Abstract
The degradation of the McpA chemoreceptor in Caulobacter crescentus accompanies the swarmer cell to the stalked-cell differentiation event. To further analyze the requirements for its degradation, we have constructed a series of strains that have deletions in the mcpA gene and in the mcpA chemotaxis operon. Internal deletions of the mcpA gene demonstrate that the highly conserved domain (signalling unit) and the methylation domains are not required for cell cycle-regulated proteolysis. The deletion of the chemotaxis operon, which is absolutely required for chemotaxis and McpA chemoreceptor methylation, has no effect on McpA proteolysis.
Proteolysis is an important mechanism in both cell differentiation and progression through the cell cycle. In Caulobacter crescentus, swarmer cells must undergo an obligate differentiation event to become sessile stalked cells before they can start to grow and divide. The differentiation of the motile swarmer cell to nonmotile stalked cell is accompanied by the proteolysis of key proteins involved in motility (3, 5, 7, 10, 17). The proteolysis of the McpA chemoreceptor (3) is accompanied by a dramatic decrease in methylesterase and methyltransferase activities (7). Unlike swarmer cells, the newly formed stalked cells are capable of initiating DNA replication. Once DNA replication is initiated, the stalked cells grow and will progress through the cell cycle to develop into predivisional cells, where the components required for motility are synthesized and targeted to the portion of the cell that will form the swarmer cell. The chemoreceptor McpA is specifically localized to the flagellated pole of the predivisional cell (2). The ability to localize chemoreceptors to the poles of the cell is not restricted to C. crescentus, as it has been shown to occur in Escherichia coli (15) and is likely to be a universal phenomenon found in other prokaryotes (8). Unlike in other prokaryotes, the chemoreceptors in C. crescentus are degraded during its life cycle; this proteolysis event plays an important part in the asymmetric distribution of the polarly localized chemoreceptors (3).
The CheR and CheB binding site, a pentapeptide (4, 20), is conserved in McpA and is located at the extreme C terminus of chemoreceptors. McpA is a methylated chemoreceptor (1), suggesting that CheR and CheB bind to the C terminus. The extreme C terminus of McpA has been shown to be required for its degradation (3). Therefore, one explanation for the observation that C-terminal deletions were no longer degraded is that they were not able to form complexes with CheR and the other proteins of the chemotaxis machinery. The methylesterase (CheB) and methyltransferase (CheR) activities are lost (7) about the same time as McpA is degraded, suggesting that there is some coordinate regulation of the chemotaxis machinery at the level of proteolysis. Because of the coincidental loss of the methylesterase, methyltransferase, and McpA, we wanted to determine what role the methylation domains (Fig. 1A) play in the degradation of McpA. We constructed internal deletions of the mcpA gene in order to avoid perturbation of the C terminus, as this has been already shown to be required for proteolysis (3). The construction of mcpA internal deletions will aid in determining whether the highly conserved signalling domain (HCD) is required for McpA proteolysis and also map the N-terminal extent of the putative degradation signal.
FIG. 1.
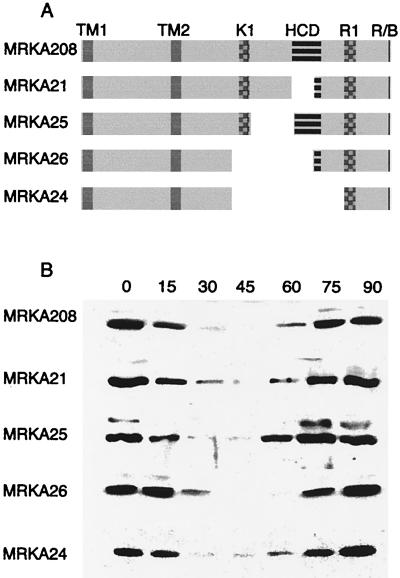
Cell cycle immunoblots of mcpA internal deletions. (A) mcpA internal deletions. The pale grey boxes denote the extent of the McpA protein present in the various chromosomal deletions of the mcpA gene in the named strains (Table 1). The transmembrane domains are denoted by TM1 and TM2 and are colored in grey. The methylation domains (checkered) are named after the K1- and R1-methylated peptides shown in the E. coli Tsr chemoreceptor (11). The HCD (striped box) is the most highly conserved domain in all chemoreceptors (13); it is involved in signalling. The gaps denote the extent of the deletions in the mcpA gene in the named C. crescentus strains. The black box at the C terminus of McpA is the CheR-CheB binding site (R/B) (4, 20). (B) Cell cycle immunoblots. Strains MRKA208, which is the wild-type strain in these experiments, MRKA21, MRKA25, MRKA26, and MRKA24 were synchronized by Percoll (Pharmacia) density centrifugation. Samples were taken at the times (minutes) indicated during the 90-min cell cycle. The same amount of cells was loaded in each lane. The cell extracts were subjected to electrophoresis on a sodium dodecyl sulfate–8% polyacrylamide gel and transferred to nitrocellulose (19); the primary antiserum was to McpA, and the secondary antiserum was anti-rabbit conjugated to horseradish peroxidase.
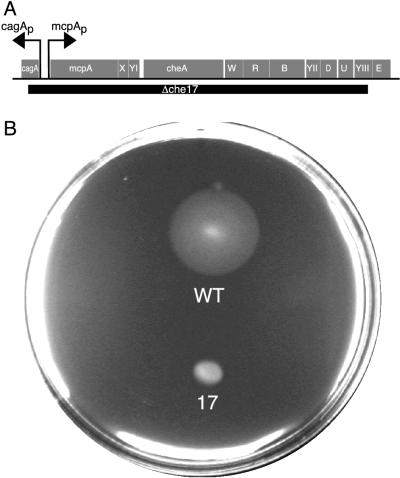
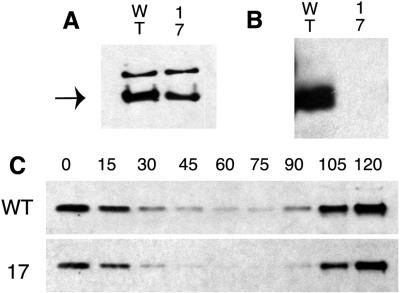
The first deletion strain, MRKA21 (Table 1), was constructed by conjugating the suicide plasmid pWA12ΔAS (Table 2) into MRKA208 by using the helper plasmid pRK600 in MT607 (Table 1). The kanamycin sucrose-sensitive transconjugants were then spread on 3% sucrose plates to obtain excision of the kanamycin-resistant plasmid from the chromosome. All sucrose-resistant colonies were subjected to Southern and immunoblot analyses, and one positive colony was named MRKA21. All subsequent internal deletions were obtained with this methodology. The MRKA21 strain had most of the HCD deleted from the mcpA gene on the chromosome (Fig. 1A). The deletion of the HCD in McpA did not prevent its proteolysis (Fig. 1B), and the degradation pattern was not significantly different from the pattern observed with the wild-type strain MRKA208 (Fig. 1B). Deletion of part of the HCD and the intervening sequence to the K1 methylation domain resulted in strain MRKA25 (Fig. 1A), which still degraded McpA (Fig. 1B). Therefore, we deleted the sequence coding for the K1 methylation domain and most of the HCD in mcpA to create strain MRKA26, as shown in Fig. 1A. Strain MRKA26 still degraded McpA (Fig. 1B). The absence of an effect on McpA degradation in the various K1 and HCD deletion derivatives of McpA suggested that a much larger deletion should be created. Hence, we deleted the entire K1 methylation domain and HCD signalling unit (Fig. 1A). The resulting deletion strain MRKA24 still degraded McpA (Fig. 1B). The only conserved cytoplasmic domain remaining that is known to interact with other chemotaxis proteins in the McpA deletion present in strain MRKA24 (Fig. 1A) was the C-terminal CheR-CheB binding site. Because C-terminal deletions that removed this CheR-CheB binding site from McpA have been shown to be stable (3), we wanted to determine whether CheR and CheB are required for McpA degradation. Because cheR and cheB reside in an operon with cheA and cheW (Fig. 2A), we decided to delete the entire operon to test whether any of the chemotaxis genes present in this operon are required for McpA degradation. Therefore, we created strain MRKA580; the Δche17 deletion removes the mcpA operon promoter and every gene in the mcpA operon except cheE (Fig. 2A). Since the mcpA operon promoter is deleted (Fig. 2A), the only gene left intact (cheE) will not be expressed. The deletion of the cagA gene will have no phenotypic effect because there is a second copy of the cagA gene in the C. crescentus genome, and deletion of the first cagA gene has no observable phenotype (M. R. K. Alley, unpublished data). The Δche17 chemotaxis deletion strain MRKA580 produces very small swarms in semisolid media (Fig. 2B). The swarmer cells do not reverse their swimming direction, and predivisional cells swim with their stalks in front, which is the predominant direction of swimming (12). The Δche17 deletion removes the mcpA gene, so we introduced the mcpA gene on a plasmid, pRMCP4, by triparental mating with MT607. The plasmid pRMCP4 is a low-copy-number plasmid (two to five copies per cell) of the IncP-1 incompatibility group. Since the number of copies of the mcpA gene on pRMCP4 are above the normal level found in the wild-type strain MRKA208, we also introduced pRMCP4 into MRKA208 as a control for any plasmid copy number effects. As shown in the immunoblot in Fig. 3A, the wild-type strain makes slightly more McpA than the Δche17 deletion strain MRKA580, which is probably due to the extra copy present on the chromosome of MRKA208. The MRKA580 strain is not able to methylate its McpA chemoreceptors (Fig. 3B), further demonstrating that the MRKA580 strain is defective in chemotaxis. No methylation of McpA was observed in extracts from the MRKA580 strain, even after a 1-month exposure to X-ray film. Therefore, the cheR gene in the mcpA operon is essential for McpA methylation. The wild-type strain bearing pRMCP4 degrades McpA (Fig. 3C), and therefore, there are no effects on McpA degradation due to the increase in levels of McpA or the presence of the IncP-1 plasmid. McpA is degraded in the Δche17 deletion strain MRKA580, suggesting that the genes in the mcpA operon are not required for McpA proteolysis and that McpA does not need a functional chemotaxis system for its degradation. Furthermore, if CheR methyltransferase and CheB methylesterase are not required for McpA proteolysis, we can postulate that the CheR-CheB binding site is involved only in McpA methylation; therefore, we can exclude its requirement in McpA degradation.
TABLE 1.
Strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH10B | F−mcrA (mrr-hsdRMS-mcrBC) ΔlacX74 φ80dlacZ ΔM15 deoR recA1 endA1 araD139 rpsL | Gibco BRL |
| MT607 | MM294 recA56 pRK2013 aphAIΩTn9 | 16 |
| C. crescentus | ||
| MRKA208 | NA1000 Δbla6 | M. R. K. Alleya |
| MRKA21 | NA1000 Δbla6 ΔmcpA8 | This study |
| MRKA24 | NA1000 Δbla6 ΔmcpA11 | This study |
| MRKA25 | NA1000 Δbla6 ΔmcpA12 | This study |
| MRKA26 | NA1000 Δbla6 ΔmcpA13 | This study |
| MRKA580 | NA1000 Δbla6 Δche17 | This study |
Unpublished data.
TABLE 2.
Plasmids
| Plasmid | Relevant characteristics and method of construction | Source or reference |
|---|---|---|
| pBGST18 | pUC18 polylinker aphAI/IncP oriT derivative of pBGS18 (18) | M. R. K. Alleya |
| pCH9 | 10.6-kb fragment in the BamHI-HindIII sites of pBGST18 | M. R. K. Alleya |
| pR6 | Chemotaxis-complementing cosmid | 1 |
| pRK290KS2 | IncP-1 replicon, pBluescript II KS polylinker | 3 |
| pCHEG5 | 5-kb BglII fragment from pR6 in the BamHI site of pBGST18 | This study |
| pMCP4 | 2.9-kb BsiWI (blunt)-XhoI fragment in the SmaI-XhoI sites of pBluescript II KS(+) (Stratagene) | This study |
| pRMCP4 | 2.9-kb XbaI-XhoI fragment from pMCP4 into the XbaI-XhoI sites of pRK290KS2 | This study |
| pCH9ΔAP | ApaI-PstI deletion of pCH9 | This study |
| pCH17 | 0.86-kb KpnI-HindI fragment from pCH9ΔAP into the KpnI-HindIII sites of pNPTS129 | This study |
| pNPTS129 | pLITMUS 29 (NEB) polylinker aphAI/IncP oriT/sacB | This study |
| pWA12ΔAS | pWA12 (1) was deleted at the SacI-ApaI sites, and the BamHI blunt-ended sacB-aphAI fragment from pMH1701 (9) was inserted into the EcoRV site. This plasmid was used to construct the MRKA21 strain. | This study |
| pCHE22ΔN | pCHE22 was deleted at the NcoI sites, and the BamHI blunt-ended sacB-aphAI fragment from pMH1701 (9) was inserted into the ScaI site. This plasmid was used to construct the MRKA24 strain. | This study |
| pCM223ΔXA | pCM223 (3) was deleted at the XmaI-ApaI sites, and the sacB gene from pIC20R-sacB (M. R. K. Alley)a was inserted into the SpeI site. This plasmid was used to construct the MRKA26 strain. | This study |
| pWA12ΔSS | pWA12 (1) was deleted at the SacI-StuI sites, and the BamHI blunt-ended sacB-aphAI fragment from pMH1701 was inserted into the EcoRV site. This plasmid was used to construct the MRKA25 strain. | This study |
Unpublished data.
FIG. 2.
The Δche17 deletion strain MRKA580 is defective in chemotaxis. (A) The grey boxes represent the open reading frames for the named genes (AJ006687). The extent of the deletion in Δche17 strains is shown by the black box. (B) A yeast extract swarm agar (0.005% yeast extract, 0.5 mM MgSO4, 0.5 mM CaCl2, 0.15% Bacto agar) plate after 48 h inoculated with equal amounts of MRKA208 (wild type [WT]) and MRKA580 (indicated by the no. 17) strains bearing the plasmid pRMCP4.
FIG. 3.
McpA methylation and cell cycle degradation in a mcpA operon deletion. (A) Immunoblot of strains MRKA208 (wild type [WT]) and MRKA580 (indicated by the no. 17) using McpA antisera. The arrow indicates the McpA protein band. (B) Immunoprecipitation of McpA in cell extracts from chloramphenicol pretreated cells labelled with [3H]methylmethionine (17). Equal numbers of counts per minute were immunoprecipitated. The cell extracts were from strains MRKA208 (WT) and MRKA580 (no. 17). (C) McpA cell cycle immunoblots of strains MRKA208 (WT) and MRKA580 (no. 17) bearing the plasmid pRMCP4. The numbers above the panel are the time points (minutes) at which the samples were taken.
The chemoreceptors are modified by methylation by CheR and CheB and form large functional complexes with CheA and CheW (6, 14). The possibility that these proteins might be involved in the stability of the McpA chemoreceptor is derived from the fact that stability of other proteins can be altered by protein complex formation. The methylesterase (CheB) and methyltransferase (CheR) activities are lost during the cell cycle (7), suggesting that the proteolysis of the chemotaxis machinery might be coordinately regulated. One possible way to coordinate the proteolysis of the chemotaxis machinery is to target the entire complex for degradation. In this study, we show that deletion of the K1 and HCD domains of McpA do not prevent its degradation (Fig. 1). When the mcpA operon was deleted, McpA was not methylated and there was no effect on the degradation of McpA. Therefore, we can assume that none of the genes in the mcpA operon are required for McpA degradation. The combined data from the internal and operon deletions would suggest that McpA is the target for its protease. Thus, McpA and the other chemoreceptors might be required for the degradation of the methylesterase and methyltransferase. To test this hypothesis, we are presently generating antisera to CheR and CheB.
Acknowledgments
This study was funded by a Wellcome Trust project grant (#044761) to M.R.K.A.
M.R.K.A. is a Royal Society Research Fellow.
REFERENCES
- 1.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley M R K, Maddock J R, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 3.Alley M R K, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 4.Barnakov A N, Barnakova L A, Hazelbauer G L. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 6.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 7.Gomes S L, Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984;178:551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- 8.Harrison D M, Skidmore J, Armitage J P, Maddock J R. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 9.Hynes M F, Quandt J, O'Connell M P, Puhler A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene. 1989;78:111–120. doi: 10.1016/0378-1119(89)90319-3. [DOI] [PubMed] [Google Scholar]
- 10.Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 11.Kehry M R, Bond M W, Hunkapiller M W, Dahlquist F W. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc Natl Acad Sci USA. 1983;80:3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koyasu S, Shirakihara Y. Caulobacter crescentus flagellar filament has a right-handed helical form. J Mol Biol. 1984;173:125–130. doi: 10.1016/0022-2836(84)90407-8. [DOI] [PubMed] [Google Scholar]
- 13.Le Moual H, Koshland D E., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S B, Signer E R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990;4:344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- 17.Shaw P, Gomes S L, Sweeney K, Ely B, Shapiro L. Methylation involved in chemotaxis is regulated during Caulobacter differentiation. Proc Natl Acad Sci USA. 1983;80:5261–5265. [PMC free article] [PubMed] [Google Scholar]
- 18.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 19.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]