Abstract
Bacteriophage λ adsorbs to its Escherichia coli K-12 host by interacting with LamB, its cell-surface receptor. We fused C-terminal portions of J, the tail fiber protein of λ, to maltose-binding protein. Solid-phase binding assays demonstrated that a purified fusion protein comprising only the last 249 residues of J could bind to LamB trimers and inhibited recognition by anti-LamB antibodies. Electron microscopy further demonstrated that the fusion protein could also bind to LamB at the surface of intact cells. This interaction prevented λ adsorption but affected only partially maltose uptake.
Bacteriophage λ adsorbs to the surface of Escherichia coli K-12 by interacting with the outer membrane protein LamB (10). LamB is a trimeric protein that forms nonspecific channels through the outer membrane (11), allowing the diffusion of small hydrophilic molecules (<600 Da). In addition, LamB is a sugar-specific porin that facilitates the diffusion of maltose and maltodextrins into the cell. Genetic analyses of LamB mutants tightly blocking phage λ adsorption allowed the identification of a series of amino acid sites, clustered mainly on three cell surface-exposed loops of the protein (reviewed in references 2 and 4). Genetic analyses of mutants of λ able to use such mutated LamB receptors (7, 14) showed that the amino acid substitutions responsible for this compensatory or suppressor effect were all located in the C-terminal portion of J, a 1,132-amino-acid protein constituting the tail fiber of λ. This suggested strongly that the C-terminal portion of J was interacting with LamB, but did not exclude totally that suppression could be due to a long-range effect and that another part of J, such as the N-terminal portion, was responsible for direct interaction with LamB.
In the present work, we directly tested the capacity of the C-terminal portion of J fused to maltose-binding protein (MBP) to bind to LamB. Genetic coupling to MBP allowed the purification of the fusion proteins by affinity chromatography with an amylose column under nondenaturing conditions, and the MBP moiety was used as a tag to reveal the fraction of MBP-J bound to LamB. We were able to demonstrate for the first time that the 20% distal portion of J was sufficient to allow binding to the LamB receptor in vitro and in vivo.
Construction, expression, and purification of the MBP-J hybrid proteins.
Genetic constructs which expressed three fusion proteins consisting of the distal portion of the J protein of λ fused to the C-terminal end of the carrier MBP were made. The shortest fusion protein, designated MBP-J/S, contained residues 884 to 1132 of J (i.e., 249 residues). The two larger MBP-J fusion proteins, designated MBP-J/M and MBP-J/L, contained 100 and 200 additional residues of J, respectively.
Purified DNA from gene J of phage λh+ (laboratory collection), coding for the wild-type J protein, was used as the template DNA in PCR performed to produce the 3′ end of the J gene. Three pairs of primers were used to generate the three double-stranded DNA fragments flanked by EcoRI and HindIII sites. The oligonucleotides of the 5′ ends (coding strands) were 5′-CG GAA TTC ATG GAG GAC ACG GAG GAA GGC-3′, 5′-CG GAA TTC GAT TAT TAC TTT TAT ATC CGC-3′, and 5′-CG GAA TTC GCG CTG GGG AAC TAC AGG CTG-3′ (the EcoRI site is underlined). The oligonucleotide of the 3′ end of J (complementary strand) in all three cases was as follows: 5′-CC AAG CTT TCA GAC CAC GCT GAT GCC CAG-3′ (the HindIII site is underlined). Oligonucleotides were synthesized by Genset (Paris, France).
The pMAL-c vector (New England Biolabs, Beverly, Mass.) used to construct the three C-terminal MBP-J fusion proteins carries a deletion of the signal sequence of MBP and therefore directs the expression (under the control of the isopropyl-β-d-thiogalactopyranoside [IPTG]-inducible ptac promoter) of the fusion proteins in the cytoplasm of the host cells. The PCR fragments were inserted downstream of the malE gene (between the EcoRI and HindIII sites). The LamB-negative mutant of E. coli K-12, JM 501 (14), was the recipient for the recombinant plasmids. The amounts of MBP-J fusion proteins MBP-J/L, MBP-J/M, and MBP-J/S expressed under IPTG-induced conditions corresponded to 17, 23, and 32%, respectively, of whole-cell proteins (data not shown). MBP-J/M and MBP-J/S were chosen for further purification. IPTG-induced cultures were broken by passage through a French press, and the MBP-J hybrid proteins were purified by affinity chromatography with an amylose column as previously described (12). Densitometric scanning of Coomassie blue-stained gels (Fig. 1) indicated that MBP-J/M and MBP-J/S were more than 90% pure. In both constructs, no major proteolytic cleavage products were detected by Western blot with anti-MBP serum (data not shown).
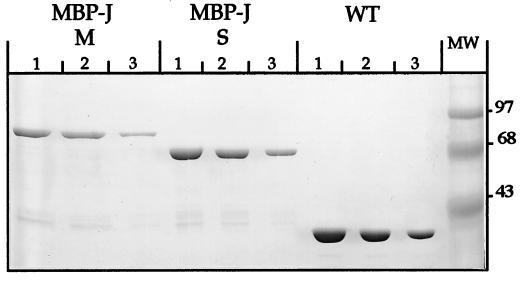
FIG. 1.
Sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis of the purified proteins. From left to right, the sets of lanes contain MBP-J/M (residues 784 to 1132 of J), MBP-J/S (residues 884 to 1132 of J) (for the two hybrid proteins, amounts were as follows: lanes 1, 15 μg; lanes 2, 10 μg; and lanes 3, 5 μg), and purified MBP (loaded as a control [lanes 1, 2, and 3 contain 30, 20, and 10 μg of protein, respectively]). The gel was stained with Coomassie brilliant blue R-250.
Interaction with purified LamB trimers in vitro.
Three different solid-phase binding assays (Western blot, dot blot, and enzyme-linked immunosorbent assay [ELISA]) demonstrated that the distal portion of the tail fiber protein J of λ was sufficient to allow binding to LamB trimers in vitro.
Dot blot and Western blot.
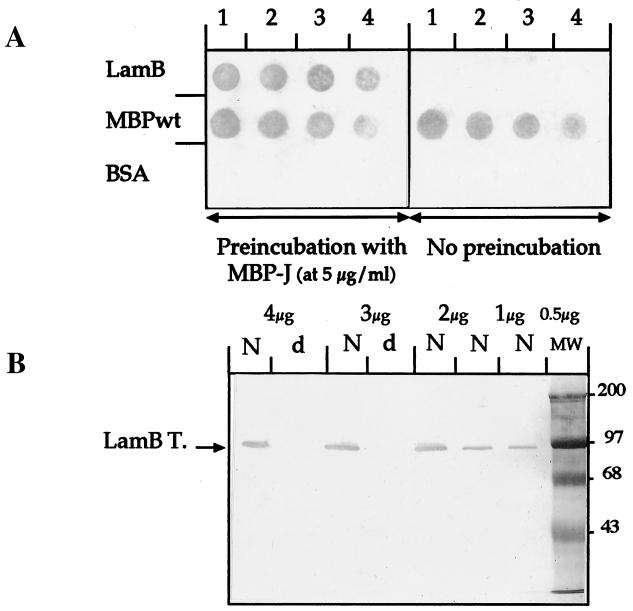
In the dot blot and the Western blot assays (Fig. 2), performed essentially as described in reference 13, the two hybrid proteins MBP-J/M and MBP-J/S yielded similar results. Only the data obtained with MBP-J/S are presented and discussed below. Nitrocellulose strips were coated with decreasing amounts of native purified LamB protein, wild-type MBP (MBPwt), or bovine serum albumin (BSA), and the nitrocellulose strips were incubated with purified MBP-J protein. As shown in Fig. 2A, the anti-MBP serum (a polyclonal antibody against purified MBP raised in rabbits [9]) reacted strongly with the spots corresponding to LamB and to MBPwt on the sheet preincubated with MBP-J but only with the spots corresponding to MBPwt on the nonpreincubated sheet. There was no nonspecific binding of BSA or MBPwt to LamB, and the anti-MBP serum did not cross-react nonspecifically with LamB or BSA. The Western blot assay further indicated that the MBP-J hybrid protein bound to LamB trimers but not to the denatured LamB monomers (Fig. 2B). This result does not rule out the idea that the hybrid protein would bind to native LamB monomers, although, to our knowledge, the existence of such a species has not been demonstrated.
FIG. 2.
Binding to LamB used to coat nitrocellulose. (A) Dot blot binding assay. Nitrocellulose was coated with decreasing amounts of either purified LamB trimers (lanes 1, 1 μg; lanes 2, 0.5 μg; lanes 3, 0.1 μg; and lanes 4, 0.05 μg), purified MBPwt (lanes 1, 20 μg; lanes 2, 10 μg; lanes 3, 5 μg; and lanes 4, 1 μg) or BSA (lanes 1, 10 μg; lanes 2, 5 μg; lanes 3, 1 μg; and lanes 4, 0.5 μg). After saturation with 2% gelatine, the sheet on the left part of the figure was preincubated with MBP-J/S at a final concentration of 5 μg/ml. The control sheet on the right part of the figure was not preincubated. (B) Western blot binding assay. Sodium dodecyl sulfate–10% gels were loaded with decreasing amounts of purified LamB trimers (from 4 to 0.5 μg as indicated on the figure). Samples were either resuspended in native loading buffer (without β-mercaptoethanol) and incubated at 37°C for 10 min (lanes N) or resuspended in denaturing loading buffer (with β-mercaptoethanol) and incubated at 100°C for 10 min (lanes d). The proteins were transferred electrophoretically onto nitrocellulose. After saturation, the sheet was preincubated with MBP-J/S (5 μg/ml). The unbound MBP-J hybrid was eliminated by extensive washes in PBS–0.1% Tween 20. We also performed the assay with a control sheet coated with the same samples but not preincubated with MBP-J hybrid. In the absence of preincubation, nothing was detected (data not shown). Lane MW, molecular weight markers. LamB T., LamB trimers.
ELISA.
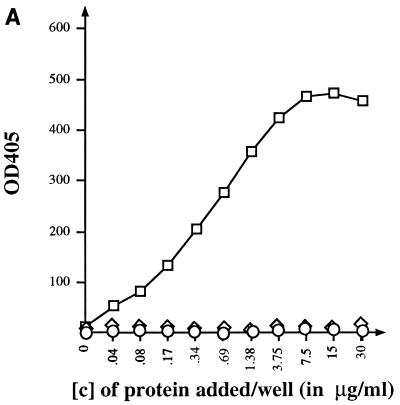
The ability of the short MBP-J fusion protein (MBP-J/S) to bind to purified LamB trimers was quantified in an ELISA. Microtitration plates (Nunc-Immuno; Inter Med, Roskilde, Denmark) were coated with a suspension of LamB trimers at a concentration of 1.5 μg/ml (100 μl per well) and incubated overnight at 37°C. The plates were washed and then saturated with 2% gelatine for 2 h at 37°C. After several washes in phosphate-buffered saline (PBS), serial dilutions of MBP-J fusion protein were added to the wells and the plates were incubated for 1 h at 37°C. The fraction of MBP-J bound to LamB was measured after reaction with anti-MBP antibody (at a final dilution of 1:1,000). Goat anti-rabbit immunoglobulin G (IgG) (heavy plus light chains [H+L]) alkaline phosphatase conjugate (final dilution, 1:1,000) and alkaline phosphatase substrate kit (Bio-Rad) were used for color detection of the LamB–MBP-J–anti-MBP complexes. As shown in Fig. 3A, MBP-J/S was found to bind tightly to LamB trimers coated to the plate. MBPwt as well as BSA did not bind to LamB.
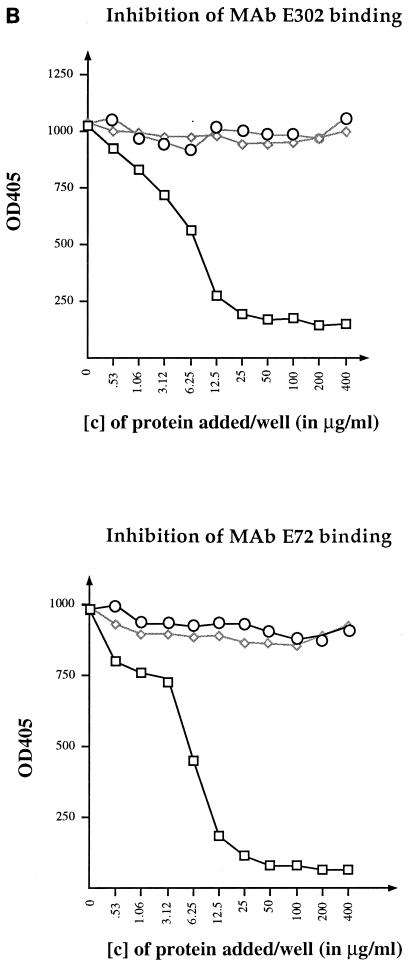
FIG. 3.
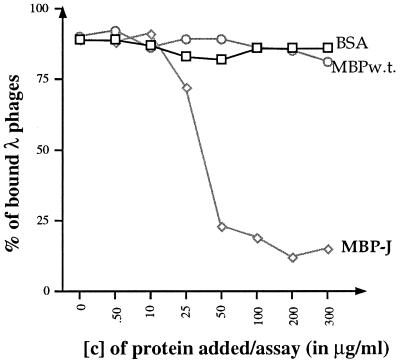
ELISA binding assays. (A) Binding of MBP-J to purified LamB trimers. Each well was coated with 150 ng of purified LamB trimers. Increasing concentrations of MBP-J/S were added. The fraction of MBP-J bound to LamB was revealed with anti-MBP antibody at a final dilution of 1/1,000. (B) Inhibition of anti-LamB antibody binding to purified LamB trimers. The plate was coated with purified LamB trimers (150 ng/well). Increasing concentrations of MBP-J/S were added to each well. The ELISA was carried out with anti-LamB mouse MAb E302 or MAb E72. □, MBP-J/S hybrid; ◊, MBPwt; ○, BSA. [c], concentration. OD405, optical density at 405 nm.
The capacity of bound MBP-J fusion protein to inhibit binding of anti-LamB monoclonal antibodies (MAbs) was also tested (Fig. 3B). For this, plates were coated with LamB trimers and incubated with serial dilutions of MBP-J, as described above. After several washes in PBS, 100 μl of anti-LamB MAb (at a final dilution of 1:500) was added and the plates were incubated at 37°C for 1 h. The LamB-MAb complexes were finally detected with goat anti-mouse IgG (H+L) alkaline phosphatase conjugate at a final dilution of 1:1,000. Two anti-LamB MAbs (E302 and E72) against purified native E. coli K-12 LamB protein, raised in mice and specific to cell surface-exposed regions of the protein, were used (5, 6). With both MAbs, a strong inhibition of binding to LamB was observed upon preincubation with MBP-J/S. One hypothesis to account for this inhibitory effect would be that the phage tail tip competes with the MAbs for the same binding site. In agreement with this hypothesis, previous genetic analyses showed that the binding sites for phages and MAbs at the surface of LamB were distinct but partially overlapping (2). However, the inhibition could also be the result of steric hindrance.
In vivo interactions between MBP-J hybrid and the LamB receptor.
We next tested the effects of MBP-J binding to intact cells on the phage receptor and the maltoporin activities of LamB and directly demonstrated binding to LamB by electron microscopy.
Inhibition of phage λ binding.
Different concentrations of purified MBP-J fusion protein (in 10-μl volumes) were first incubated at room temperature for 20 min with bacterial suspensions of the LamB-positive strain P4X8 (in 100-μl volumes). Then 10 μl of a dilution of phage λ, containing approximately 500 PFU, per mixture was added, and the incubation was continued for 20 min. The mixtures were finally centrifuged at 10,000 × g in a microcentrifuge for 5 min. The number of infectious phage particles still present in the supernatants (i.e., the unbound phage) was determined by spreading onto indicator bacteria (Luria broth-grown P4X8). As shown in Fig. 4, an almost complete inhibition of phage λ binding was obtained when MBP-J/S was added at a final concentration of 50 μg/ml. In contrast, BSA and MBPwt had no effect on phage binding.
FIG. 4.
In vivo inhibition of phage λ binding. The one-step extended host range derivative of wild-type λ phage (λh° [laboratory collection]) was used in this assay. Sensitivity to λ was assayed by spot tests on lawns of the LamB-positive E. coli K-12 strain, P4X8, plated by the tryptone overlay method (1). Plates were incubated overnight at 37°C. The fraction of λ phages bound to the bacteria was determined by subtracting the number of infectious phage particles still present in the supernatants (the fraction not bound to the bacterial pellet) from the total number of infectious phages initially added in the assay. Purified MBPwt and BSA were used as negative controls.
In vivo maltose uptake.
We measured maltose uptake, as previously described (2), at 2 μM maltose, a concentration at which LamB is limiting for transport (3, 8). After growth, the bacteria were preincubated on ice with 50 μg of MBPwt, MBP-J hybrid, or BSA, for 1 h. The mixtures were further incubated at room temperature for 30 min before the addition of 14C-labelled maltose (specific activity, 40 μCi/μmol). The assay revealed that binding of MBP-J/S led only to a partial (approximately two-thirds) reduction of maltose uptake (data not shown). This result may indicate either that binding of MBP-J to LamB only partially affected access of the sugar to its binding site within the channel or, more simply, that one-third of the LamB molecules did not have MBP-J bound to them. This preliminary observation should be further confirmed by using liposomal or other in vitro reconstituted systems.
Electron microscopy.
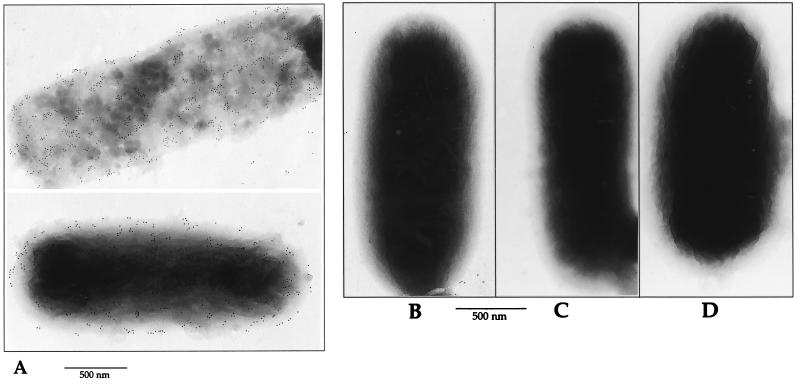
Electron microscopic analyses were performed on the LamB-positive strain P4X8 and its LamB-negative derivative (14). Bacteria were suspended in PBS at a concentration of 5 × 109 bacteria/ml. A drop (20 to 50 μl) of this suspension was placed on a sheet of Parafilm. A formvar-coated nickel grid was placed on the drop for 2 min and then, sequentially, onto drops of the following reagents at room temperature: PBS–0.1% glutaraldehyde for 5 min, PBS–50 mM NH4Cl for 5 min, PBS–1% BSA–1% normal goat serum for 5 min, MBP-J/S hybrid protein at a concentration of 5 μg/ml (or the same concentration of MBPwt) for 20 to 30 min, and anti-MBP antiserum (dilution, 1/100) for 30 min. After five washes, IgG (H+L) anti-rabbit immunoglobulin-gold conjugate was added and the incubation was continued for 10 min. After several washes, the grids were dried and then coated with 3 nm of carbon in a vacuum evaporator. Specimens were examined with a Philips CM12 transmission electron microscope, under standard conditions (80 to 120 kV). As illustrated in Fig. 5, MBP-J/S hybrid protein (Fig. 5A) but not MBPwt (Fig. 5B) bound to the surface of the LamB-positive strain P4X8. As shown by Fig. 5C and D, there was no nonspecific binding of MBP-J/S or MBPwt to the LamB-negative derivative of P4X8 (14).
FIG. 5.
Electron microscopy. Electron micrographs of bacterial cells after immunogold labelling with anti-MBP serum. Bacteria producing the wild-type LamB protein (P4X8) were preincubated with either MBP-J/S hybrid (A) or MBPwt as a negative control (B). The same assay was also performed on the LamB-negative derivative of P4X8, designated P4X8-LamBneg. (C and D) P4X8-LamBneg preincubated with MBP-J/S and MBPwt, respectively.
Acknowledgments
Jiang Wang was supported by a grant from Association Franco-Chinoise pour la Recherche Scientifique et Technique (PRA BT no. 97-04) and a grant from Centre National de la Recherche Scientifique.
We thank Colin Tinsley for careful reading of the manuscript. We thank Pierre Gounon, Station Centrale de Microscopie Electronique, Institut Pasteur Paris (Paris, France) for immunoelectron microscopy. Station Centrale de Microscopie Electronique is supported by Institut Pasteur.
REFERENCES
- 1.Braun-Breton C, Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor in lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981;148:845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charbit A, Gehring K, Nikaido H, Ferenci T, Hofnung M. Maltose transport and starch binding in phage resistant point mutants of maltoporin functional and topological implications. J Mol Biol. 1988;201:487–496. doi: 10.1016/0022-2836(88)90630-4. [DOI] [PubMed] [Google Scholar]
- 3.Charbit A, Wang J, Michel V, Hofnung M. A cluster of charged and aromatic residues in the C-terminal portion of maltoporin participates in sugar binding and uptake. Mol Gen Genet. 1998;260:185–192. doi: 10.1007/s004380050884. [DOI] [PubMed] [Google Scholar]
- 4.Charbit A, Werts C, Michel V, Klebba P E, Quillardet P, Hofnung M. A role for residue 151 of LamB in bacteriophage lambda adsorption: possible steric effect of amino acid substitutions. J Bacteriol. 1994;176:3204–3209. doi: 10.1128/jb.176.11.3204-3209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desaymard C, Debarbouillé M, Jolit M, Schwartz M. Mutations affecting antigenic determinants of an outer membrane protein of Escherichia coli. EMBO J. 1986;5:1383–1388. doi: 10.1002/j.1460-2075.1986.tb04371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabay J, Schwartz M. Monoclonal antibodies as a probe for structure and function of Escherichia coli outer membrane protein. J Biol Chem. 1982;257:6627–6630. [PubMed] [Google Scholar]
- 7.Hofnung M, Jezierska A, Braun-Breton C. lamB mutations in E. coli K12: growth of λ host range mutants and effects of nonsense suppressors. Mol Gen Genet. 1976;145:207–213. doi: 10.1007/BF00269595. [DOI] [PubMed] [Google Scholar]
- 8.Klebba P E, Newton S M C, Charbit A, Michel V, Perrin D, Hofnung M. Further genetic analysis of the C-terminal external loop region in Escherichia coli maltoporin. Res Microbiol. 1997;148:375–387. doi: 10.1016/S0923-2508(97)83868-5. [DOI] [PubMed] [Google Scholar]
- 9.Martineau P, Guillet J-G, Leclerc C, Hofnung M. Expression of heterologous peptides at two permissive sites of the MalE protein: antigenicity and immunogenicity of foreign B and T-cell epitopes. Gene. 1992;113:35–46. doi: 10.1016/0378-1119(92)90667-e. [DOI] [PubMed] [Google Scholar]
- 10.Randall-Hazelbauer L, Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973;116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirmer T, Keller T A, Wang Y-F, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 12.Szmelcman S, Clement J M, Jehanno M, Schwartz O, Montagnier L, Hofnung M. Export and one-step purification from Escherichia coli of a MalE-CD4 hybrid protein that neutralizes HIV in vitro. J Acquir Immune Defic Syndr. 1990;3:859–872. [PubMed] [Google Scholar]
- 13.Wang J, Michel V, Hofnung M, Charbit A. Cloning of the j gene of bacteriophage lambda, expression and solubilization of the J protein: first in vitro studies on the interactions between J and LamB, its cell surface receptor. Res Microbiol. 1998;149:611–624. doi: 10.1016/s0923-2508(99)80009-6. [DOI] [PubMed] [Google Scholar]
- 14.Werts C, Michel V, Hofnung M, Charbit A. Adsorption of bacteriophage lambda on the LamB protein of E. coli K12: point mutations in gene J of lambda responsible for extended host range. J Bacteriol. 1994;176:941–947. doi: 10.1128/jb.176.4.941-947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]