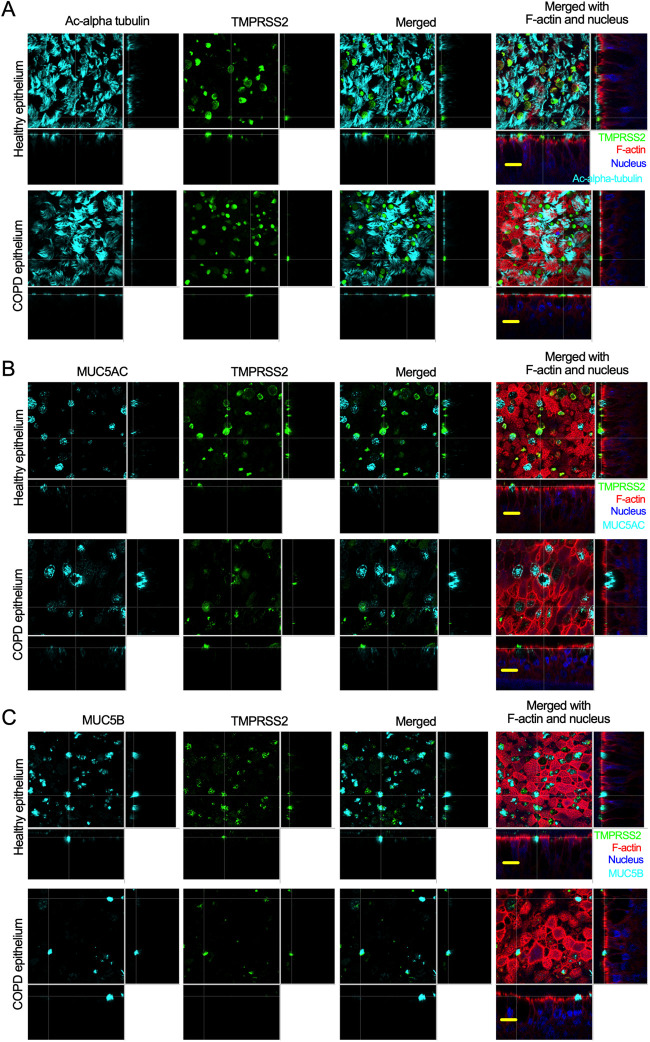
TMPRSS2 is an important host cofactor for SARS-CoV-2 entry into target cells (4, 11, 76). We visualized TMPRSS2 in the apical site of the airway epithelium by staining with anti-TMPRSS2 and found that TMPRSS2 expression hardly overlapped with cilium (Fig. 3A). We then tested the expression of TMPRSS2 along with that of the goblet cell markers MUC5AC and MUC5B. Indeed, TMPRSS2 overlapped with MUC5AC (Fig. 3B) and MUC5B (Fig. 3C). Therefore, it appears that both TMPRSS2 and ACE2 are expressed on the same cell surface (Fig. 2 and 3). These results indicate that goblet cells are an easy target of SARS-CoV-2 infection in the respiratory epithelium.
FIG 3.
TMPRSS2 is expressed at higher levels in goblet cells. (A) The airway epithelia (described in Fig. 1) were stained for cilia (anti-acetyl-alpha tubulin, cyan), TMPRSS2 (anti-TMPRSS2, green), and F-actin (rhodamine phalloidin, red); the nuclei were stained with DAPI (blue). Bar =15 μm. (B) The airway epithelia were stained for the identification of MUC5AC+ goblet cells (anti-MUC5AC, cyan), TMPRSS2 (anti-TMPRSS2, green), F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). Bar =15 μm. (C) The airway epithelia were stained to identify MUC5B+ goblet cells (anti-MUC5B, cyan), TMPRSS2 (anti-TMPRSS2, green), F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). Bar = 15 μm.
SARS-CoV-2 preferentially infects goblet cells.
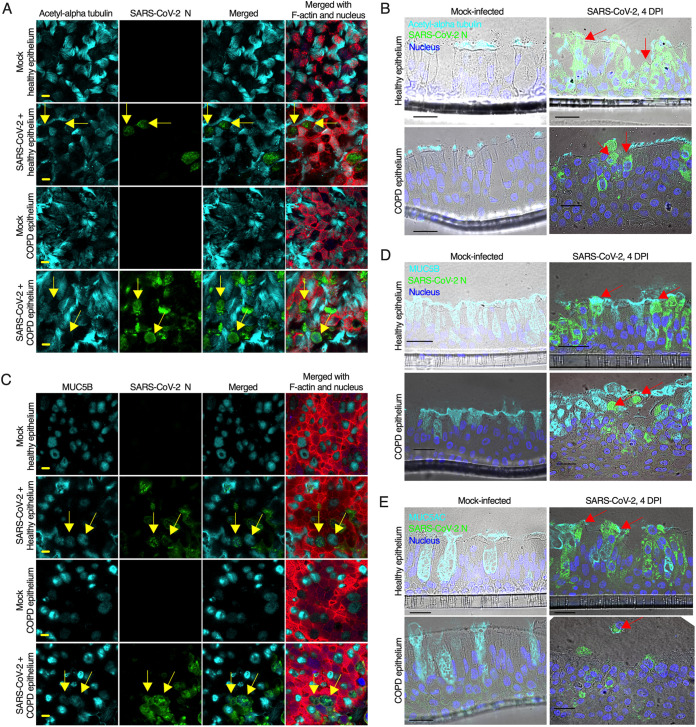
Although we confirmed that SARS-CoV-2 entry receptors are expressed at higher levels on nonciliated goblet cells, a number of previous studies have suggested that SARS-CoV-2 targets ciliated cells (77, 78). Therefore, we first examined whether SARS-CoV-2 infects nonciliated goblet cells. We infected the apical side of the airway epithelium with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1. At 4 days postinfection (dpi), we fixed the cells and stained them for SARS-CoV-2 nucleoprotein (N) and a ciliated cell marker. The results revealed that SARS-CoV-2 infects both healthy and COPD epithelium and causes a substantial cytopathic effect (CPE) (Fig. S3A and B). SARS-CoV-2 infection was higher in the COPD epithelium than in the healthy epithelium, which is addressed later in the article. Although in some cases, the virus-induced extensive CPE made it difficult to distinguish SARS-CoV-2 cell tropism, we focused on multiple random areas with less CPE but virus infection. We found that SARS-CoV-2 infects nonciliated cells in both healthy and COPD epithelium (Fig. 4A). We also used a second detection method to visualize viral and cellular markers in cross-sections of the epithelium. Immunohistochemistry-based staining confirmed the extensive CPE induced by SARS-CoV-2 in both healthy and COPD epithelium. Apparently, SARS-CoV-2 infected both ciliated and nonciliated epithelial cells in the airway epithelium (Fig. 4B). Using staining strategies similar to those described before, we found that SARS-CoV-2 infects MUC5B-positive (Fig. 4C and D) and MUC5AC-postive (Fig. 4E) goblet cells. To determine whether SARS-CoV-2 infects basal cells, we stained the sectioned epithelium for P63 and the SARS-CoV-2 spike (S) protein. We did not observe any overlap between the SARS-CoV-2 S protein and the basal cell marker (Fig. S4). These results suggest that SARS-CoV-2 preferentially infects nonciliated goblet cells.
FIG 4.
SARS-CoV-2 infects goblet cells. (A) The airway epithelia were mock-infected or infected with SARS-CoV-2 at an MOI of 0.1. At 4 days postinfection (dpi), the epithelia were fixed, permeabilized, and stained for the identification of cilia (anti-acetyl-alpha tubulin, cyan), SARS-CoV-2 N (anti-N, green), F-actin (rhodamine phalloidin, red), and nuclei (DAPI, blue). The arrows indicate SARS-CoV-2-infected nonciliated cells. (B) At 4 dpi, the epithelia (described in panel A) were stained for the ciliated cell marker acetyl-alpha tubulin (anti-Ac-alpha tubulin, cyan), SARS-CoV-2 N (anti-N, green), and F-actin (rhodamine phalloidin, red); the nuclei were stained with DAPI (blue). The arrows indicate nonciliated infected cells. (C) At 4 dpi, the epithelia (described in panel A) were embedded in paraffin, sectioned, and stained for the detection of MUC5B-positive goblet cells (anti-MUC5B, cyan) and SARS-CoV-2 N (anti-N, green); the nuclei were stained with DAPI (blue). The arrows indicate SARS-CoV-2-infected nonciliated MUC5B-positive goblet cells. (D) At 4 dpi, the sectioned epithelia were stained for the goblet cell marker MUC5B (anti-MUCB, cyan) and SARS-CoV-2 N (anti-N, green); the nuclei were also stained with DAPI (blue). The arrows indicate SARS-CoV-2-infected MUC5B-positive goblet cells. (E) At 4 dpi, the sectioned epithelia were stained for the goblet cell marker MUC5AC (anti-MUC5AC, cyan) and SARS-CoV-2 N (anti-N, green); the nuclei were also stained with DAPI (blue). The arrows indicate SARS-CoV-2-infected MUC5AC-positive goblet cells. (A and B) Bar = 5 μm. (C, D, and E) Bar = 20 μm. See also Fig. S3 and S4.
SARS-CoV-2 infection in airway epithelium recapitulates lung disease pathophysiology.
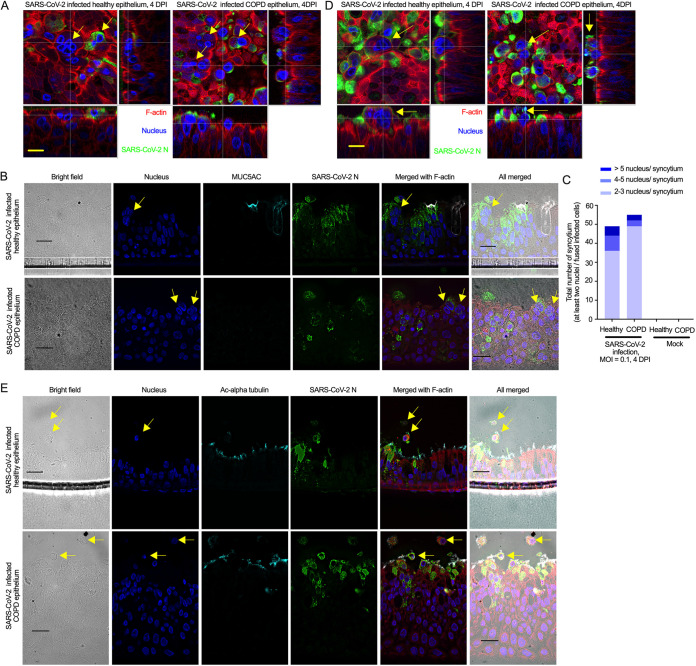
To determine whether SARS-CoV-2 infection in the airway epithelium recapitulates the virus-induced pathophysiology in the lung, we examined the infected epithelium under a confocal microscope and found that SARS-CoV-2 infection causes substantial damage to the apical site of the infected epithelium, as confirmed by extensive CPE in both healthy and COPD epithelium (Fig. 4 and Fig. S3). We also found substantial secretion in the SARS-CoV-2-infected airway epithelium based on mucus staining; however, we did not quantify the mucus secretion in either mock or infected airway epithelium. Based on image analysis, we observed apical damage of the SARS-CoV-2-infected epithelium, including loss of cellular junctions, loss of ciliary damage, substantial mucus production, and the protraction of nuclei, which are all common features observed in SARS-CoV-2-infected lungs (Fig. 4 and Fig. S3) (79). Additionally, we investigated whether SARS-CoV-2-infected cells in the epithelium might form syncytia (multinucleated cells), a hallmark of SARS-CoV-2 infection in the lung (80) that had also been reported for SARS-CoV (29). Indeed, we found that SARS-CoV-2-infected cells formed syncytia in both healthy and COPD epithelium (Fig. 5A and B). Importantly, SARS-CoV-2 infection causes more syncytia in the infected COPD epithelium than in the mock-infected epithelium (Fig. 5C).
FIG 5.
SARS-CoV-2 induces syncytia and cell sloughing. (A) The airway epithelia (mock or infected; described in Fig. 4) were stained for SARS-CoV-2 N (anti-N, green) and F-actin (rhodamine phalloidin, red); the nuclei were also stained with DAPI (blue). The arrows indicate syncytia. Bar = 5 μm. (B) At 4 dpi, the sectioned epithelia were stained for the goblet cell marker MUC5AC (anti-MUC5AC, cyan) and SARS-CoV-2 N (anti-N, green), and the nuclei were also stained with DAPI (blue). The arrows indicate syncytia. Bar = 20 μm. (C) At 4 dpi, the number of syncytia (at least two nuclei in the fused infected cells) were quantified in the infected airway epithelium (healthy or COPD) (at least two independent donors). The distribution of nuclei in the syncytium is indicated by different colors in the same bar. The data were obtained from two independent experiments. No statistical analysis was done. (D) At 4 dpi, the airway epithelium was stained for SARS-CoV-2 N (anti-N, green) and F-actin (rhodamine phalloidin, red), and the nuclei were stained with DAPI (blue). The arrows indicate cell sloughing. Bar = 5 μm. (E) At 4 dpi, the sectioned epithelium was stained for cilia (anti-acetyl-alpha tubulin, cyan) and SARS-CoV-2 N (anti-N, green), and the nuclei were stained with DAPI (blue). The arrows indicate cell sloughing. Bar = 20 μm.
Cell sloughing has been reported from lung autopsy findings of SARS-CoV-2-infected patients (79). Therefore, we examined whether SARS-CoV-2 infection in our epithelium model recapitulates cell sloughing. Indeed, we found that SARS-CoV-2 induces cell sloughing in both healthy and COPD epithelium as confirmed by two independent methods, immunofluorescence and immunohistochemistry (IHC) (Fig. 5D and E). These results demonstrate that hallmark pathological features of SARS-CoV-2 are recapitulated in the infected airway epithelium model.
SARS-CoV-2 replicates higher and exacerbates pathophysiology in COPD epithelium.
To determine whether SARS-CoV-2 replicates better in the COPD epithelium, we determined the titer of SARS-CoV-2 in the apical wash of the infected epithelium and found that SARS-CoV-2 replication was increased by almost a log in the COPD compared to the healthy epithelium (Fig. S5). This substantially higher replication in the COPD epithelium was unique to SARS-CoV-2, as neither SARS-CoV nor MERS-CoV replication increased by10-fold (Fig. S5). When the apical shedding of the coronaviruses at 96 h postinfection (hpi) were presented relative to the initial viral presence at 1 hpi, SARS-CoV-2 shedding was 1.5 times higher in COPD than healthy donors (Fig. 6A). As goblet cells in the COPD epithelium harbor the host cell entry factors for both SARS-CoV-2 and SARS-CoV, we wanted to determine whether goblet cells harbor MERS-CoV entry receptor dipeptidyl-peptidase 4 (DPP4). DPP4 expression by goblet cells in the respiratory system has previously been reported (81). Our result also suggested that DPP4 expression was detected in the goblet cells (positive cells with distinct morphology similar to goblet cells) (Fig. S6). Surprisingly, MERS-CoV replication was lower than that of other coronaviruses in both healthy and COPD airway epithelium. Whether disparity in the entry receptor use by different coronaviruses causes the differences in the viral replications in the airway epithelium remains to be determined. However, we found that MERS-CoV infection caused higher syncytium formation than SARS-CoV infection in both healthy and COPD epithelium (Fig. 6B and C). Interestingly, SARS-CoV infection induced less syncytium in the healthy epithelium than in the infected COPD epithelium (Fig. 6B and C). Whether syncytium formation is directly related to the cell type specificity is yet to be determined. In line with virus replication results, severe pathophysiology (e.g., syncytium formation) was observed in the infected COPD airway epithelium by all the coronaviruses tested (Fig. 6A to C). In addition, severe cytopathic effect of SARS-CoV-2 was observed in COPD epithelium compared to healthy epithelium determined by the staining signaling of SARS-CoV-2 nucleoprotein (Fig. 6D). Although we can infer that higher SARS-CoV-2 replication in the COPD airway epithelium may correspond with severe disease exacerbation, the mechanism of the disease exacerbation remains to be determined. However, we found squamous metaplasia in SARS-CoV-2-infected COPD epithelium, which was rather infrequently found in the SARS-CoV-2-infected healthy epithelium (Fig. 1I; Fig. 4B, D, and E; Fig. S7) (82). Squamous metaplasia is known to increase bronchial wall thickening as seen in bronchitis (52, 72, 83). As tracheobronchitis is one of the most common histopathological features in COVID-19 disease fatalities (84), we evaluated whether SARS-CoV-2 infection increases the height of the epithelium. First, there was a substantial increase in squamous metaplasia in the COPD epithelium due to SARS-CoV-2 infection (Fig. S7A to C). Second, the increased metaplasia apparently changed the morphology of the nonciliated goblet cells in the infected COPD epithelium (Fig. S7C). Whether this change in the goblet cell morphology impacted the mucus hyperplasia remains to be determined. Third, in contrast to the healthy epithelium, SARS-CoV-2 induced higher squamous metaplasia in the infected COPD epithelium and caused a substantial increase in the height of the epithelium (Fig. S7D).
FIG 6.
SARS-CoV-2 replicates at a higher rate than SARS-CoV and MERS-CoV in the infected COPD epithelium. (A) At 1 h postinfection (hpi) or 96 hpi, the apical wash of the SARS-CoV-2-, SARS-CoV-, or MERS-CoV-infected airway epithelium (MOI = 0.1) was collected, and SARS-CoV-2, SARS-CoV, or MERS-CoV titration was performed based on the 50% tissue culture infective dose (TCID50) (see Fig. S5). (B) At 96 hpi, the infected airway epithelium was fixed, permeabilized, and stained for viral protein (N protein of SARS-CoV/SARS-CoV-2 detection using N-specific monoclonal antibody, N protein of MERS-CoV detection using N-specific monoclonal antibody). F-actin and the nucleus were stained by rhodamine phalloidin and DAPI, respectively. White arrows indicate syncytia in the infected airway epithelium. The images represent at least five random positions of the infected airway epithelium. (C) The syncytium formation was quantified in the infected airway epithelium. The distribution of nuclei in the syncytium is indicated by different colors in the same bar. The data were obtained from an independent experiment. (D) The apical side of the SARS-CoV-2-infected epithelium was stained for N and imaged under a fluorescence microscope. For determining the extent of cytopathic effect (CPE), the N stained area was measured using ImageJ software. The CPE-based severity between SARS-CoV-2-infected healthy and COPD epithelium is plotted. The error bars represent the SEMs. The statistical significance was determined by unpaired two-tailed t tests. The data were obtained from two independent experiments.
DISCUSSION
Here, we have shown that the COPD epithelium model recapitulates the bronchial biophysical and pathophysiological characteristics of COPD, such as goblet cell hyperplasia and squamous metaplasia (52, 70, 72, 85). The significantly higher number of MUC5B-positive cells in the COPD airway epithelium is in line with the COPD clinical feature of predominant MUC5B mucin in the sputum (67, 69). A previous report suggested the presence of altered ciliated cells in COPD airway epithelium (85), which was not observed in our model. The duration of cell differentiation may be the reason, as we differentiated NHBE cells for 4 weeks that may require for obtaining pseudostratified epithelium terminal differentiation (including ciliogenesis) (49, 85). We used our previously published culture method to generate multicellular diversity and physiologically functioning airway epithelium that resembles the airway surface in vivo (49–51, 86). One of the limitations in the ALI culture research is that passaging of primary NHBE cells may impact on their ability to differentiate into airway epithelium. We demonstrated that primary NHBE cells obtained after four passages without using any additional supplements can still be differentiated into human airway epithelium (50). In a separate study, we confirmed that passaging NHBE cells up to four times has an insignificant effect on the whole-genome transcriptome by comparing transcriptome profiles of each passage of cells (data not shown). The ability to expand primary cells that also form fully differentiated mucociliary epithelium reduces repeat sample collections from patients where samples are difficult to obtain and limited, such as infants and deceased patients (50, 87). Our results demonstrate that primary NHBE cells from either healthy adults or high-risk adults (those with COPD) can be passaged up to four times, and normal epithelial phenotypic features are maintained in passaged primary NHBE cells.
One emergent question is why the human-to-human transmission of SARS-CoV-2 is much higher than that of SARS-CoV, although both viruses share ACE2 as a cell surface receptor and use TMPRSS2 to facilitate their entry into the host cell (4, 11). The SARS-CoV-2 spike protein has an additional furin cleavage site that is absent in SARS-CoV, and it is hypothesized that furin cleavage facilitates human-to-human transmission (11, 88). We found that SARS-CoV-2 infects goblet cells due to the higher expression of ACE2 and TMPRSS2; however, further studies are needed to confirm whether SARS-CoV-2’s preferential infection in goblet cell contributes to higher person-to-person transmission. The major function of goblet cells in the lung epithelium is mucin production to trap pathogens, dust, and particles, which are cleared by a process known as mucociliary clearance (89). The possibility that SARS-CoV-2 infects goblet cells could explain the presence of viral RNA in sputum (31) and might explain the easy transmission of the virus from person to person via direct contact or droplet fomite transmission. There may be disagreement about whether aerosol transmission played a major role in SARS-CoV-2 spread in general (20), but we believe both droplets and direct contact are the most likely contributors to person-to-person transmission (17, 29, 30). Recent studies have also suggested that goblet cells are permissive to SARS-CoV-2 infection (90, 91). We also found that SARS-CoV-2 infected ciliated cells but not basal cells in the airway epithelium. As SARS-CoV infection is limited to ciliated cells (12), we think that SARS-CoV-2 infection in goblet cells may provide higher stability in the environment, which in turn helps its higher transmission than its predecessor SARS-CoV (92). Additionally, influenza A virus infects goblet cells in addition to ciliated cells (93), whereas respiratory syncytial virus (RSV) infects only ciliated cells (94). Further experiments are necessary to establish virus-specific preferential cell tropism in the lung and determine the differences in the respiratory virus-induced pathogenicity and transmissibility.
Our airway epithelium model recapitulated some SARS-CoV-2-induced lung histopathological findings, such as prominent nucleoli (79), cytoplasmic vacuolation, multinucleated giant cells (95), squamous metaplasia (84) and epithelial cell sloughing, which is the most common finding (96). SARS-CoV-2 infection in the bronchial epithelium models induced a substantial amount of cell syncytium formation and cell sloughing, which indicates that tracheobronchial cells are highly associated with virus-induced pathogenesis. Therefore, our data suggest that bronchial cells are important in SARS-CoV-2 pathogenesis (84). In patients with COPD, goblet cell hyperplasia is a common clinical manifestation, and an increasing number of goblet cells result in more mucus production (71). Because we have shown that SARS-CoV-2 replicated at a higher rate in the COPD epithelium, we believe that goblet cell hyperplasia is responsible for this phenomenon and may explain why COPD patients are at higher risk for severe outcomes of COVID-19. Our data also suggest that high-risk adults (those with COPD) are vulnerable to severe disease outcomes from SARS-CoV and MERS-CoV infections. Surprisingly, we found that the MERS-CoV replication rate was lower than those of SARS-CoV and SARS-CoV-2 in the bronchial airway epithelium. MERS-CoV uses DPP4 for entry into host cells, while both SARS-CoV and SARS-CoV-2 use ACE2 and TMRPSS2 (4, 97). Despite goblet cells harboring receptors for all these viruses, MERS-CoV’s lower replication may indicate a disparity in the receptor use by these viruses for their efficient entry. However, MERS-CoV infection-induced disease exacerbation can be explained by the observed higher syncytium formation in the bronchial airway epithelium. We primarily focused our observations on SARS-CoV-2 infection-induced pathobiology in healthy and COPD epithelium. Interestingly, we found that SARS-CoV-2 infection increased squamous metaplasia in the COPD epithelium. Squamous metaplasia in the columnar epithelium corelates with increased severity of airway obstruction in COPD (98). Although squamous metaplasia is one of the histopathological findings in the SARS-CoV-2-infected patient fatalities (84), our results provide evidence for the first time that SARS-CoV-2 infection increases squamous metaplasia, which may result in bronchitis in infected COPD patients. In fact, tracheobronchitis is one of the most common histopathological features in COVID-19 disease fatalities (84). However, donor-to-donor variation can be possible in squamous metaplasia development of the COPD airway epithelium, as we found squamous metaplasia in the airway epithelium generated from NHBE cells of multiple independent COPD donors. No SARS-CoV-2 infectious virions were detected in the basal medium of the infected healthy and COPD epithelium (tested up to 4 dpi), which might suggest that the initial virus-induced damage was limited at the apical site. Further studies utilizing in vitro systems such as human-airway-on-a-chip (99, 100) need to confirm our findings. In addition, our airway epithelium model lacks both resident and infiltrating immune cells (dendritic cells, macrophages, lymphocytes, etc.) and is devoid of the endothelial layer and thus cannot be used to study the impact of immune responses on SARS-CoV-2 infections. However, we can provide inferences based on the detection of immunoregulatory factors (chemokines and cytokines), but these hypotheses remain to be confirmed. However, COPD patients can have an inadequate immune response, which may worsen the inflammatory lung microenvironment and disease severity (64, 101, 102). Although this study does not have the scope to determine whether a defective immune response played a major role in higher SARS-CoV-2 infection in COPD, this study has established two important aspects of why COPD patients are vulnerable to severe SARS-CoV-2-induced disease pathophysiology. First, the COPD bronchial airway model recapitulates goblet cell hyperplasia. Second, SARS-CoV-2 receptor ACE2 and its cofactor TMPRSS2 are expressed substantially more on goblet cells than on healthy cells. Indeed, our results showed that SARS-CoV-2-induced viral titer, syncytium formation, and extent of CPE were higher in COPD than healthy donors. We acknowledge that the definitive differences between goblet cell hyperplasia and defective immune response in the SARS-CoV-2-induced severe pathophysiology in COPD bronchial airway epithelium is beyond the scope of this study. We also acknowledge that our results were obtained from two healthy donors and three COPD donors. However, we believe the airway epithelium model provides an excellent tool for demystifying some of the SARS-CoV-2 pathophysiological features, which may lead to identifying novel targets for therapeutic development.
In conclusion, we investigated SARS-CoV-2 entry receptor and cofactor expression in the bronchial airway epithelium models of healthy adults and high-risk adults (those with COPD) and found that goblet cells preferentially harbor both entry receptor ACE2 and cofactor TMPRSS2. Our studies revealed that SARS-CoV-2 infection of goblet cells leads to virus-induced syncytium formation and cell sloughing in the airway epithelium, which may be responsible for the severe virus-induced lung pathophysiology. We found that SARS-CoV-2 replicates better in COPD airway epithelium, likely due to COPD-associated goblet cell hyperplasia. Thus, we postulate that goblet cells play a critical role in SARS-CoV-2 infection of the lung and are responsible for more severe outcomes of SARS-CoV-2 infection in COPD patients.
MATERIALS AND METHODS
Cells.
Primary normal human bronchial epithelial (NHBE) cells of healthy adults (two donors) and high-risk adults (those with COPD) (three donors, deidentified) were obtained from Kristina Bailey at the University of Nebraska Medical Center (UNMC), Omaha, NE, under an approved material transfer agreement (MTA) between the University of North Dakota (UND) and UNMC. The protocol for obtaining cells was reviewed by the UNMC institutional review board (IRB) and was determined not to constitute human subject research (no. 318-09-NH). A549 cells (a human lung epithelial cell line, ATCC-185) were obtained from Peter Collins at the National Institutes of Health (NIH), Bethesda, MD. Vero cells used for 50% tissue culture infection dose (TCID50) assay were a resource of the Feldmann Lab at the Rocky Mountain Laboratories, Hamilton, MT.
Virus.
The SARS-CoV-2 isolate nCoV-WA1-2020 (MN985325.1) was kindly provided by the CDC as Vero passage 3 (74). The virus was propagated once in Vero E6 cells in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) supplemented with 2% fetal bovine serum (FBS; Thermo Fisher Scientific), 1 mM l-glutamine (Thermo Fisher Scientific), 50 U/mL penicillin, and 50 μg/mL streptomycin (Thermo Fisher Scientific) (virus isolation medium). The virus stock used in this study was 100% identical to the initially deposited GenBank sequence (MN985325.1); sequencing did not detect any virus stock contaminants. MERS-CoV (HCoV-EMC/2012, Vero passage 6) was kindly provided by the Department of Viroscience, Erasmus Medical Center, Rotterdam, The Netherlands, and propagated once in Vero E6 cells in DMEM (Sigma-Aldrich). SARS-CoV (Urbani strain, GenBank accession no. AY278741) provided by Heinz Feldmann, Rocky Mountain Laboratories (RML), National Institute of Allergy and Infectious Diseases (NIAID), NIH.
Biosafety statement.
Work with SARS-CoV-2 was performed in the high-level biocontainment facilities at the RML, NAID, NIH, Hamilton, MT. All infectious work followed standard operating procedures (SOPs) approved by the institutional biosafety committee.
Cell culture.
We passaged the primary NHBE cells (passage 1) three times before differentiating them (passage 4) into a pseudostratified epithelium. For each passage, the cells were grown in a 100-mm culture dish (Corning, Inc.) precoated with PurCol (Cell Systems, 5005). The cells were maintained in airway epithelial cell (AEC) growth medium (PromoCell; C20601) containing AEC supplements (PromoCell), 2% penicillin/streptomycin (Thermo Fisher Scientific, 15140122), and 1% amphotericin B (Thermo Fisher Scientific, 15290026) (complete AEC medium) at 37°C in an incubator with 5% CO2. The cells were grown to 90% confluence, and the medium was changed every other day. Confluent monolayers of cells were dissociated with 5 mL of TrypLE (Thermo Fisher Scientific, 12604021) and pelleted, and one-third of the cells were reseeded in a culture dish containing complete AEC medium for passaging. A549 cells were grown in F-12 medium (Thermo Fisher Scientific, 11765054) with 10% HyClone FBS (GE Healthcare, SH3007103), 2% penicillin/streptomycin, and 1% amphotericin B.
Air-liquid interface (ALI) culture.
Transwell insert (6.5 mm) with 0.4-μm-pore polyester membrane inserts (Corning, Inc., 3470) were coated with PureCol for 20 min before cell seeding. NHBE cells (5 × 104) suspended in 200 μL of complete AEC medium were seeded in the apical part of the Transwell insert. Subsequently, 500 μL of complete AEC medium was added to the basal part of the Transwell insert. When the cells formed a confluent layer on the Transwell insert, the AEC medium was removed from the apical part, and PneumaCult-ALI basal medium (Stemcell Technologies, 05002) with the required supplements (Stemcell Technologies), 2% penicillin/streptomycin, and 1% amphotericin B (complete ALI basal medium) was added to the basal part. The ALI medium in the basal part was changed every other day, and the apical surface was washed with 1× Dulbecco’s phosphate-buffered saline (DPBS) (Thermo Fisher Scientific) once per week initially but more frequently when more mucus was observed on later days. However, the thickness of the mucus was not determined. All the cells were differentiated for at least 4 weeks at 37°C in an incubator with 5% CO2.
Transportation of Transwell inserts containing airway epithelium.
The differentiated airway epithelia for 22 days were transported in transportation medium at 4°C via overnight FedEx biological material shipment to the Rocky Mountain Laboratories (RML, Hamilton, MT). The semisolid transportation medium was prepared by adding agarose to the complete AEC medium. Upon receipt, the Transwell inserts containing airway epithelia were immediately transferred into new plates with complete AEC medium in the basal part and maintained before infection as described in the section on ALI culture.
Virus infection.
Virus infection was conducted in the high-level biocontainment facilities at RML, NIAID, NIH. The 4-week-differentiated airway epithelia were washed with 200 μL of 1× PBS to remove mucus and infected on the apical site with either SARS-CoV-2, SARS-CoV, or MERS-CoV at an MOI of 0.1 in 1× PBS for 1 h (at 37°C with 5% CO2). For mock infection, the assigned Transwell inserts were similarly incubated with 1× PBS without virus. The viral inoculum was then removed, and the epithelia were washed twice with 200 μL of 1× PBS. Then, 200 μL of apical wash and basal medium were collected for virus titration. Fresh ALI medium (1,000 μL) with supplements was added to the basal part of each Transwell insert, and the apical part was kept empty. Mock-infected and virus-infected Transwell inserts were incubated for 4 days at 37°C in an incubator with 5% CO2.
50% Tissue culture infective dose (TCID50).
Vero cells were seeded on 96-well plates to confluence, and SARS-CoV-2, SARS-CoV, or MERS-CoV endpoint titration was performed. The cells were inoculated with 10-fold dilutions of the supernatants collected during the experiment in DMEM, 2% FBS, l-glutamine, and penicillin-streptomycin (P/S). The cells were incubated for 7 days, and the cytopathic effect (CPE) was scored under a microscope. The TCID50 was calculated using the Reed-Muench formula (103).
Virus-infected sample collection.
At 4 dpi, 200 μL of 1× PBS was added to the apical site of the Transwell insert and incubated for several minutes, and the apical wash was collected for virus titration. Similarly, basal medium was collected for virus titration. All the samples were stored at −80°C until titration. For paraformaldehyde (PFA) fixation, 200 μL of 4% PFA was added at the apical site of the Transwell inserts and incubated for 30 min, and the Transwell inserts were further maintained overnight in 4% PFA before being transported outside the biosafety level 4 (BSL4) lab according to the standard protocol of RML. For protein and RNA sample collections, 200 μL of 1× PBS was added to the apical side of each Transwell insert, and cells were scraped with a miniscraper, collected into a cryovial, and pelleted by being spun down with a table-top bench centrifuge. For the protein samples, 100 μL of 2× SDS buffer was added to the cryovials containing the cell pellet, and the tube was boiled for 10 min. After a quick spin, the samples were transferred into new tubes and removed from the BSL4 laboratory using a standard RML protocol. For the RNA samples, the cell pellets were resuspended in 600 μL of RLT buffer (Qiagen) and transported out of the laboratory following the RML protocol with 600 μL of ethanol. RNA was extracted using a Qiagen RNeasy minikit (Qiagen, 74104) and eluted in 50 μL of RNase-free water. All the samples were shipped to the Mehedi lab at UND using an appropriate overnight biological shipment procedure.
Hematoxylin and eosin (H&E) staining.
Four days postinfection, NHBE-ALI and COPD-ALI mock and infected Transwell inserts were fixed with 4% PFA for 24 h. The membrane was cut off using a scalpel and embedded in a paraffin mold. Sections (5 μm) were cut using a microtome. H&E staining was performed by two 5-min Histo-Clear incubations followed by three 5-min incubations with 100% ethanol, one 5-min incubation with 95% ethanol, and one 5-min incubation with 70% ethanol. Subsequently, the samples were washed with tap water for 2 min, incubated with 4% acidified Harris hematoxylin for 2 min, washed with tap water for 2 min, and incubated with 0.5% lithium carbonate for 20 s. The samples were subsequently washed with tap water for 5 min, incubated with eosin Y solution with pholoxine for 2 min, and then subjected to a series of ethanol incubations: incubation with 70% ethanol for 5 min, incubation with 95% ethanol for 5 min, and three 5-min incubations with 100% ethanol. Finally, two 5-min incubations with Histo-Clear were performed, and coverslips were then placed using a permanent mounting medium.
Alcian blue staining.
The 5-μm sectioned ALI-NHBE and COPD mock and infected samples were also subjected to alcian blue staining. The sections were deparaffinized as described in the section on H&E staining using Histo-Clear and ethanol. The cells were then washed with tap water for 2 min, incubated in 3% acetic acid for 3 min, and stained with alcian blue for 30 min. The sections were subsequently rinsed with 3% acetic acid for 10 s to help prevent nonspecific staining. The sections were washed under tap water for 10 min and then rinsed for 2 min in Milli-Q water. The sections were counterstained with nuclear fast red solution for 5 min and washed under tap water for 5 min. The sections were subsequently dehydrated and cleared with a gradient of alcohol and Histo-Clear as in the protocol used for H&E staining. The slides were mounted with permanent mounting medium and cover slipped.
Fluorescence imaging.
The apical site of the airway epithelium was washed with PBS, and both the apical and basal parts were fixed with 4% paraformaldehyde (PFA) (Polysciences, Inc., 18814-10) and blocked with 10% goat serum (Vector Laboratories, S-1000-020) solution in immunofluorescence (IF) washing buffer (130 mM NaCl2, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% bovine serum albumin [BSA], 0.2% Triton X-100, and 0.05% Tween 20) for 1 h. The Transwell inserts were then incubated with the following primary antibodies (Abs) (alone or in combination) in IF washing buffer overnight at 4°C: anti-ZO-1 rabbit polyclonal (1:200) (Thermo Fisher Scientific, 40-2200), anti-E-cadherin rabbit monoclonal (1:200) (Cell Signaling Technologies, 3195S), anti-MUC5B rabbit monoclonal (1:500) (Atlas Antibodies, HPA008246), anti-MUC5AC mouse polyclonal (1:100) (Abnova, H00004586-A01), anti-MUC5AC rabbit monoclonal (1:200) (Cell Signaling Technology, 61193), anti-acetyl-alpha-tubulin rabbit monoclonal (1:500) (Cell Signaling Technologies, 5335), anti-TMPRSS2 mouse monoclonal (1:50) (Santa Cruz Biotechnology, sc-515727), anti-SARS CoV-2 nucleocapsid mouse monoclonal (1:10) (Thermo Fisher Scientific, MA1-7404), anti-MERS-CoV nucleoprotein (N) mouse monoclonal (1:100) (Sino Biological, Inc., HB12SE1903), and anti-ACE2 mouse monoclonal (1:20) (R&D Systems, MAB933). The next day, the inserts were washed with the IF washing buffer and incubated with an anti-mouse Alexa Flour 488-conjugated Ab (1:200) (Thermo Fisher Scientific, A-11029) and/or anti-rabbit Alexa Flour 647 (1:200) (Thermo Fisher Scientific, A-21245) in IF washing buffer for 3 h in the dark at 4°C. The cells were washed twice with PBS and incubated with rhodamine phalloidin (1:500) (Cytoskeleton, Inc., PHDR1) in IF washing buffer for 30 min at room temperature in the dark. After two washes with PBS, the cell nuclei were stained with NucBlue fixed cell stain ReadyProbes (Thermo Fisher Scientific, R37606) for 30 min in the dark at room temperature. The epithelium was mounted on Tech-Med microscope slides (Thomas Scientific) using ProLong-Gold antifade mounting medium (Thermo Fisher Scientific, P36390). Images were captured using a confocal laser-scanning microscope (Olympus FV3000) enabled with a 60× lens objective. The 405-nm laser was used to excite the DAPI (4′,6-diamidino-2-phenylindole) signal for nucleus detection, the 488-nm laser was used to excite Alexa Flour 488 for MUC5AC, ACE2, TMPRSS2, or SARS CoV-2 nucleocapsid protein detection, the 561-nm laser was used to excite rhodamine phalloidin for F-actin detection, and the 640-nm laser was used to excite Alexa Flour 647 for MUC5B, acetyl-alpha-tubulin, E-cadherin, ZO-1, or MUC5AC detection. At least two random fields were selected per sample and imaged. The images were processed with Imaris software version 9.5.1 (Oxford Instruments Group) and used for the conversion of Z-stack images (.oir format) to.tiff format and for additional image postprocessing. Separately, the tiling of Z-stack (3 × 3) images was captured using a Leica DMi8 microscope followed by image processing using a 3D deconvolution image processing module in LASX software (Leica Microsystems) associated with the microscope. We used nine independent images to quantify the total cell number based on F-actin (Texas Red channel) and goblet cells using the Alexa Flour 647 channel (anti-MUC5B) with the Fiji multipointer option. Similarly, we quantified the MUC5AC+ cells using at least three confocal images. Twelve independent images obtained from six independent Transwell inserts from both healthy and COPD samples were used to determine ACE2-stained areas. Initially, all Z-stack images were converted to.tiff format using IMRIS software, and then ACE2 expression was determined using the area fraction option of ImageJ. Cells which were stained by both ACE2 (green, Alexa Flour 488) and MUC5AC/MUC5B (cyan, Alexa Flour 647) were considered ACE2-expressed MUC5AC (ACE2+MUC5AC+) cells and ACE2-expressed MUC5B (ACE2+MUC5B+) cells, respectively. Cells were counted manually to determine ACE2+MUC5AC+cells and ACE2+MUC5B+cells by using the Grid option of IMARIS software, and individual cells were determined by F-actin (red, rhodamine phalloidin) staining signal. Data were obtained from two independent images of healthy and COPD samples. The SARS-CoV-2 nucleoprotein-stained area fraction of healthy and COPD samples was measured using ImageJ software. The results obtained from a total of six independent images (212 μm by 212 μm/image) were derived from two each of healthy and COPD samples. All images were first converted into.tiff format and then transported into ImageJ software. RGB images were converted into 8-bit images. Then the image threshold was checked. To confirm the unbiased threshold, the auto threshold option was applied. The error bar represents the standard errors of the mean (SEMs). The statistical significance was determined by unpaired two-tailed t tests.
Immunohistochemistry.
Healthy and COPD airway epithelia (mock or infected) were sectioned into 5-μm sections as described previously for immunohistochemistry. Before staining, the antigen retrieval process was performed using R-buffer A in a Retriever 2100 instrument. The slides were allowed to cool in the buffer overnight and washed three times in PBS with Tween 20 (PBST; 0.05% Tween 20) for 10 min. Using a Pap pen, a hydrophobic barrier was drawn around the tissue. Once the hydrophobic barrier was dry, the tissue was incubated with blocking buffer (10% goat serum in PBS) for 2 h in a humidified, light-protected chamber. After incubation with blocking buffer, the tissue was immediately incubated with the following primary antibodies overnight at 4°C in a humidified, light-protected chamber: mouse monoclonal anti-SARS-CoV/SARS-CoV-2 N (1:50) (Thermo Fischer Scientific, MA1-7404) for the detection of SARS-CoV/SARS-CoV-2 nucleoprotein (N), rabbit polyclonal anti-SARS-CoV-2 spike (S) protein (1:100) (Thermo Fischer Scientific, PA5-81795) for the detection of SARS-CoV2 S protein, mouse monoclonal anti-MERS-CoV nucleoprotein (N) protein (1:100) (Sino Biological, Inc., HB12SE1903) for the detection of MERS-CoV N protein, acetylated-alpha-tubulin (1:500) (Cell Signaling Technologies) for the staining of ciliated cells, MUC5AC (1:500) (Cell Signaling Technologies, 5535) and MUC5B (1:500) (Atlas Antibodies, HPA008246) for the staining of goblet cells, and P63 (1:100) (Abcam Inc., ab735) for the staining of basal cells. The next day, the slides were washed three times with PBST and then incubated for 45 min with anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific) and anti-rabbit Alexa Fluor 647 (Thermo Fisher Scientific) secondary antibodies in a humidified, light-protected chamber. The slides were subsequently washed three times with PBST, incubated with rhodamine phalloidin (1:100) (Cytoskeleton, Inc.) for 30 min at room temperature in a humidified, light-protected chamber, and washed three times with PBST. The nuclei were then stained by incubation with NucBlue fixed cell stain ReadyProbes (Thermo Fisher Scientific) for 5 min at room temperature in a humidified, light-protected chamber. After incubation with the nuclear dye, the slides were washed once with PBST and twice with deionized (DI) water, and a cover glass was then placed on the tissue section using ProLong-Gold antifade mounting medium (Thermo Fisher Scientific). The slides were scanned using a Leica DMi8 inverted fluorescence microscope with a 63× oil objective, and the images were further processed using a 3D deconvolution image processing module in the LASX software (Leica Microsystems) associated with the microscope.
Ciliary beat frequency (CBF).
Cilia on the apical surface of the cells in the differentiated epithelial layer (after 4 weeks of differentiation) were visualized in phase-contrast mode with a Leica DMi8 microscope with a 20× lens objective and an attached environmental control chamber (37°C with 5% CO2) (Leica Microsystems). For each Transwell insert, six different random fields were recorded for approximately 2.1 s at 120 frames per second. The images were captured at 37°C and analyzed using the Sisson-Ammons Video Analysis (SAVA) system version 2.1.15 to determine the CBF (Hz) (Ammons Engineering).
Transepithelial electrical resistance (TEER).
The permeability of the differentiated epithelial layer (after 4 weeks of differentiation) was determined by measuring the TEER using an epithelial volt-ohm meter (EVOM2, World Precision Instruments, Inc.). The EVOM2 was calibrated according to the manufacturer’s instructions, and the STX2 electrode was sterilized with 70% ethanol before use. The internal electrode (smaller in size) was placed in the apical part of each Transwell insert (PBS was added during the TEER reading), and the external electrode (larger in size) was placed in the basal part of the Transwell insert, which contained ALI basal medium, to measure the membrane voltage and resistance of the epithelial layer. An empty Transwell insert (filled with PBS) containing no NHBE cells was used to correct for the background resistance. Three readings were taken for each Transwell insert. The TEER value of each sample was calculated by subtracting the background value.
Quantitative real-time PCR (qRT-PCR).
Airway epithelia cultured on 6.5-mm Transwell membranes were washed and treated for 1 min at room temperature with RLT buffer (Qiagen) with 1% β-mercaptoethanol (Sigma-Aldrich). The cells were scraped using a cell scraper, collected into a QIAshredder tube, and centrifuged at 15,000 rpm and 4°C for 3 min. The eluate was used for the extraction of total RNA using a total DNA/RNA extraction kit (Qiagen), and DNase I treatment was performed to remove DNA from the samples according to the manufacturer’s instructions. We also followed a similar approach for the extraction of RNA from A549 cells. The RNA concentration was determined with an Epoch microplate spectrophotometer (BioTek). A total of 500 ng of RNA was used for first-strand cDNA synthesis (Thermo Fisher Scientific) using Oligo(dT) primers (Thermo Fisher Scientific). qRT-PCR was performed using TaqMan assays (ACE2: Hs1085333_m1 and ACTB: Hs99999903_m1, for calibration) (Thermo Fisher Scientific) with the CFX384 real-time PCR system (Bio-Rad), and fold changes were calculated to determine the relative expression levels.
Western blotting.
The airway epithelium cultured on 6.5-mm Transwell membranes was washed with PBS, scraped out of all the cells, and pelleted by centrifugation at 10,000 rpm for 5 min. The cell pellet was incubated with 1× LDS loading buffer (Thermo Fisher Scientific) with proteinase inhibitor (Roche), transferred into a QIAshredder microcentrifuge tube, and centrifuged for 3 min at 15,000 rpm in a tabletop centrifuge. The elusion from the QIAshredder was collected and stored in a freezer. The protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, 23225). For the detection of ACE2, total protein (30 μg) was separated on 4 to 12% Bis-tris SDS polyacrylamide gels (reducing) and then subjected to dry blot transfer onto polyvinylidene difluoride (PVDF) membranes according to the manufacturer’s instructions (Life Technologies). The PVDF membrane was imaged using an Odyssey CLX system (Li-Cor Biosciences). ACE2 was detected by Western blotting using anti-ACE2 goat polyclonal antibody (R&D Systems, AF933) and corresponding donkey anti-goat IRDye 800 secondary antibodies (Li-Cor Biosciences, 926-32214). For the loading control, alpha-tubulin was detected by anti-alpha-tubulin mouse monoclonal antibody (Sigma-Aldrich, T6199) and the corresponding goat anti-mouse IRDye 680 secondary antibodies (Li-Cor Biosciences, 926-68070). Image Studio version 5.2 (LI-COR Biosciences) was used to quantify the protein signal.
Epithelial height measurement.
We measured the epithelial height for the determination of the increased squamous metaplasia-based height increment on the IHC images using the scale feature of the LASX software (Leica Microsystems). The plastic membrane of the Transwell insert was not included in the height measurement. We used at least four independent slides for the measurement, with at least three independent reads per slide.
Statistical analysis.
Parameters such as the number of replicates, number of independent experiments, mean ± standard deviation (SD), mean ± standard error of mean (SEM), and statistical significance are reported in the figures and figure legends. A P value of less than 0.05 was considered to indicate significance. Where appropriate, the statistical tests and post hoc statistical analysis methods are noted in the figure legends or Materials and Methods. *, P < 0.05; **, P < 0.01, ***, P < 0.001; NS, not significant.
ACKNOWLEDGMENTS
We thank the Microscopy Core (UND, Grand Forks), funded by NIH P20GM103442 of the INBRE program, for providing access to an Olympus FV300 confocal microscope and Swojani Shrestha for the technical support provided. Histological services were provided by the Histology Core (UND, Grand Forks), funded by NIH P20GM113123, NIH DaCCoTA CTR, NIH U54GM128729, and UNDSMHS funds. We also thank the Imaging Core (UND, Grand Forks), funded by NIH P20GM113123, NIH U54GM128729, and UNDSMHS funds, for the IMARIS image analyses.
This work was funded by NIH/NIGMS P20GM113123 and the VA grant 101-BX005413 and partially by the Intramural Research Program, NIAID, NIH.
M.M., J.O., and S.N.T. conceived the project and designed the experiments. K.L.B. provided the primary cells as well as training and guidance on primary cell differentiation. M.M., J.O., and S.N.T. performed the experiments. S.N.T. generated the confocal images. F.F. and H.F. performed the virus infection experiments in the BSL-4 laboratory. J.O., B.A.D., and K.J. generated the IHC images, B.A.D. performed the H&E and alcian blue staining, and M.M., J.O., and S.N.T. wrote the paper. M.M. and H.F. edited the paper.
Footnotes
Supplemental material is available online only.
Contributor Information
Masfique Mehedi, Email: masfique.mehedi@und.edu.
Gabriel I. Parra, US Food and Drug Administration.
REFERENCES
- 1.Bayati A, Kumar R, Francis V, McPherson PS. 2021. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem 296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. 2008. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res 18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia HP, Look DC, Hickey M, Shi L, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB, Jr. 2006. Infection of human airway epithelia by SARS coronavirus is associated with ACE2 expression and localization. Adv Exp Med Biol 581:479–484. doi: 10.1007/978-0-387-33012-9_85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li MY, Li L, Zhang Y, Wang XS. 2020. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi F, Qian S, Zhang S, Zhang Z. 2020. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH II, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, et al. 2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To KF, Lo AW. 2004. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol 203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. 2020. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, HCA Lung Biological Network . 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Rostami MR, Leopold PL, Mezey JG, O’Beirne SL, Strulovici-Barel Y, Crystal RG. 2020. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med 202:219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung NHL. 2021. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol 19:528–545. doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morawska L, Cao J. 2020. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int 139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tharayil A, Rajakumari R, Mozetic M, Primc G, Thomas S. 2022. Contact transmission of SARS-CoV-2 on fomite surfaces: surface survival and risk reduction. Interface Focus 12:20210042. doi: 10.1098/rsfs.2021.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Lu X, Lun W, Li T, Rao B, Wang D, Wu D, Qiu F, Yang Z, Lu J. 2021. Transmission dynamics of SARS-CoV-2 in a mid-size city of China. BMC Infect Dis 21:793. doi: 10.1186/s12879-021-06522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul A, Bhattacharjee JK, Pal A, Chakraborty S. 2021. Emergence of universality in the transmission dynamics of COVID-19. Sci Rep 11:18891. doi: 10.1038/s41598-021-98302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez A, Srigley J. 2022. Transmission of SARS-CoV-2: still up in the air. Lancet 399:519. doi: 10.1016/S0140-6736(21)02794-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, Ayeh SK, Chidambaram V, Karakousis PC. 2021. Modes of transmission of SARS-CoV-2 and evidence for preventive behavioral interventions. BMC Infect Dis 21:496. doi: 10.1186/s12879-021-06222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. 2021. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 397:1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prather KA, Marr LC, Schooley RT, McDiarmid MA, Wilson ME, Milton DK. 2020. Airborne transmission of SARS-CoV-2. Science 370:303–304. doi: 10.1126/science.abf0521. [DOI] [PubMed] [Google Scholar]
- 24.Firle C, Steinmetz A, Stier O, Stengel D, Ekkernkamp A. 2022. Aerosol emission from playing wind instruments and related COVID-19 infection risk during music performance. Sci Rep 12:8598. doi: 10.1038/s41598-022-12529-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon KS, Park JI, Park YJ, Jung DM, Ryu KW, Lee JH. 2020. Evidence of long-distance droplet transmission of SARS-CoV-2 by direct air flow in a restaurant in Korea. J Korean Med Sci 35:e415. doi: 10.3346/jkms.2020.35.e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong SCY, Kwong RT, Wu TC, Chan JWM, Chu MY, Lee SY, Wong HY, Lung DC. 2020. Risk of nosocomial transmission of coronavirus disease 2019: an experience in a general ward setting in Hong Kong. J Hosp Infect 105:119–127. doi: 10.1016/j.jhin.2020.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNeill VF. 2022. Airborne transmission of SARS-CoV-2: evidence and implications for engineering controls. Annu Rev Chem Biomol Eng 13:123–140. doi: 10.1146/annurev-chembioeng-092220-111631. [DOI] [PubMed] [Google Scholar]
- 29.Franks TJ, Chong PY, Chui P, Galvin JR, Lourens RM, Reid AH, Selbs E, McEvoy CP, Hayden CD, Fukuoka J, Taubenberger JK, Travis WD. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol 34:743–748. doi: 10.1016/s0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farooqi FI, Morgan RC, Dhawan N, Dinh J, Yatzkan G, Michel G. 2020. Airway hygiene in COVID-19 pneumonia: treatment responses of 3 critically ill cruise ship employees. Am J Case Rep 21:e926596. doi: 10.12659/AJCR.926596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. 2020. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 33.Dhand R, Li J. 2020. Coughs and sneezes: their role in transmission of respiratory viral infections, including SARS-CoV-2. Am J Respir Crit Care Med 202:651–659. doi: 10.1164/rccm.202004-1263PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afewerky HK. 2020. Pathology and pathogenicity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Exp Biol Med (Maywood) 245:1299–1307. doi: 10.1177/1535370220942126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. 2020. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for COVID-19 . 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. 2020. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology 296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, Wang H, Shen H, Li Z, Geng J, Han H, Cai J, Li X, Kang W, Weng D, Lu Y, Wu D, He L, Yao K. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol 200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. 2020. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 42:505–514. doi: 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi Y, Lagniton PN, Ye S, Li E, Xu R-H. 2020. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci 16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin W, Cao W, Zhou G, Wang L, Sun J, Zhu A, Wang Z, Zhou Y, Liu X, Li Y, Zhong N, Zhao J, Liu L, Ran P. 2021. Analysis of pathological changes in the epithelium in COVID-19 patient airways. ERJ Open Res 7:00690-2020. doi: 10.1183/23120541.00690-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimura S, Andoh Y, Haraguchi M, Shirato K. 1996. Continuity of airway goblet cells and intraluminal mucus in the airways of patients with bronchial asthma. Eur Respir J 9:1395–1401. doi: 10.1183/09031936.96.09071395. [DOI] [PubMed] [Google Scholar]
- 44.Lippi G, Henry BM. 2020. Chronic obstructive pulmonary disease is associated with severe coronavirus disease 2019 (COVID-19). Respir Med 167:105941. doi: 10.1016/j.rmed.2020.105941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Lian N, Deng Y, Lin S. 2020. The impact of COPD and smoking history on the severity of COVID-19: a systemic review and meta-analysis. J Med Virol 92:1915–1921. doi: 10.1002/jmv.25889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CDC. 2020. People with certain medical conditions. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html. Accessed 25 October 2020.
- 47.Sin DD. 2020. COVID-19 in COPD: a growing concern. EClinicalMedicine 26:100546. doi: 10.1016/j.eclinm.2020.100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung JM, Niikura M, Yang CWT, Sin DD. 2020. COVID-19 and COPD. Eur Respir J 56:2002108. doi: 10.1183/13993003.02108-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osan JK, DeMontigny BA, Mehedi M. 2021. Immunohistochemistry for protein detection in PFA-fixed paraffin-embedded SARS-CoV-2-infected COPD airway epithelium. STAR Protoc 2:100663. doi: 10.1016/j.xpro.2021.100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rayner RE, Makena P, Prasad GL, Cormet-Boyaka E. 2019. Optimization of normal human bronchial epithelial (NHBE) cell 3D cultures for in vitro lung model studies. Sci Rep 9:500. doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pawlina W. 2016. Chapter 19: Respiratory system p 671–697. In Pawlina W (ed), Rehistology: a text book and atlas with correlated cell and molecular biology, 7th ed. Wolters Kluwer, Philadelphia, PA. [Google Scholar]
- 52.Rigden HM, Alias A, Havelock T, O'Donnell R, Djukanovic R, Davies DE, Wilson SJ. 2016. Squamous metaplasia is increased in the bronchial epithelium of smokers with chronic obstructive pulmonary disease. PLoS One 11:e0156009. doi: 10.1371/journal.pone.0156009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitsett JA. 2018. Airway epithelial differentiation and mucociliary clearance. Ann Am Thorac Soc 15:S143–S148. doi: 10.1513/AnnalsATS.201802-128AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caramori G, Di Gregorio C, Carlstedt I, Casolari P, Guzzinati I, Adcock IM, Barnes PJ, Ciaccia A, Cavallesco G, Chung KF, Papi A. 2004. Mucin expression in peripheral airways of patients with chronic obstructive pulmonary disease. Histopathology 45:477–484. doi: 10.1111/j.1365-2559.2004.01952.x. [DOI] [PubMed] [Google Scholar]
- 55.Ma R, Wang Y, Cheng G, Zhang HZ, Wan HY, Huang SG. 2005. MUC5AC expression up-regulation goblet cell hyperplasia in the airway of patients with chronic obstructive pulmonary disease. Chin Med Sci J 20:181–184. [PubMed] [Google Scholar]
- 56.Huang X, Guan W, Xiang B, Wang W, Xie Y, Zheng J. 2022. MUC5B regulates goblet cell differentiation and reduces inflammation in a murine COPD model. Respir Res 23:11. doi: 10.1186/s12931-021-01920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, Radicioni G, Kesimer M, Chua M, Dang H, Livraghi-Butrico A, Ehre C, Doerschuk CM, Randell SH, Matsui H, Nagase T, O'Neal WK, Boucher RC. 2019. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am J Respir Crit Care Med 199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiyokawa H, Morimoto M. 2020. Notch signaling in the mammalian respiratory system, specifically the trachea and lungs, in development, homeostasis, regeneration, and disease. Dev Growth Differ 62:67–79. doi: 10.1111/dgd.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crystal RG. 2014. Airway basal cells. The “smoking gun” of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 190:1355–1362. doi: 10.1164/rccm.201408-1492PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang BY, Gil J, Kaufman D, Gan L, Kohtz DS, Burstein DE. 2002. P63 in pulmonary epithelium, pulmonary squamous neoplasms, and other pulmonary tumors. Hum Pathol 33:921–926. doi: 10.1053/hupa.2002.126878. [DOI] [PubMed] [Google Scholar]
- 61.Persson BD, Jaffe AB, Fearns R, Danahay H. 2014. Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS One 9:e102368. doi: 10.1371/journal.pone.0102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polosukhin VV, Cates JM, Lawson WE, Zaynagetdinov R, Milstone AP, Massion PP, Ocak S, Ware LB, Lee JW, Bowler RP, Kononov AV, Randell SH, Blackwell TS. 2011. Bronchial secretory immunoglobulin a deficiency correlates with airway inflammation and progression of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 184:317–327. doi: 10.1164/rccm.201010-1629OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higham A, Quinn AM, Cancado JED, Singh D. 2019. The pathology of small airways disease in COPD: historical aspects and future directions. Respir Res 20:49. doi: 10.1186/s12931-019-1017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brusselle GG, Joos GF, Bracke KR. 2011. New insights into the immunology of chronic obstructive pulmonary disease. Lancet 378:1015–1026. doi: 10.1016/S0140-6736(11)60988-4. [DOI] [PubMed] [Google Scholar]
- 65.Barnes PJ. 2013. New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat Rev Drug Discov 12:543–559. doi: 10.1038/nrd4025. [DOI] [PubMed] [Google Scholar]
- 66.Kirkham S, Sheehan JK, Knight D, Richardson PS, Thornton DJ. 2002. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J 361:537–546. doi: 10.1042/0264-6021:3610537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirkham S, Kolsum U, Rousseau K, Singh D, Vestbo J, Thornton DJ. 2008. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 178:1033–1039. doi: 10.1164/rccm.200803-391OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lillehoj EP, Kato K, Lu W, Kim KC. 2013. Cellular and molecular biology of airway mucins. Int Rev Cell Mol Biol 303:139–202. doi: 10.1016/B978-0-12-407697-6.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silva MA, Bercik P. 2012. Macrophages are related to goblet cell hyperplasia and induce MUC5B but not MUC5AC in human bronchus epithelial cells. Lab Invest 92:937–948. doi: 10.1038/labinvest.2012.15. [DOI] [PubMed] [Google Scholar]
- 70.Kim V, Oros M, Durra H, Kelsen S, Aksoy M, Cornwell WD, Rogers TJ, Criner GJ. 2015. Chronic bronchitis and current smoking are associated with more goblet cells in moderate to severe COPD and smokers without airflow obstruction. PLoS One 10:e0116108. doi: 10.1371/journal.pone.0116108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaykhiev R. 2019. Emerging biology of persistent mucous cell hyperplasia in COPD. Thorax 74:4–6. doi: 10.1136/thoraxjnl-2018-212271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reid AT, Veerati PC, Gosens R, Bartlett NW, Wark PA, Grainge CL, Stick SM, Kicic A, Moheimani F, Hansbro PM, Knight DA. 2018. Persistent induction of goblet cell differentiation in the airways: therapeutic approaches. Pharmacol Ther 185:155–169. doi: 10.1016/j.pharmthera.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 73.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB Jr. 2005. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science 369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH 3rd, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schafer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schaefer IM, Padera RF, Solomon IH, Kanjilal S, Hammer MM, Hornick JL, Sholl LM. 2020. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol 33:2104–2114. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giacca M, Bussani R, Schneider E, Zentilin L, Collesi C, Ali H, Braga L, Secco I, Volpe MC, Colliva A. 2020. Persistence of viral RNA, widespread thrombosis and abnormal cellular syncytia are hallmarks of COVID-19 lung pathology. medRxiv doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyerholz DK, Lambertz AM, McCray PB, Jr. 2016. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol 186:78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borczuk AC, Salvatore SP, Seshan SV, Patel SS, Bussel JB, Mostyka M, Elsoukkary S, He B, Del Vecchio C, Fortarezza F, Pezzuto F, Navalesi P, Crisanti A, Fowkes ME, Bryce CH, Calabrese F, Beasley MB. 2020. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol 33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Randell SH. 2006. Airway epithelial stem cells and the pathophysiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc 3:718–725. doi: 10.1513/pats.200605-117SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, Bullock H, Goldsmith CS, Silva-Flannery L, Seixas JN, Reagan-Steiner S, Uyeki T, Denison A, Bhatnagar J, Shieh WJ, Zaki SR, COVID-19 Pathology Working Group . 2020. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis 26:2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gohy S, Carlier FM, Fregimilicka C, Detry B, Lecocq M, Ladjemi MZ, Verleden S, Hoton D, Weynand B, Bouzin C, Pilette C. 2019. Altered generation of ciliated cells in chronic obstructive pulmonary disease. Sci Rep 9:17963. doi: 10.1038/s41598-019-54292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 87.Wolf S, Perez GF, Mukharesh L, Isaza N, Preciado D, Freishtat RJ, Pillai D, Rose MC, Nino G. 2017. Conditional reprogramming of pediatric airway epithelial cells: a new human model to investigate early-life respiratory disorders. Pediatr Allergy Immunol 28:810–817. doi: 10.1111/pai.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogers DF. 2003. The airway goblet cell. Int J Biochem Cell Biol 35:1–6. doi: 10.1016/s1357-2725(02)00083-3. [DOI] [PubMed] [Google Scholar]
- 90.Hao S, Ning K, Kuz CA, Vorhies K, Yan Z, Qiu J. 2020. Long-term modeling of SARS-CoV-2 infection of in vitro cultured polarized human airway epithelium. mBio 11:e02852-20. doi: 10.1128/mBio.02852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mulay A, Konda B, Garcia G, Jr, Yao C, Beil S, Villalba JM, Koziol C, Sen C, Purkayastha A, Kolls JK, Pociask DA, Pessina P, de Aja JS, Garcia-de-Alba C, Kim CF, Gomperts B, Arumugaswami V, Stripp BR. 2021. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep 35:109055. doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. 2020. Aerosol and surface stability of HCoV-19 (SARS-CoV-2) compared to SARS-CoV-1. medRxiv doi: 10.1101/2020.03.09.20033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA 101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 76:5654–5666. doi: 10.1128/jvi.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Falasca L, Nardacci R, Colombo D, Lalle E, Di Caro A, Nicastri E, Antinori A, Petrosillo N, Marchioni L, Biava G, D'Offizi G, Palmieri F, Goletti D, Zumla A, Ippolito G, Piacentini M, Del Nonno F. 2020. Postmortem findings in Italian patients with COVID-19: a descriptive full autopsy study of cases with and without comorbidities. J Infect Dis 222:1807–1815. doi: 10.1093/infdis/jiaa578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA. 2020. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Araya J, Cambier S, Markovics JA, Wolters P, Jablons D, Hill A, Finkbeiner W, Jones K, Broaddus VC, Sheppard D, Barzcak A, Xiao Y, Erle DJ, Nishimura SL. 2007. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest 117:3551–3562. doi: 10.1172/JCI32526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Synowiec A, Szczepański A, Barreto-Duran E, Lie LK, Pyrc K. 2021. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a systemic infection. Clin Microbiol Rev 34:e00133-20. doi: 10.1128/CMR.00133-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, Moller R, Hoagland D, Oishi K, Horiuchi S, Uhl S, Blanco-Melo D, Albrecht RA, Liu WC, Jordan T, Nilsson-Payant BE, Golynker I, Frere J, Logue J, Haupt R, McGrath M, Weston S, Zhang T, Plebani R, Soong M, Nurani A, Kim SM, Zhu DY, Benam KH, Goyal G, Gilpin SE, Prantil-Baun R, Gygi SP, Powers RK, Carlson KE, Frieman M, tenOever BR, Ingber DE. 2021. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng 5:815–829. doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee J, Taneja V, Vassallo R. 2012. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res 91:142–149. doi: 10.1177/0022034511421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rovina N, Koutsoukou A, Koulouris NG. 2013. Inflammation and immune response in COPD: where do we stand? Mediators Inflamm 2013:413735. doi: 10.1155/2013/413735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. American J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 to S7. Download spectrum.00459-22-s0001.pdf, PDF file, 1.1 MB (1.2MB, pdf)