Abstract
The opdA (prlC) gene of Salmonella enterica serovar Typhimurium and Escherichia coli encodes the metalloprotease oligopeptidase A (OpdA). We report that opdA is cotranscribed with a downstream open reading frame, yhiQ. Transcription of this operon is induced after a temperature shift (30 to 42°C), and this induction depends on the heat shock sigma factor encoded by the rpoH (htpR) gene.
The Salmonella enterica serovar Typhimurium opdA gene encodes the metalloprotease oligopeptidase A (OpdA) (17). opdA is a homolog of the Escherichia coli prlC gene, a site of suppressors of the localization defect conferred by certain signal sequence mutations (2, 4, 14, 15). OpdA is also required for the proteolytic processing of a phage P22 protein in vivo (5), and it can degrade the cleaved lpp signal peptide in vitro (10). Comparison of the OpdA amino acid sequence with other protein sequences indicated that it is a member of a subfamily of Zn metalloproteases with representatives in both animals and fungi (3). This family includes the mitochondrial intermediate peptidase, which removes a small peptide from certain proteins that are transported through the mitochondrial membranes (9). In this study, the opdA sequence has been extended to include a downstream open reading frame (ORF) of unknown function, yhiQ. We show that opdA and yhiQ form an operon, that transcription of this operon is induced by temperature upshift, and that this induction is dependent on RpoH (ς32 or ςH).
Nucleotide sequence of serovar Typhimurium yhiQ.
The sequence of the opdA gene (2) suggested the presence of another ORF beginning downstream of the opdA ORF. To characterize this ORF, we determined the nucleotide sequence of an additional 997 bp beyond the end of opdA (Fig. 1). This sequence (GenBank accession no. AF137028) contains an ORF encoding a protein of either 253 amino acids (if the GTG at nucleotides [nt] 7 to 9 is used as the translational start) or 221 amino acids (if ATG at nt 103 to 105 is used as the translational start). Given the strong conservation of amino acid sequence between the YhiQ homologs in the region of the ORF between the GTG start codon and the first AUG codon, it seems likely that the protein is translated starting with GTG. There is a possible transcription terminator at nt 34 to 62. Downstream from the yhiQ ORF, there are two repetitive extragenic palindromic (REP) sequences (13) in inverted orientation, which have the potential to form a large stem-loop. A potential ribosome binding site precedes both of the possible translation starts, and the codon usage of the ORF is consistent with other expressed Salmonella genes (18). In preliminary experiments, a ∼28-kDa protein was produced when yhiQ was transcribed in vivo from a phage T7 promoter, indicating that yhiQ may be translated in vivo.
FIG. 1.
DNA sequence and translation of serovar Typhimurium yhiQ. The sequence shown begins immediately after the end of the opdA ORF at nt 2645 in GenBank sequence accession no. M84574. The double underline indicates a potential transcriptional terminator: 6-bp stem, 6-nt loop, and seven T's in the next 11 nt. Either GTG (nt 7 to 9) or ATG (nt 103 to 105) could serve as the translation start for YhiQ. A potential ribosome binding site for the ATG start site is indicated by the single underline. A potential ribosome binding site for the GTG start is present in the opdA coding region not shown in Fig. 1. Dashed lines identify two REP sequences in inverted orientation.
Comparison of the YhiQ sequence with the sequence databases identified three similar hypothetical proteins of unknown function from E. coli (94% identity), Haemophilus influenzae (68% identity), and Neisseria gonorrhoeae (41% identity). The E. coli yhiQ gene is located immediately downstream from prlC, the E. coli homolog of opdA. There are no REPs in the E. coli sequence, but there is a potential stem-loop structure (13-bp stem, 4-nt loop) after the E. coli yhiQ ORF (12). In H. influenzae, the putative homologs of opdA and yhiQ are unlinked. Clearly, opdA and yhiQ are not always associated since the genome of Synechocystis sp. contains an opdA homolog (accession no. D90916) but does not encode a YhiQ-related ORF.
opdA and yhiQ constitute an operon.
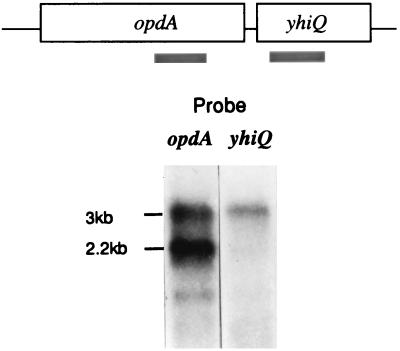
Because there was no obvious promoter preceding yhiQ, Northern blot analysis was carried out on RNA isolated from TN4465 (leuBCD485 opdA10::MudI1734 containing plasmid pCM138, which carries both opdA and yhiQ) to determine if yhiQ and opdA were cotranscribed. As shown in Fig. 2, the opdA-specific probe hybridized to two transcripts; one of 2.2 kb and the other of 3 kb. However, the yhiQ-specific probe hybridized only to the 3-kb transcript. The sizes of these transcripts are consistent with the lengths of the opdA ORF, 2,078 bp, and the combined opdA and yhiQ ORFs, 2,858 bp. These results show that opdA is the first gene in a two-gene operon. The two transcripts may result either from occasional read-through of the potential transcription terminator between opdA and yhiQ or from the degradation of the longer transcript to yield the shorter one.
FIG. 2.
Northern blot analysis of opdA RNA. Total RNA was isolated from TN4465 (1), treated with RNase-free DNase, fractionated by agarose-formaldehyde gel electrophoresis, and vacuum blotted to a nylon membrane (11). Half the membrane was hybridized to a radiolabeled 1.36-kb opdA-specific DNA fragment, and the other half was hybridized to a 0.55-kb yhiQ-specific fragment. These DNA probes were generated by PCR amplification and labeled with [α-32P]dATP. Hybridization was conducted at 60°C in 20 mM sodium phosphate–5× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate)–7% sodium dodecyl sulfate–2× Denhardt's solution–100 μg of denatured salmon sperm DNA per ml for 18 h (11). The membrane was washed extensively, concluding with a final wash in 0.1× SSC–1% sodium dodecyl sulfate at 60°C. Bands were visualized by autoradiography. The positions of the 3-kb message containing both opdA and yhiQ and of a 2.2-kb message containing only opdA are indicated. A third, weaker band may represent a partially degraded fragment of the opdA message.
The opdA operon is a ς32-dependent heat shock operon.
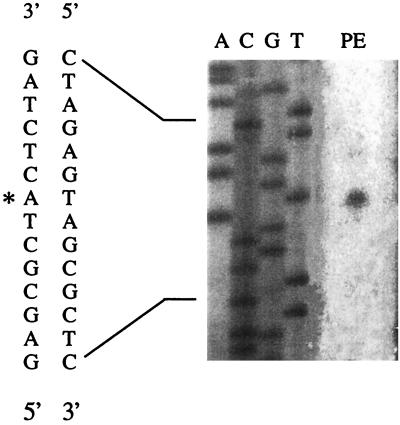
A near-consensus ς32-dependent promoter sequence is present upstream from the start of translation of opdA (Fig. 3) (2, 4). To determine if this sequence could be the opdA promoter, the start of transcription was determined by primer extension analysis. The initial nucleotide in the mRNA was found to be an A, 55 bp 5′ to the start of OpdA translation (Fig. 4). This result is consistent with the identification of the ς32 promoter sequence as the functional opdA promoter.
FIG. 3.
Comparison of opdA promoter region and consensus ς32 promoter. The consensus sequence is from reference 8. The double underline indicates the 5′ end of the opdA mRNA as determined by primer extension analysis and corresponds to nt 548 in GenBank sequence accession no. M84574.
FIG. 4.
Identification of the transcriptional start site for OpdA. Strain TN4465 containing pCM138 was grown overnight at 30°C, diluted 1:100, and incubated at 30°C to an optical density at 600 nm of 0.4. The culture was then shifted to 42°C for 10 min, and total RNA was isolated by using Qiagen's RNeasy Mini kit. A 25-nt primer complementary to nt 603 to 627 of GenBank sequence accession no. M84574 was used for extension, and the product is shown in lane PE. Lanes A, C, G, and T contain sequence reactions generated with the same primer. Radioactivity was detected by autoradiography and, in addition, by the use of a Molecular Dynamics Storm 860 PhosphorImager. An asterisk indicates the transcription start in the DNA sequence shown on the left.
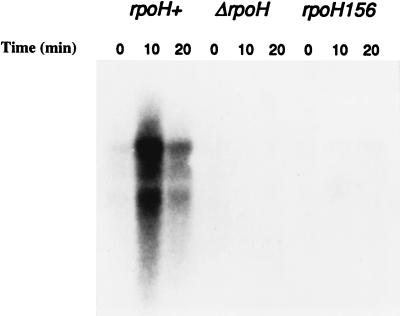
To determine if expression of the opdA operon was dependent on ς32, Northern blot analysis was done with RNA isolated from strains carrying an opdA plasmid and lacking functional RpoH. Plasmid pCM138, containing the entire opdA operon, was transformed into E. coli strains with three different rpoH alleles: rpoH wild type (SC122), rpoH156 (K165) (6), and ΔrpoH30::Kan (CAG9333) (19). Cells were grown at 30°C and shifted to 42°C, and total RNA was isolated from samples taken at various times after the shift. The RNA was fractionated by agarose-formaldehyde gel electrophoresis and transferred to a nylon membrane as described above. The membrane was probed with a radiolabeled opdA probe. As shown in Fig. 5, opdA transcription increases after a temperature upshift, and this transient induction requires the rpoH gene product, ς32. Thus, opdA and yhiQ are indeed previously unidentified heat shock genes.
FIG. 5.
RpoH-dependent heat shock induction of opdA transcription. E. coli strains containing pCM138 and one of three different rpoH alleles (rpoH wild type, rpoH156, or ΔrpoH30::Kan) were grown at 30°C and shifted to 42°C, and samples were taken at the indicated times after the shift. Total RNA was isolated, separated by agarose-formaldehyde gel electrophoresis, and transferred to a nylon membrane. The membrane was probed with a radiolabeled opdA probe.
The role of OpdA and YhiQ in the cell's response to stress is unknown. Strain TN3101, which contains a polar opdA::MudI1734 insertion, showed no defect in growth rate at 30, 37, or 42°C. There is also no obvious correlation between the predicted molecular weight of either OpdA or YhiQ and any of the identified heat shock-induced proteins in the E. coli protein index of VanBogelen et al. (16). YhiQ appears to be unnecessary for OpdA function, since plasmid pCM258, which carries opdA but not yhiQ, was able to complement all known defects conferred by an insertion mutation in opdA.
Several other protease genes in E. coli are known to be part of the heat shock regulon, and it seems likely that the degradation of irremediably misfolded proteins is a major component of the heat shock response (7). Only one natural macromolecular substrate for OpdA has been identified (phage P22 gp7), and this protein is not degraded but rather is specifically processed by OpdA. It is not clear whether OpdA participates directly in the degradation of misfolded proteins or whether it plays a more specialized role in the heat shock response. Perhaps the identification of cellular substrates for OpdA will clarify its functional role. The function of YhiQ remains completely mysterious.
Acknowledgments
We thank Tina Knox for performing the primer extension experiment and John Cronan and Carol Gross for providing strains.
This work was supported by a grant (AI10333) from the National Institute for Allergy and Infectious Diseases.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Conlin C A, Miller C G. Cloning and nucleotide sequence of opdA, the gene encoding oligopeptidase A in Salmonella typhimurium. J Bacteriol. 1992;174:1631–1640. doi: 10.1128/jb.174.5.1631-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlin C A, Miller C G. Dipeptidyl carboxypeptidase and oligopeptidase A from Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1995;248:567–579. doi: 10.1016/0076-6879(95)48036-6. [DOI] [PubMed] [Google Scholar]
- 4.Conlin C A, Trun N J, Silhavy T J, Miller C G. Escherichia coli prlC encodes an endopeptidase and is homologous to the Salmonella typhimurium opdA gene. J Bacteriol. 1992;174:5881–5887. doi: 10.1128/jb.174.18.5881-5887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conlin C A, Vimr E R, Miller C G. Oligopeptidase A is required for normal phage P22 development. J Bacteriol. 1992;174:5869–5880. doi: 10.1128/jb.174.18.5869-5880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper S, Ruettinger T. A temperature sensitive nonsense mutation affecting the synthesis of a major protein of Escherichia coli K12. Mol Gen Genet. 1975;139:167–176. doi: 10.1007/BF00264696. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 8.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 9.Isaya G, Kalousek F. Mitochondrial intermediate peptidase. Methods Enzymol. 1995;248:556–567. doi: 10.1016/0076-6879(95)48035-8. [DOI] [PubMed] [Google Scholar]
- 10.Novak P, Dev I K. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988;170:5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Sofia H J, Burland V, Daniels D L, Plunkett G R, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern M J, Ames G F, Smith N H, Robinson E C, Higgins C F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 14.Trun N J, Silhavy T J. Characterization and in vivo cloning of prlC, a suppressor of signal sequence mutations in Escherichia coli K12. Genetics. 1987;116:513–521. doi: 10.1093/genetics/116.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trun N J, Silhavy T J. PrlC, a suppressor of signal sequence mutations in Escherichia coli, can direct the insertion of the signal sequence into the membrane. J Mol Biol. 1989;205:665–676. doi: 10.1016/0022-2836(89)90312-4. [DOI] [PubMed] [Google Scholar]
- 16.VanBogelen R A, Sankar P, Clark R L, Bogan J A, Neidhardt F C. The gene-protein database of Escherichia coli: edition 5. Electrophoresis. 1992;13:1014–1054. doi: 10.1002/elps.11501301203. [DOI] [PubMed] [Google Scholar]
- 17.Vimr E R, Green L, Miller C G. Oligopeptidase-deficient mutants of Salmonella typhimurium. J Bacteriol. 1983;153:1259–1265. doi: 10.1128/jb.153.3.1259-1265.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada K, Wada Y, Doi H, Ishibashi F, Gojobori T, Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991;19(Suppl.):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y N, Kusukawa N, Erickson J W, Gross C A, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor ς32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]