ABSTRACT
Wastewater-based epidemiology (WBE) has been widely used to track levels of SARS-CoV-2 infection in the community during the COVID-19 pandemic. Due to the rapid expansion of WBE, many methods have been used and developed for virus concentration and detection in wastewater. However, very little information is available on the relative performance of these approaches. In this study, we compared the performance of five commonly used wastewater concentration methods for the detection and quantification of pathogenic viruses (SARS-CoV-2, norovirus, rotavirus, influenza, and measles viruses), fecal indicator viruses (crAssphage, adenovirus, pepper mild mottle virus), and process control viruses (murine norovirus and bacteriophage Phi6) in laboratory spiking experiments. The methods evaluated included those based on either ultrafiltration (Amicon centrifugation units and InnovaPrep device) or precipitation (using polyethylene glycol [PEG], beef extract-enhanced PEG, and ammonium sulfate). The two best methods were further tested on 115 unspiked wastewater samples. We found that the volume and composition of the wastewater and the characteristics of the target viruses greatly affected virus recovery, regardless of the method used for concentration. All tested methods are suitable for routine virus concentration; however, the Amicon ultrafiltration method and the beef extract-enhanced PEG precipitation methods yielded the best recoveries. We recommend the use of ultrafiltration-based concentration for low sample volumes with high virus titers and ammonium levels and the use of precipitation-based concentration for rare pathogen detection in high-volume samples.
IMPORTANCE As wastewater-based epidemiology is utilized for the surveillance of COVID-19 at the community level in many countries, it is crucial to develop and validate reliable methods for virus detection in sewage. The most important step in viral detection is the efficient concentration of the virus particles and/or their genome for subsequent analysis. In this study, we compared five different methods for the detection and quantification of different viruses in wastewater. We found that dead-end ultrafiltration and beef extract-enhanced polyethylene glycol precipitation were the most reliable approaches. We also discovered that sample volume and physico-chemical properties have a great effect on virus recovery. Hence, wastewater process methods and start volumes should be carefully selected in ongoing and future wastewater-based national surveillance programs for COVID-19 and beyond.
KEYWORDS: enteric viruses, environmental virology, human respiratory viruses, public health surveillance, sewage concentration
The COVID-19 pandemic has so far resulted in 410 million confirmed cases and 5.8 million deaths worldwide (1). The illness is caused by SARS-CoV-2, an enveloped, spherical coronavirus with a single-stranded RNA (ssRNA) genome that is 30 kb in size (2). COVID-19 causes a wide range of symptoms, including flu-like symptoms, such as fever and chills, cough, fatigue, headache, and the loss of taste and smell (3). The symptoms are often mild, and the infected individuals may be asymptomatic; however, they can still infect others (4). These cases are usually undetected, which largely contributes to the rapid spread of the disease (5).
Wastewater-based epidemiology (WBE) has been successfully used for the surveillance of chemicals and pathogens, including several human viruses at the community level (6–8). Since the beginning of the COVID-19 pandemic, many countries have trialed the usefulness of WBE for tracking SARS-CoV-2 at the community level. As all infected individuals, including asymptomatic and presymptomatic cases, shed the virus in their feces, the monitoring of wastewater can be a useful addition to COVID-19 surveillance (9). Several studies have shown that the RNA of the virus can be readily detected and quantified in wastewater and that WBE can be used as an early warning system and a predictive tool for COVID-19 monitoring (9–12). Hence, many countries have implemented WBE as a component of their COVID-19 surveillance and decision-making portfolio (13–18).
Although WBE is a cost-effective approach supporting the understanding of viral disease spread, it has its limitations. A major factor affecting the use of WBE is the robust detection of the target virus in the samples. As the viral RNA is usually present at low concentrations in wastewater, the samples need to be concentrated prior to nucleic acid extraction and the quantification of the viral target using quantitative or digital PCR (19). Many methods have been used for wastewater concentration for SARS-CoV-2 RNA quantification, including ultracentrifugation, filtration, ultrafiltration, adsorption and precipitation-based approaches (20–22). However, the viral recovery using these methods is usually not assessed (23–25).
There are many factors to consider when selecting a concentration method for WBE, including the availability of equipment, cost, time available for sample processing, optimal sample volume, etc. As ultracentrifugation is time-consuming and requires expensive equipment, it has not been implemented in routine WBE surveillance. Electronegative and electropositive membrane filtration methods seem a viable alternative; however, extracting viral nucleic acids from a membrane filter can be challenging (26). Furthermore, the filter membranes are subject to clogging if the samples have a high turbidity. Nonetheless, precipitation- and ultrafiltration-based approaches are more robust and versatile and hence may be more suitable for many WBE applications (27–29).
In this study, we evaluated the usefulness of wastewater concentration methods for the surveillance of pathogenic viruses, namely, SARS-CoV-2, influenza, measles virus (MeV), norovirus (NoV), rotavirus (RoV), fecal indicator viruses, such as crAssphage, human mastadenoviruses (AdV), and pepper mild mottle virus (PMMoV), and potential process control viruses, including Phi6 bacteriophage and murine norovirus. We explored the viral recoveries using five concentration methods (Table 1): polyethylene glycol (PEG) precipitation (used in the Welsh and Northern Ireland surveillance programs), a modified PEG method with an initial elution with beef extract (30), ammonium sulfate (AS) precipitation (used for the national SARS-CoV-2 wastewater surveillance program in England), dead-end ultrafiltration using Amicon filters (used in the Scottish monitoring program), and tangential flow ultrafiltration with the InnovaPrep (IP) device, designed for wastewater testing. We also explored the trade-off between increasing sample volume and the efficiency of viral recovery. For verification, we used the two best-performing methods on neat (i.e., unspiked) wastewater samples.
TABLE 1.
Experimental setup and sample number for spiking experimenta
| Method code | Description | Water type | Water vol (mL) | Supernatant vol (mL) | Replicates (×) |
|---|---|---|---|---|---|
| A: PEG | Polyethylene glycol (PEG) precipitation | Distilled water | 20 50 200 |
15 37.5 150 |
3 3 3 |
| Distilled water—spiked | 20 50 200 |
15 37.5 150 |
3 2 3 |
||
| Wastewater | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| Wastewater—spiked | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| B: BE-PEG | Elution with beef extract (BE)—PEG precipitation | Distilled water | 20 50 200 |
15 37.5 150 |
3 3 3 |
| Distilled water—spiked | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| Wastewater | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| Wastewater—spiked | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| C: AS | Ammonium sulphate (AS) precipitation | Distilled water | 20 50 200 |
15 37.5 150 |
3 3 3 |
| Distilled water—spiked | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| Wastewater | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| Wastewater—spiked | 20 50 200 |
15 37.5 150 |
3 3 3 |
||
| D: AM | Amicon (AM) filtration | Distilled water | 20 50 |
15 20 |
3 3 |
| Distilled water—spiked | 20 50 |
15 20 |
3 3 |
||
| Wastewater | 20 50 |
15 20 |
3 3 |
||
| Wastewater—spiked | 20 50 |
15 20 |
3 3 |
||
| E: IP | InnovaPrep (IP) filtration | Distilled water—spiked | 20 50 |
15 37.5 |
3 3 |
| Wastewater—spiked | 20 50 |
15 37.5 |
3 3 |
Water volume is the volume of the sample taken and centrifuged to clarify the samples from solid matter. Supernatant volume refers to the volume of the clarified solution concentrated.
RESULTS
Virus recovery in deionized water and wastewater: laboratory spiking experiment.
In this experiment, deionized and wastewater samples were spiked with SARS-CoV-2, influenza (flu) A/B, NoVGII, RoV, and MeV. Due to the overall high level of PMMoV, AdV, and crAssphage in wastewater, no spiking was performed for these viral targets. The nonspiked deionized water samples were negative for the target viruses, suggesting no cross-contamination. The unspiked wastewater samples were positive for SARS-CoV-2 and RoV; however, the levels were 1 to 2 orders of magnitude lower than those of the spiked virus concentrations and were deemed negligible during analysis.
The generalized linear model identified sample starting volume, concentration method, and water type as significant predictors of viral recovery (Table 2). The model coefficients reveal that sample starting volume was a significant negative correlate of viral recovery; the Amicon method had the highest mean recovery; and wastewater had reduced recovery over deionized water.
TABLE 2.
Generalized linear model with gamma residuals (link = log) predicting recovery of 11 viruses in the liquid phasea
| Variable | Model estimate |
|---|---|
| Intercept | 3.759 (0.078)*** |
| Vol | –0.007 (0.001)*** |
| Method: AS | –1.245 (0.092)*** |
| Method: BE-PEG | –0.368 (0.092)*** |
| Method: IP | –1.55 (0.101)*** |
| Method: PEG | –1.569 (0.094)*** |
| Water type: WW | –0.848 (0.057)*** |
| AICb | 4,748.24 |
| R squared | 0.597 |
| Adjusted R squared | 0.594 |
Pellet methods were excluded from the model due to nonstandard starting volumes. The model results include the variable coefficient on a log scale indicating its effect (positive numbers indicating increased recovery), followed by the standard error in parentheses and a significance code (***, P < 0.001) rounded to three decimal places. The Amicon method and deionized water (DW) are predicted using the intercept.
AIC, Akaike information criterion.
(i) Greater recovery of spiked virus in deionized water compared to wastewater regardless of concentration method. Deionized water had greater median recovery of spiked virus (7.64%) compared to wastewater (5.23%), which was found to be significant when comparing mean log10 transformed viral recovery values (Fig. 1a; Welch two-sample t test [log y]: t = 5.5, df = 578, P < 0.001). Greater recovery from deionized water over wastewater was seen in all viruses except for Phi6 and RoV (Fig. 1b). The highest recoveries (38 to 100%) were observed for all viruses when the Amicon ultrafiltration method was used on deionized water samples. These results suggest that wastewater contains other chemicals or materials which reduce the efficiency of the concentration and extraction steps or the qPCR amplification process.
FIG 1.
Greater recovery of spiked viruses, influenza A/B viruses (flu-A/B), measles virus (MeV), murine norovirus (MNV), SARS-CoV-2 (N1), norovirus GII (NoVGII), bacteriophage phi6 (Phi6), and rotavirus (RoV) in deionized water (DW) compared to wastewater (WW). Data derived from all concentration methods. (a) All spiked virus recovery results combined. (b) Recovery by individual virus. Boxes depict the 25th, 50th, and 75th percentile ranges after omitting outliers greater or less than ±1.5× the interquartile range (IQR), which is shown by the whiskers.
(ii) Viral recovery improved with a reduced starting volume of wastewater. The lowest starting volume (15 mL) had the greatest median viral recovery (4.85%), followed by 37.5 mL (3.84%) and then the largest volume (150 mL; 1.75%; Fig. 2a). The mean log10 transformed viral recovery was significantly different between groups (analysis of variance [ANOVA] [log y]: F value = 36.25, P < 0.001), as well as all pairwise comparisons (Fig. 2c; pairwise t tests with pooled standard deviation [SD]; P < 0.05; Holm adjustment method). The negative trend between volume and recovery was seen in the recovery of all individual viruses. Although MNV and RoV had higher median recovery for 37.5-mL samples compared to 15-mL aliquots (Fig. 2b), the difference was not significant (P > 0.05).
FIG 2.
Recovery for human mastadenovirus (AdV), crAssphage (CrAss), influenza A/B virus (Flu-A/B), measles virus (MeV), murine norovirus (MNV), SARS-CoV-2 (N1), norovirus GII (NoVGII), bacteriophage phi6 (Phi6), pepper mild mottle virus (PMMoV), and rotavirus (RoV) as a function of starting volume of wastewater. Data derived from all concentration methods. (a) All recovery results combined. (b) Recovery separated by virus with a variable y scale (recovery percentage). (c) Existence of any significant differences in the tested volumes. (To analyze which volume is significantly better for viral recovery, panel c is to be analyzed in conjunction with panel ‘a and panel b). The P values (Holm adjustment method) of pairwise comparisons were calculated between extraction volumes with two sample t tests with pooled standard deviations (***, P < 0.001; **, P < 0.01; *, P < 0.05). Comparisons were made with an ANOVA after log10 transformation of recovery, followed by pairwise t tests; Pairwise comparisons found significant differences between all volumes with the Holm adjustment method (P < 0.05).
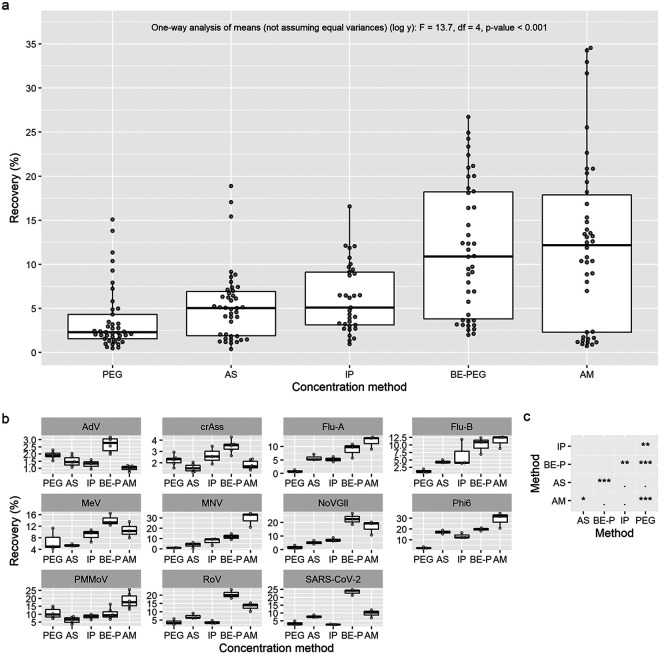
(iii) BE-PEG and Amicon concentration methods have the greatest viral recovery. Different concentration methods had a major effect on viral recovery (Fig. 3); Amicon and modified PEG (BE-PEG) methods had the highest median recovery (12.2% and 10.9%, respectively), followed by IP (5.1%) and then AS (5.0%) and PEG methods (2.3%). The variance of the Amicon method was significantly different from all other methods (P < 0.05), so a Welch ANOVA was selected (one-way analysis of means not assuming equal variances), which found significant differences between the method’s mean log10 transformed recovery (one-way analysis of means [not assuming equal variances] [log y]: F = 13.7, df = 4, P < 0.001). Post hoc pairwise comparisons with t tests found that the Amicon method’s viral recovery was not significantly different from that of the IP method due to the greater variance observed in the Amicon results, whereas BE-PEG method recoveries were significantly different from all other methods except Amicon (Fig. 3c; P value Holm adjustment method). The performance of the Amicon method compared to other methods varied highly between viruses, performing well for flu, MNV, and PMMoV while poorly for AdV and CrAss (Fig. 3b). The BE-PEG method, on the other hand, was consistently better than other methods (excluding Amicon) for individual virus recovery. These results suggest the BE-PEG method provides the greatest and most consistent viral recovery.
FIG 3.
Influence of concentration methods on virus recovery from wastewater at a sample starting volume of 15 mL. (a) All recovery results with starting volumes of 15 mL combined. (b) Recovery separated by virus with a variable y scale. (c) Existence of any significant differences among concentration methods. (To analyze which volume is significantly better for viral recovery, panel c is to be analyzed in conjunction with panel a and panel b). The P values of pairwise comparisons of method recovery were calculated using t tests without pooled standard deviations (***, P < 0.001; **, P < 0.01; *, P < 0.05; -., P > 0.05; P value Holm adjustment method). Comparisons were made with an ANOVA after log10 transformation of recovery, followed by pairwise t tests (c); BE-PEG and Amicon methods had the highest median recovery, but due to Amicon method’s greater variance, pairwise comparisons with IP (third-highest median recovery) were only significantly different for BE-PEG (P < 0.05; panel c).
(iv) Solid fraction may contain few virus particles. The pellet recovered after the first centrifugation step (after spiking) had significantly lower viral recovery than the BE-PEG and PEG concentrates (Fig. 4a; (BE-PEG) Welch two-sample t test [log y]: t = 16.9, df = 147, P < 0.001; (PEG) Welch two-sample t test [log y]: t = 7, df = 55, P < 0.001). The pellet had consistently lower recovery of all individual viruses (Fig. 4b). These results suggest that a greater proportion of spike virus is suspended in the liquid of a sample compared to the solid fraction.
FIG 4.
Viral recovery in the pellet from the first centrifugation step (10,000 × g, 10 min, 4°C) in the viral extraction procedure. The sample solid fraction (pellet) has significantly lower viral recovery than the concentrated sample; thus, removal via centrifugation will likely increase the median viral recovery of a concentrated sample. (a) All recovery results with starting volumes of 50 mL combined. (b) Recovery separated by virus with a variable y scale. Comparisons were made with a Welch two-sample t test after log10 transformation of recovery.
Virus recovery in neat wastewater samples.
(i) Amicon and BE-PEG concentration methods varied in recovery depending on the target virus. The two best-performing wastewater concentration methods, the Amicon and BE-PEG methods, were tested on neat, unspiked wastewater samples collected at 13 wastewater treatment plants (Table 1). The 115 samples included grab and 24-h composite samples and showed great variations in physicochemical characteristics (Table 1).
We found significantly greater Phi6 (process control virus) recoveries when the BE-PEG method was used (4.51%) than for the Amicon method (0.77%). We did not find any influenza B, HIV, or hepatitis B/C viruses; however, crAssphage, SARS-CoV-2, flu A, EV, EVD68, NoV GI/GII, and measles virus were detected. The viruses detected sporadically (flu A: 0.88% detection rate with Amicon and 0% with BE-PEG; measles virus: 1.75% with Amicon and 2.63% with BE-PEG) were excluded from further analysis.
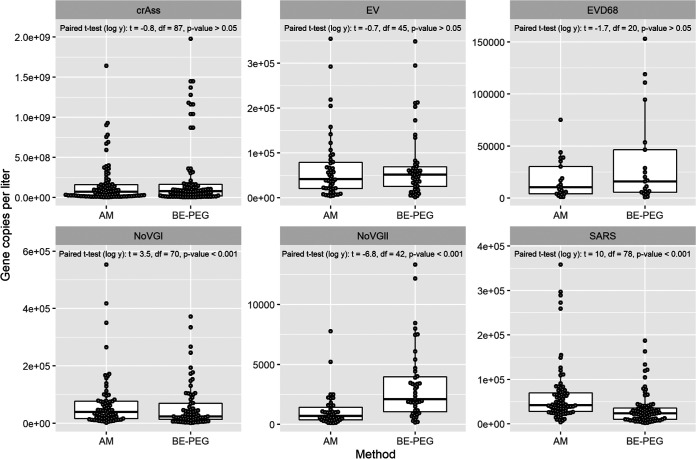
The statistical analyses revealed that the BE-PEG and Amicon methods had various performances depending on the target virus (Fig. 5). Median gene copies (gc) per liter were similar between methods when targeting crAssphage (Amicon: 7 × 107 gc/L; BE-PEG: 7.7 × 107 gc/L), EV (Amicon: 4.2e × 104 gc/L; BE-PEG: 5.2 × 104 gc/L), and EVD68 (Amicon: 1.0 × 104 gc/L; BE-PEG: 1.6 × 104 gc/L), and mean log10 transformed recoveries were not found to differ significantly between methods (paired t test; P > 0.05). Conversely, the Amicon method recovered greater median concentrations when targeting SARS-CoV-2 (Amicon: 4.2 × 104 gc/L; BE-PEG: 2.4 × 104 gc/L) and NoVGI (Amicon: 3.9 × 104 gc/L; BE-PEG: 2.4 × 104 gc/L), while the BE-PEG method recovered greater median concentrations when targeting NoVGII compared to the Amicon method (Amicon: 7 × 102 gc/L; BE-PEG: 2.1 × 103 gc/L), differences that were found to be significant when comparing mean log10 transformed concentrations (paired t test; P < 0.001). These conflicting results suggest that neither method is consistently better than the other.
FIG 5.
Comparison of viral recovery for Amicon and BE-PEG concentration methods tested on neat, unspiked wastewater samples collected at 13 wastewater treatment plants. Statistical comparisons were made using paired t tests after log transformation of the gene copies per liter. Recovery of crAss, enterovirus (EV), and enterovirus D68 (EVD68) could not be assumed to have differing means, while SARS-CoV-2 and norovirus GI (NoVGI) had significantly greater mean recovery with the Amicon method, and NoVGII had significantly greater mean recovery with the BE-PEG method.
(ii) The Amicon method performs comparatively better than BE-PEG as concentrations of ammonium increase. The linear mixed effects model (lmm) analysis found that ammonium concentration had a significant positive effect (P < 0.01) on method performance, suggesting that the Amicon method is better suited for samples with high concentrations of ammonium inhibitors. However, none of the other chemistry variables had significantly different effects on the performance of the Amicon and BE-PEG methods (P > 0.05), and the intercept was not significant (see Fig. S10 in the supplemental material).
DISCUSSION
In this study, we compared the performance of wastewater concentration methods currently used for COVID-19 monitoring in the United Kingdom (13) for the detection of different human viruses. Overall, the results suggested that all methods are suitable for routine viral surveillance; however, there were significant differences in their ability to recover viruses.
The first step of all methods we compared was the elimination of solid matter using centrifugation. This step is commonly used prior to virus concentration in wastewater samples, especially prior to ultrafiltration, as solid matter may cause membrane clogging (23, 31). However, virus particles may be attached to the solid matter in samples with high conductivity and/or with high levels of organic matter (32) and hence be excluded from virus concentration, resulting in low viral titers. In this study, we found that only a small proportion of the viruses attach to the solid particles in the pellet fraction, similar to our previous study where negligible amounts of SARS-CoV-2 RNA was recovered in the pellets of wastewater from six treatment plants in the United Kingdom (11). However, other studies suggest that up to 23% of SARS-CoV-2 can be recovered from the pellet fraction (33, 34). The contradictory findings suggest that there may be substantial differences in the virus adsorbing capacity to solid matter in different wastewater samples. Therefore, the presence of virus in the solid fraction cannot be ruled out. We note that higher viral loads may be found in the primary settling tank at wastewater treatment plants, as the concentration of solids is much greater (>100-fold) than in influent wastewater.
We found consistently higher viral recoveries from deionized water than in wastewater, regardless of the concentration method applied. Furthermore, increased wastewater volumes also had a negative impact on the recoveries of all viruses; however, the PEG method was less affected by volume increase than the AS, BE-PEG, Amicon, and IP methods. This suggests that organic matter (e.g., polysaccharides, ribonucleases) and other inhibitors of extraction and PCR coconcentrate with viruses (35). The Phi6 bacteriophage (used as a process control) also showed high variations in the neat/raw wastewater samples. Similar observations were made in other studies using transmissible gastroenteritis coronavirus (36), F-specific RNA (FRNA) bacteriophages (9, 37) and mengovirus (38) as a process control. Due to inconsistent wastewater matrices having different effects on viral recovery, a representative process control virus should always be used for virus concentration efficacy evaluation and control (39). In future WBE programs, it is likely that a multivirus process control will be needed to cover the range of viral types targeted.
It is important to note that we observed differences in the recoveries of different viruses, which may be due to differences in the structure, shape, size, or genetic material and their culturing, inactivation, and degradation processes. Due to environmental factors (e.g., temperature, physical pressure), viruses may become inactivated, releasing the genetic material in the environment where the genome size and structure would affect viral stability. The effect of viral properties on recoveries in wastewater should be addressed in future studies.
We found significant differences in the performance and applicability of different sample process methods (Table 3). Overall, the Amicon ultrafiltration concentration method gave high viral recoveries; however, the Amicon method showed great variations between viruses and replicates. Similar recoveries were observed using centrifugation-based ultrafiltration for process control viruses, such as murine hepatitis virus (56.0% ± 32.3%) (34), MS2 bacteriophage (33.3 ± 15.6%) (40), human coronavirus OC43 strain, (24% ± 2%) (41), and SARS-CoV-2 (25.9% to 65.3%) (42). In our study, Amicon filtration was only suitable for low sample volumes, up to 20 mL. However, the sample volume may be increased to up to 60 mL if samples are filtered through 0.45- and/or 0.2-μm filters to eliminate debris (41, 43). Another disadvantage of Amicon filtration was that the volume of the resulting concentrate was also inconsistent, and it may be too high for certain RNA/DNA extraction methods. Furthermore, depending on the sample characteristics, ultrafiltration times also varied from sample to sample, resulting in long centrifugation times, which may have a negative effect on virus integrity and recovery.
TABLE 3.
Comparison and ranking of the methods used for virus concentration in wastewatera
| Method | Viral recovery (%) | Sample vol (mL) | Concn factor (×) | Effective vol (mL) | Benchtop time/sample (min) | Overall time/sample (h) | Cost/sample (£) |
|---|---|---|---|---|---|---|---|
| PEG | 3.3 | 150 | 1500 | 6 | 50 | 18 | <1 |
| BE-PEG | 13.2 | 150 | 1500 | 6 | 60 | 18 | <1 |
| AS | 6.2 | 150 | 1500 | 6 | 40 | 3 | <1 |
| IP | 6.2 | 37.5 | 375 | 1.5 | 20 | 1 | 25 |
| Amicon | 14.1 | 15 | 150 | 0.6 | 30 | 1–2 | 8 |
The effective volume is the proportional volume of the original wastewater sample assayed by RT-qPCR (4 μL/reaction).
The IP ultrafiltration method enabled rapid viral concentration from up to 50 mL of wastewater; however, the viral recoveries were lower than those of the Amicon ultrafiltration. A previous study also found that centrifugation-based ultrafiltration outperforms the IP method for the recovery of murine hepatitis virus (33). Similarly, high recoveries were observed when process control viruses, such as bovine coronavirus (16.8% to 53.2%) and MS2 bacteriophage (53.6%) were concentrated (44). However, another study showed significantly lower recoveries for bovine coronavirus (5.5%) and bovine respiratory syncytial virus (7.6%) (45) when using IP concentration, suggesting that the water type and composition affect viral recovery. The IP method was also useful as a secondary concentration method for a large volume of wastewater for the human coronavirus OC43 strain, resulting in 48% ± 2% recoveries (21); however, it failed to concentrate poliovirus (0.32% recovery) (46). Due to the simplicity and rapid process, IP methods have been used for SARS-CoV-2 surveillance in wastewater in the United States (12, 45). Nonetheless, the IP consumables are more expensive than the reagents and consumables used in the other methods, and hence the method may not be feasible for routine mass testing.
From the precipitation-based concentration methods, the BE-PEG method gave the best viral recoveries, similar to those for Amicon filtration, whereas the PEG and AS precipitations were less efficient for viral recovery. PEG precipitation has been widely used for viral recovery from large volumes of water and wastewater samples for decades (24, 25, 31, 47), and the addition of beef extract-sodium nitrate elution has also been shown to enhance viral recovery (30, 48). PEG induces the precipitation of viruses from the solution by reducing the solubility of proteins, while the beef extract enhances the precipitation by allowing the viral particles to bind it, which explains the recovery superiority of BE-PEG over PEG precipitation (49). The sodium nitrate added increases conductivity and hence assists in the detachment of virus particles from organic matter (50). Studies also showed that precipitation-based methods outperform IP concentration for the recovery of poliovirus, crAssphage, and SARS-CoV-2, suggesting that this process is robust and more resilient to organic matter than ultrafiltration (12, 51), whereas others found that PEG precipitation performs similarly to Amicon ultrafiltration, with both methods resulting in approximately 40% SARS-CoV02 recoveries (29). The precipitation-based methods are very easy to perform, and the associated reagents are inexpensive and usually available in bulk quantities. The bench time required for these methods is short, and many samples can be handled at once; however, the incubation times, especially for the PEG methods, significantly increase the overall processing time (29). Another disadvantage of precipitation-based methods is the potential resuspension of the pellet concentrate after centrifugation, and hence this step should be performed without delays.
We also used the two best-performing methods, Amicon and BE-PEG, for mass testing of wastewater samples. Both methods successfully detected most of the target viral sequences, except for influenza B virus and hepatitis B/C viruses and HIV. The influenza B infection rates were very low in the study area during wastewater sampling, which explains the lack of viruses in the samples. However, for viral genomes associated with the viral families Flaviviridae, Hepadnavididae, and Retroviridae in wastewater using sequencing-based approaches (52–55), these viruses degrade rapidly in the environment, and hence we were not able to identify hepatitis B/C viruses and HIV in the samples. The higher viral detection rates observed using the BE-PEG method (150 mL sample volume) compared to the Amicon method (15 mL sample volume) suggested that the precipitation of higher sample volumes enables the detection of rare viral targets. The quantitative analysis found no significant differences in the measured concentrations, suggesting that both methods are suitable for abundant targets. The only notable difference in the two methods was their performance at high ammonium levels, suggesting that the Amicon method may be preferable when catchments have high concentrations of ammonium, including those with high levels of agricultural runoff entering the sewage network.
Conclusions and future work. We found that all five methods are suitable for wastewater testing for virus quantification using quantitative PCR (qPCR), with the Amicon ultrafiltration and beef extract elution-PEG precipitation methods performing the best. However, great variations between viruses and replicates in viral recoveries were observed using the Amicon method. The Amicon method is suitable for small sample volumes with high ammonium levels and therefore should be used when abundant viruses are quantified, whereas the precipitation method may be used for early detection when levels are low and for rare pathogens. We also found that the composition and volume of wastewater significantly affects virus recovery, and therefore sample volume should be carefully chosen. Future work may include pretreatment procedures (e.g., increased salinity) to enhance viral recovery from large volumes of sewage and the better understanding of the effects of virus characteristics on viral recoveries.
MATERIALS AND METHODS
Wastewater collection and chemical analysis.
For the spiking experiment, raw sewage was collected on 5 November 2021 at wastewater treatment plant 1 (WWTP1). The wastewater was stored at 4°C until use. For surveillance, raw sewage samples were collected at 13 treatment plants in England four times a week over a 2-week period from 12 November to 25 November 2021 (n = 115; Table 4). The samples were transported to the laboratory, chilled overnight, and stored at 4°C. The sample process started within 24 h after the samples were taken. The sample pH, turbidity, electrical conductivity, and ammonium and orthophosphate ion concentrations were measured as described previously (11).
TABLE 4.
Wastewater sampling sites and physicochemical properties of the wastewater samples analyzed in this studya
| Site | Type | n | pH | Turbidity (NTU) | Conductivity (μS/cm) | Ammonium (mg/L) | Phosphate (mg/L) |
|---|---|---|---|---|---|---|---|
| WWTP1 | Grab | 1 | 7.12 | 10.6 | 2145 | 5.0 | 2.06 |
| WWTP2 | Grab/comp | 8 | 7.45 ± 0.05 | 53.5 ± 12.3 | 889 ± 126 | 25.3 ± 7.9 | 2.15 ± 0.51 |
| WWTP3 | Grab | 1 | 7.61 | 202.0 | 1,212 | 5.1 | 3.09 |
| WWTP4 | Grab/comp | 9 | 7.52 ± 0.06 | 95.1 ± 18.7 | 1,644 ± 260 | 30.0 ± 5.0 | 2.90 ± 0.33 |
| WWTP5 | Comp | 10 | 7.57 ± 0.04 | 101.3 ± 8.7 | 1,236 ± 91 | 45.2 ± 5.9 | 2.56 ± 0.23 |
| WWTP6 | Grab/comp | 9 | 7.59 ± 0.09 | 64.3 ± 11.3 | 1,225 ± 264 | 38.9 ± 16.4 | 2.68 ± 0.64 |
| WWTP7 | Grab/comp | 9 | 7.57 ± 0.07 | 103.6 ± 15.7 | 1,208 ± 98 | 26.0 ± 6.0 | 2.39 ± 0.31 |
| WWTP8 | Comp | 9 | 7.37 ± 0.06 | 134.3 ± 20.5 | 1,076 ± 251 | 24.8 ± 7.8 | 2.11 ± 0.51 |
| WWTP9 | Grab/comp | 9 | 7.46 ± 0.08 | 53.1 ± 11.9 | 1,042 ± 105 | 28.3 ± 6.3 | 3.47 ± 0.36 |
| WWTP10 | Grab/comp | 7 | 7.51 ± 0.05 | 117.3 ± 21.4 | 1,175 ± 109 | 32.3 ± 8.8 | 3.13 ± 0.42 |
| WWTP11 | Grab/comp | 9 | 7.60 ± 0.07 | 103.8 ± 30.9 | 1,458 ± 150 | 33.8 ± 4.9 | 2.61 ± 0.26 |
| WWTP12 | Grab/comp | 9 | 7.56 ± 0.06 | 119.3 ± 13.3 | 2,069 ± 801 | 35.1 ± 7.8 | 3.04 ± 0.33 |
| WWTP13 | Grab/comp | 8 | 7.39 ± 0.10 | 61.3 ± 10.1 | 1,686 ± 249 | 26.1 ± 6.5 | 2.40 ± 0.53 |
| WWTP14 | Grab/comp | 17 | 7.62 ± 0.04 | 102.8 ± 19.3 | 1,089 ± 117 | 37.8 ± 4.6 | 2.92 ± 0.43 |
Comp, 24-h composite sample. Values represent means ± standard error of the mean (SEM).
Virus spiking.
Virus spiking was performed in a biosafety level 2/containment level 2 (BSL2/CL2) laboratory using a class II biosafety cabinet. Sample bottles were disinfected using 70% industrial methylated spirit (IMS) prior to removal from the cabinet.
For spiking, we used heat-inactivated SARS-CoV-2 (kindly provided by Andrew Weightman, Cardiff University), inactivated influenza A/California/07/2009 (H1N1), B/Lee/40, rotavirus SA11 (RoV) cultures (kindly provided by Eleanor Gaunt, University of Edinburgh), norovirus GII (NoVGII) in diluted and filtered fecal matter from a patient with confirmed norovirus infection (kindly provided by Lydia Drumwright, University of Cambridge), and measles virus (MeV) in the form of a vaccine (VWR International, USA). We also used non-human viruses which are commonly used as process controls, namely, Phi6 bacteriophage and murine norovirus (MNV), which we cultured in-house as described in Kevill et al. (51).
Wastewater collected from WWTP1 (Table 1) and deionized water samples (3 L each) were spiked with each virus to reach a final concentration of approximately 104 to 105 genome copies (gc)/mL per virus type. The same volumes of neat (i.e., unspiked) deionized water and wastewater samples were also prepared. A 0.5-mL aliquot of each water type (deionized water, deionized water plus viral spikes, wastewater, wastewater plus viral spikes) was saved in triplicate for direct RNA/DNA extraction to determine the concentration of each virus in the samples. The water samples were mixed by shaking and then aliquoted to 20, 50, and 200 mL for each method (Table 1). Overall, 143 samples were generated and processed as described below.
The rest of the wastewater samples collected at 13 locations in England were spiked only with Phi6 bacteriophage as a process control to estimate virus recovery.
Wastewater concentrations.
All wastewater concentration processes were performed in a BSL2/CL2 laboratory using a class II biosafety cabinet. Sample bottles were disinfected using 70% IMS prior to removal from the cabinet.
Method A: polyethylene glycol precipitation (PEG method). The PEG precipitation method described previously for SARS-CoV-2 detection in wastewater (56) was used with small modifications. The samples were centrifuged at 10,000 × g at 4°C for 10 min to clarify the solutions. The pellets from the 50-mL samples were subject to direct RNA/DNA extraction, whereas the rest of the pellets were discarded. The pH of the supernatants (Table 1) was adjusted to 7 to 7.5 and then were mixed with PEG 8000 and NaCl to reach the final concentrations of 10% and 2%, respectively. The solutions were mixed by inverting several times and incubated at 4°C for 16 h. The mixture was then centrifuged at 10,000 × g at 4°C for 30 min. The resulting pellet was subject to RNA/DNA extraction.
Method B: modified PEG precipitation (BE-PEG method). This method implemented a beef extract elution to detach viruses from solid matter prior to PEG precipitation (30). The samples were mixed with Lab Lemco beef extract (Oxoid, USA) and sodium nitrate to reach the final concentrations of 3% and 2 M, respectively. The pH of the mixture was adjusted to 5.5, and then it was incubated at 50 rpm at room temperature for 30 min. The PEG precipitation protocol was then followed, as described above (Table 1). The first centrifugation pellets from the 50-mL samples were subjected to RNA/DNA extraction, whereas the rest of the pellets were discarded. This method was also used for the concentration of the surveillance samples (200 mL each).
Method C: ammonium sulfate precipitation (AS method). The AS precipitation method described previously for SARS-CoV-2 detection in wastewater was used (51). In brief, the samples were centrifuged at 10,000 × g at 4°C for 10 min to clarify the solutions. The supernatants (Table 1) were mixed with AS to reach a final concentration of 40%. The solutions were mixed by inverting them several times and were incubated at 4°C for 1 to 2 h. The mixture was then centrifuged at 10,000 × g at 4°C for 30 min. The resulting pellet was subjected to RNA/DNA extraction.
Method D: ultrafiltration using the Amicon Ultra centrifugal filters (Amicon method). The samples were centrifuged at 4,000 × g at 4°C for 10 min to clarify the solutions. The pellets were discarded, whereas the supernatants (Table 1) were transferred to 10-kDa Amicon Ultra-15 centrifugal filters (Merck Life Science UK Ltd., Watford, UK). The samples were centrifuged at 5,000 × g for 30 to 60 min to reach a final volume of 200 to 500 μL. The filtrates were discarded. This method was also used for the concentration of the surveillance samples (20 mL each).
Method E: ultrafiltration using the InnovaPrep system (IP method). The samples were mixed with 5% Tween 20 (Sigma-Aldrich, USA) to reach the final concentration of 0.05% Tween. The mixtures were then centrifuged at 10,000 × g at 4°C for 10 min to clarify the solutions. The pellets were discarded, whereas the supernatants (Table 1) were filtered using the InnovaPrep (IP) concentrating pipette with 0.05-μm polysulfone (PS) hollow fiber filter tips (CP Select, USA). The tips were changed between samples. Samples were eluted in 25 mM Tris elution fluid (CP Select, USA), which contains 0.075% Tween 20.
Viral RNA/DNA extraction.
Viral RNA/DNA of both concentrated and unconcentrated samples were extracted using the NucliSens extraction system (bioMérieux, France) on a Kingfisher 96 Flex system (Thermo Scientific, USA) as described previously (51, 56). In brief, the samples were mixed and incubated with NucliSens lysis buffer for 10 min, followed by the addition of NucliSens magnetic silica beads with a 10-min incubation to allow the viral nucleic acids to bind to the beads. The binding was followed by washing steps using NucliSens wash buffers 1 to 3 and a final elution of RNA/DNA in NucliSens wash buffer 3. The final volume of the eluent was 100 μL.
Viral RNA/DNA quantification.
The viral RNA/DNA were quantified with reverse transcriptase quantitative PCR (RT-qPCR) (RNA targets) and with qPCR (DNA targets). All qPCRs were performed using a QuantStudio Flex 6 real-time PCR machine (Applied Biosystems, Inc., USA). The primers and probes for the target viruses have been used and validated previously (51, 56). Primers, probes, standards, and reaction conditions are detailed in Table S1. For quantification, a dilution series of DNA/RNA standards incorporating the target sequence was used. Each reaction plate contained multiple nontemplate controls, which were negative throughout the study, suggesting no cross-contamination. Assay limit of detection and limit of quantification values and RT-qPCR quality control data are summarized in Table S2.
For all samples, the RNA targets were quantified with probe-based assays using the TaqMan viral 1-step RT-qPCR master mix (Applied Biosystems, Inc., USA) with 1 μg bovine serum albumin (BSA) in the reaction mixes. Additionally, 16 nmol MgSO4 was added to the reaction mix for the SARS-CoV-2, Phi6, and influenza targets. Duplex RT-qPCR assays were used for the SARS-CoV-2 N1 gene fragment and the Phi6 phage (51) for influenza A and B (57) and for NoVGII and MNV (58), respectively. Singleplex assays were used for measles virus (59) and for rotavirus (RoV) with a commercial primer and probe mix (Primerdesign, UK). Adenovirus and crAssphage were quantified using the QuantiFast SYBR and the QuantiNova Probe qPCR reagents, respectively, with 1 μg BSA in the reaction mix, as described previously (30, 60).
The neat, unspiked samples from the surveillance study were also tested for human immunodeficiency virus (HIV) and hepatitis B and C viruses using a commercial triplex assay following the manufacturer’s protocols (Primerdesign, UK). Additionally, a duplex assay was used for the quantification of enteroviruses (EV) (Public Health Wales, personal communication) and enterovirus D68 (EVD68) (61), and a singleplex assay was used for norovirus GI (NoVGI) (58). When the Phi6 control recovery was <0.1%, the quantification was repeated with 2 μL sample/reaction; however, the reduced volumes did not affect recovery rates, suggesting little inhibition.
Data analysis.
The initial qPCR data analysis and quality control were performed using the QuantStudio Flex 6 real-time PCR software v1.7.1 (Applied Biosystems, Inc., USA). The viral concentrations were expressed as gc/μL RNA/DNA extract. The viral concentrations (gc/L) in wastewater determined by concentration methods were calculated as
| (1) |
The viral concentrations (gc/L) in the original unconcentrated samples were calculated as
| (2) |
Virus recoveries were calculated as
| (3) |
Subsequent data analysis and statistical tests were carried out in R (62). For the samples from the laboratory spiking experiment, sample starting volume (15 mL, 37.5 mL, and 150 mL), concentration method (PEG, AS, IP, BE-PEG, and Amicon), and water type (wastewater [WW], and deionized water [DW]) were selected as factors and covariates of viral recovery. The selected features were assessed in combination as predictors of viral recovery using a generalized linear model (glm) with the response variable modeled as a gamma distribution with a logarithmic link function following an assessment of the right skewed response variable distribution (equation 4; see Fig. S1 for residual plots).
| (4) |
where Recoveryi is the individual value of N total observations in the response variable and i relates to the individual observations of each variable. exp[…] indicates an exponential function of the product within the square brackets. α0 is the intercept, α1,…α3 are the fixed-effects coefficient estimates, and ϵ is the error between the model prediction and observation i of the response variable.
Following the glm, comparisons of individual features were visualized, and statistical tests were performed. For statistical tests, the recovery percentile was log10 transformed to meet assumptions of a Gaussian distribution (see Fig. S2 to S6 for quantile-quantile [qq] plots). Equality of variances were tested with F tests. Statistical comparisons of features with two levels and nonequal variance were made with Welch two sample t tests. Comparisons with three or more levels with equal variance were made with a one-way ANOVA, followed by pairwise two-sample t tests with pooled standard deviations (SD), adjusting P values with the Holm-Bonferroni (H-B) method (63). Comparisons with three or more levels and nonequal variance were made with a Welch ANOVA (one-way comparison of means), followed by pairwise two-sample t tests without pooled SD, adjusting P values with the H-B method. Paired tests were not selected due to missing data created by removal of undetermined results and sample removal during qPCR quality control. Water type was compared with only spiked viruses due to the lack of naturally present virus in deionized water. Starting volume was compared using only wastewater to simulate typical interactions seen in wastewater projects and not using the Amicon or IP methods due to differing volumes. Method comparison was made using 15-mL wastewater sample volumes which were used in all concentration methods, including the Amicon and IP methods.
For the neat wastewater samples, viral recoveries with the Amicon and BE-PEG methods were compared for six viruses (crAssphage, EV, EVD68, SARS-CoV-2, NoVGI, and NoVGII). The recovery in gene copies per liter was log10 transformed to meet assumptions for parametric analysis, and then statistical comparisons were made using paired t tests. Residual qq plots can be found in Fig. S7. Concentrations of ammonium, electrical conductivity, and orthophosphate have been shown to correlate with human populations and thus relate to flow over short periods of time flow (13). Therefore, the variability in flow had to be removed to assess the comparative effect of water chemistry variables on each concentration method. To remove this variability, the viral concentrations (gc/L) recovered by the Amicon method were divided by the viral concentrations recovered by the BE-PEG method. As each sample was paired, both sample methods were influenced by the same variability in flow, which was removed through this division. A linear mixed effects model (lmm) was then used to assess the comparative effect of wastewater chemistry and quality variables (turbidity, pH, orthophosphate, electrical conductivity, and ammonium) on the proportion of each concentration method’s viral recovery, utilizing random effects for the target virus to borrow strength in a combined assessment of all viruses (equation 6). Prior to the model being fit, the dependent variable was power transformed using the Box-Cox method (equations 5 and 6) (64), with lambda (λ) selected by maximizing the log-likelihood of a multiple linear regression with all predictors used in the final linear mixed effects model. The residual plots of the final model can be found in Fig. S8.
| (5) |
where y is the variable to be power transformed and λ is selected through maximizing the log-likelihood of a multiple linear regression with all predictors used in the final linear mixed effects model (equation 6).
| (6) |
where y(λ)ij is the individual value of N total observations in the response variable after power transformation. j relates to group in the factor variable modeled with random effects, and the maximum j is equal to the number of levels in the target virus variable; β0 is the intercept, and β1,…,β5 are the fixed-effect coefficient estimates; γj is the random-effect coefficients in group j; and ε is the error between the model prediction and observation i in group j (model parameters were fit independently of equation 4).
Data availability.
The full script and data are provided in a dedicated repository (https://github.com/CameronPellett/Spiked-virus-concentration-Bangor and https://github.com/CameronPellett/Chem-Con-Bangor-Bath).
ACKNOWLEDGMENTS
The work was supported under the C215.3 Wastewater Based Epidemiology Programme within the UK Government Accelerated Capability Environment (ACE). The Centre for Environmental Biotechnology Project was funded though the European Regional Development Fund (ERDF) by the Welsh government.
We thank Daphne Beniston and Andrew Singer (Accelerated Capability Environment, Homeland Security, UK) for their help organizing the project. We thank Public Health Wales for providing enterovirus primer and probe sequences and the staff at Anglian Water, Northumbrian Water, Southern Water, Thames Water, United Utilities, Yorkshire Water, and Welsh Water/Dŵr Cymru for their support with wastewater sampling. We also thank Andrew Weightman (Cardiff University, UK), Lydia Drumwright (University of Cambridge, UK), and Eleanor Gaunt (University of Edinburgh. UK) for providing the inactivated SARS-CoV-2, norovirus, rotavirus, and influenza virus stocks and Ian Goodfellow (University of Cambridge, UK) for providing the original MNV stock.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Kata Farkas, Email: fkata211@gmail.com.
Sébastien P. Faucher, McGill University
REFERENCES
- 1.WHO. 2020. WHO Coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 15 February 2022.
- 2.Bhat EA, Khan J, Sajjad N, Ali A, Aldakeel FM, Mateen A, Alqahtani MS, Syed R. 2021. SARS-CoV-2: insight in genome structure, pathogenesis and viral receptor binding analysis: an updated review. Int Immunopharmacol 95:107493. doi: 10.1016/j.intimp.2021.107493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascarella G, Strumia A, Piliego C, Bruno F, del Buono R, Costa F, Scarlata S, Agrò FE. 2020. COVID‐19 diagnosis and management: a comprehensive review. J Intern Med 288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran DP, Topol EJ. 2021. The proportion of SARS-CoV-2 infections that are asymptomatic: a systematic review. Ann Intern Med 174:655–662. doi: 10.7326/M20-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A, Solanki R, Sivadas N. 2021. Estimation of undetected symptomatic and asymptomatic cases of COVID‐19 infection and prediction of its spread in the USA. J Med Virol 93:3202–3210. doi: 10.1002/jmv.26897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xagoraraki I. 2020. Can we predict viral outbreaks using wastewater surveillance? Environ Eng 146:01820003. doi: 10.1061/(ASCE)EE.1943-7870.0001831. [DOI] [Google Scholar]
- 7.Feng L, Zhang W, Li X. 2018. Monitoring of regional drug abuse through wastewater-based epidemiology: a critical review. Sci China Earth Sci 61:239–255. doi: 10.1007/s11430-017-9129-x. [DOI] [Google Scholar]
- 8.Choi PM, Tscharke BJ, Donner E, O’Brien JW, Grant SC, Kaserzon SL, Mackie R, O’Malley E, Crosbie ND, Thomas KV, Mueller JF. 2018. Wastewater-based epidemiology biomarkers: past, present and future. Trends Analyt Chem 105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- 9.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. 2020. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett 7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJ, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. 2020. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillary LS, Farkas K, Maher KH, Lucaci A, Thorpe J, Distaso MA, Gaze WH, Paterson S, Burke T, Connor TR, McDonald JE, Malham SK, Jones DL. 2021. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res 200:117214. doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai Y, Davis A, Jones D, Lemeshow S, Tu H, He F, Ru P, Pan X, Bohrerova Z, Lee J. 2021. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci Total Environ 801:149757. doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wade MJ, lo Jacomo A, Armenise E, Brown MR, Bunce JT, Cameron GJ, Fang Z, Farkas K, Gilpin DF, Graham DW, Grimsley JMS, Hart A, Hoffmann T, Jackson KJ, Jones DL, Lilley CJ, McGrath JW, McKinley JM, McSparron C, Nejad BF, Morvan M, Quintela-Baluja M, Roberts AMI, Singer AC, Souque C, Speight VL, Sweetapple C, Walker D, Watts G, Weightman A, Kasprzyk-Hordern B. 2022. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J Hazard Mater 424:127456. doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshaviah A, Hu XC, Henry M. 2021. Developing a flexible national wastewater surveillance system for COVID-19 and beyond. Environ Health Perspect 129:45002. doi: 10.1289/EHP8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrestha S, Yoshinaga E, Chapagain SK, Mohan G, Gasparatos A, Fukushi K. 2021. Wastewater-based epidemiology for cost-effective mass surveillance of COVID-19 in low-and middle-income countries: challenges and opportunities. Water (Basel) 13:2897. doi: 10.3390/w13202897. [DOI] [Google Scholar]
- 16.Gibas C, Lambirth K, Mittal N, Juel MAI, Barua VB, Roppolo Brazell L, Hinton K, Lontai J, Stark N, Young I, Quach C, Russ M, Kauer J, Nicolosi B, Chen D, Akella S, Tang W, Schlueter J, Munir M. 2021. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci Total Environ 782:146749. doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar M, Joshi M, Shah AV, Srivastava V, Dave S. 2021. First wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: a perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. medRxiv. doi: 10.1016/j.scitotenv.2021.148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald SF, Rossi G, Low AS, McAteer SP, O’Keefe B, Findlay D, Cameron GJ, Pollard P, Singleton PTR, Ponton G, Singer AC, Farkas K, Jones D, Graham DW, Quintela-Baluja M, Tait-Burkard C, Gally DL, Kao R, Corbishley A. 2021. Site specific relationships between COVID-19 cases and SARS-CoV-2 viral load in wastewater treatment plant influent. Environ Sci Technol 55:15276–15286. doi: 10.1021/acs.est.1c05029. [DOI] [PubMed] [Google Scholar]
- 19.Haramoto E, Kitajima M, Hata A, Torrey JR, Masago Y, Sano D, Katayama H. 2018. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res 135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed W, Payyappat S, Cassidy M, Harrison N, Besley C. 2020. Sewage-associated marker genes illustrate the impact of wet weather overflows and dry weather leakage in urban estuarine waters of Sydney, Australia. Sci Total Environ 705:135390. doi: 10.1016/j.scitotenv.2019.135390. [DOI] [PubMed] [Google Scholar]
- 21.McMinn BR, Korajkic A, Kelleher J, Herrmann MP, Pemberton AC, Ahmed W, Villegas EN, Oshima K. 2021. Development of a large volume concentration method for recovery of coronavirus from wastewater. Sci Total Environ 774:145727. doi: 10.1016/j.scitotenv.2021.145727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu D, Huang Z, Luo J, Zhang X, Sha S. 2020. Primary concentration—the critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci Total Environ 747:141245. doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusiñol M, Martínez-Puchol S, Forés E, Itarte M, Girones R, Bofill-Mas S. 2020. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr Opin Environ Sci Health 17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farkas K, Hillary LS, Malham SK, McDonald JE, Jones DL. 2020. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr Opin Environ Sci Health 17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Rosa G, Bonadonna L, Lucentini L, Kenmoe S, Suffredini E. 2020. Coronavirus in water environments: occurrence, persistence and concentration methods: a scoping review. Water Res 179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, Haramoto E, Gyawali P, Korajkic A, McMinn BR, Mueller JF, Simpson SL, Smith WJM, Symonds EM, Thomas KV, Verhagen R, Kitajima M. 2020. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ 739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y, Ellenberg RM, Graham KE, Wigginton KR. 2016. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol 50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 28.Corpuz MVA, Buonerba A, Vigliotta G, Zarra T, Ballesteros F, Campiglia P, Belgiorno V, Korshin G, Naddeo V. 2020. Viruses in wastewater: occurrence, abundance and detection methods. Sci Total Environ 745:140910. doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailepessov D, Arivalan S, Kong M, Griffiths J, Low SL, Chen H, Hapuarachchi HC, Gu X, Lee WL, Alm EJ, Thompson J, Wuertz S, Gin K, Ching NL, Wong JCC. 2022. Development of an efficient wastewater testing protocol for high-throughput country-wide SARS-CoV-2 monitoring. Sci Total Environ 826:154024. doi: 10.1016/j.scitotenv.2022.154024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farkas K, Cooper DM, McDonald JE, Malham SK, de Rougemont A, Jones DL, Rougemont A, de Jones DL, de Rougemont A, Jones DL. 2018. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci Total Environ 634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 31.Ikner LA, Gerba CP, Bright KR. 2012. Concentration and recovery of viruses from water: a comprehensive review. Food Environ Virol 4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- 32.Petala M, Dafou D, Kostoglou M, Karapantsios T, Kanata E, Chatziefstathiou A, Sakaveli F, Kotoulas K, Arsenakis M, Roilides E, Sklaviadis T, Metallidis S, Papa A, Stylianidis E, Papadopoulos A, Papaioannou N. 2021. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. case study: the city of Thessaloniki in Greece. Sci Total Environ 755:142855. doi: 10.1016/j.scitotenv.2020.142855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forés E, Bofill-Mas S, Itarte M, Martínez-Puchol S, Hundesa A, Calvo M, Borrego CM, Corominas LL, Girones R, Rusiñol M. 2021. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci Total Environ 768:144786. doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed W, Bivins A, Simpson SL, Smith WJM, Metcalfe S, McMinn B, Symonds EM, Korajkic A. 2021. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Sci Total Environ 799:149386. doi: 10.1016/j.scitotenv.2021.149386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrader C, Schielke A, Ellerbroek L, Johne R. 2012. PCR inhibitors: occurrence, properties and removal. J Appl Microbiol 113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 36.Mlejnkova H, Sovova K, Vasickova P, Ocenaskova V, Jasikova L, Juranova E. 2020. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int J Environ Res Public Health 17:5508–5509. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haramoto E, Malla B, Thakali O, Kitajima M. 2020. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci Total Environ 737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randazzo W, Cuevas-Ferrando E, Sanjuan R, Domingo-Calap P, Sanchez G. 2020. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int J Hyg Environ Health 230:113621. doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennechart-Collette C, Martin-Latil S, Guillier L, Perelle S. 2015. Determination of which virus to use as a process control when testing for the presence of hepatitis A virus and norovirus in food and water. Int J Food Microbiol 202:57–65. doi: 10.1016/j.ijfoodmicro.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 40.Balboa S, Mauricio-Iglesias M, Rodríguez S, Martínez-Lamas L, Vasallo FJ, Regueiro B, Lema JM. 2021. The fate of SARS-CoV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19 . Sci Total Environ 772:145268. doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Mazurowski L, Dewan A, Carine M, Haak L, Guarin TC, Dastjerdi NG, Gerrity D, Mentzer C, Pagilla KR. 2022. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Sci Total Environ 817:152958. doi: 10.1016/j.scitotenv.2022.152958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonçalves J, Koritnik T, Mioč V, Trkov M, Bolješič M, Berginc N, Prosenc K, Kotar T, Paragi M. 2021. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci Total Environ 755:143226. doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crits-Christoph A, Kantor RS, Olm MR, Whitney ON, Al-Shayeb B, Lou YC, Flamholz A, Kennedy LC, Greenwald H, Hinkle A, Hetzel J, Spitzer S, Koble J, Tan A, Hyde F, Schroth G, Kuersten S, Banfield JF, Nelson KL. 2021. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio 12:e02703-20. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juel MAI, Stark N, Nicolosi B, Lontai J, Lambirth K, Schlueter J, Gibas C, Munir M. 2021. Performance evaluation of virus concentration methods for implementing SARS-CoV-2 wastewater based epidemiology emphasizing quick data turnaround. Sci Total Environ 801:149656. doi: 10.1016/j.scitotenv.2021.149656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, Thompson H, Keeling D, Mitchell J, Gonzalez D. 2020. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res 186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falman JC, Fagnant-Sperati CS, Kossik AL, Boyle DS, Meschke JS. 2019. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food Environ Virol 11:20–31. doi: 10.1007/s12560-018-09364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bofill-Mas S, Rusiñol M. 2020. Recent trends on methods for the concentration of viruses from water samples. Curr Opin Environ Sci Health 16:7–13. doi: 10.1016/j.coesh.2020.01.006. [DOI] [Google Scholar]
- 48.Lewis GD, Metcalf TG. 1988. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl Environ Microbiol 54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machado-Moreira B, Monteiro S, Santos R, Martinez-Murcia A, Rajkovic A, Smigic N, Richards KG, Abram F, Burgess CM. 2020. Impact of beef extract used for sample concentration on the detection of Escherichia coli DNA in water samples via qPCR. J Microbiol Methods 168:105786. doi: 10.1016/j.mimet.2019.105786. [DOI] [PubMed] [Google Scholar]
- 50.Cao H, Tsai FTC, Rusch KA. 2010. Salinity and soluble organic matter on virus sorption in sand and soil columns. Ground Water 48:42–52. doi: 10.1111/j.1745-6584.2009.00645.x. [DOI] [PubMed] [Google Scholar]
- 51.Kevill JL, Pellett C, Farkas K, Brown MR, Bassano I, Denise H, McDonald JE, Malham SK, Porter J, Warren J, Evens NP, Paterson S, Singer AC, Jones DL. 2022. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Sci Total Environ 808:151916. doi: 10.1016/j.scitotenv.2021.151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greninger AL, DeRisi JL. 2015. Draft genome sequences of ciliovirus and brinovirus from San Francisco wastewater. Genome Announc 3:e00651-15. doi: 10.1128/genomeA.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q, Zhao H, Du B. 2017. Bacteria and bacteriophage communities in bulking and non-bulking activated sludge in full-scale municipal wastewater treatment systems. Biochem Eng J 119:101–111. doi: 10.1016/j.bej.2016.12.017. [DOI] [Google Scholar]
- 54.Brisebois E, Veillette M, Dion-Dupont V, Lavoie J, Corbeil J, Culley A, Duchaine C. 2018. Human viral pathogens are pervasive in wastewater treatment center aerosols. J Environ Sci (China) 67:45–53. doi: 10.1016/j.jes.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCall C, Wu H, Miyani B, Xagoraraki I. 2020. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res 184:116160. doi: 10.1016/j.watres.2020.116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farkas K, Hillary LS, Thorpe J, Walker DI, Lowther JA, McDonald JE, Malham SK, Jones DL. 2021. Concentration and quantification of SARS-CoV-2 RNA in wastewater using polyethylene glycol-based concentration and qRT-PCR. Methods Protocols 4:17–19. doi: 10.3390/mps4010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu B, Kirby MK, Davis WG, Warnes C, Liddell J, Liu J, Wu K-H, Hassell N, Benitez AJ, Wilson MM, Keller MW, Rambo-Martin BL, Camara Y, Winter J, Kondor RJ, Zhou B, Spies S, Rose LE, Winchell JM, Limbago BM, Wentworth DE, Barnes JR. 2021. Multiplex real-time reverse transcription PCR for influenza A virus, influenza B virus, and severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 27:1821–1830. doi: 10.3201/eid2707.210462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farkas K, Malham SK, Peters DE, de Rougemont A, McDonald JE, de Rougemont A, Malham SK, Jones DL. 2017. Evaluation of two triplex one-step qRT-PCR assays for the quantification of human enteric viruses in environmental samples. Food Environ Virol 9:342–349. doi: 10.1007/s12560-017-9293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hummel KB, Lowe L, Bellini WJ, Rota PA. 2006. Development of quantitative gene-specific real-time RT-PCR assays for the detection of measles virus in clinical specimens. J Virol Methods 132:166–173. doi: 10.1016/j.jviromet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 60.Farkas K, Adriaenssens EM, Walker DI, McDonald JE, Malham SK, Jones DL. 2019. Critical evaluation of crAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ Virol 11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poelman R, Schölvinck EH, Borger R, Niesters HGM, van Leer-Buter C. 2015. The emergence of enterovirus D68 in a Dutch University Medical Center and the necessity for routinely screening for respiratory viruses. J Clin Virol 62:1–5. doi: 10.1016/j.jcv.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 63.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70. [Google Scholar]
- 64.Box GEP, Cox DR. 1964. An analysis of transformations. J R Stat Soc: Series B (Methodological) 26:211–243. doi: 10.1111/j.2517-6161.1964.tb00553.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01102-22-s0001.pdf, PDF file, 0.8 MB (858.8KB, pdf)
Data Availability Statement
The full script and data are provided in a dedicated repository (https://github.com/CameronPellett/Spiked-virus-concentration-Bangor and https://github.com/CameronPellett/Chem-Con-Bangor-Bath).