ABSTRACT
The environmental pathogenic bacterium Chromobacterium violaceum kills Gram-positive bacteria by delivering violacein packed into outer membrane vesicles, but nothing is known about its contact-dependent competition mechanisms. In this work, we demonstrate that C. violaceum utilizes a type VI secretion system (T6SS) containing multiple VgrG proteins primarily for interbacterial competition. The single T6SS of C. violaceum contains six vgrG genes, which are located in the main T6SS cluster and four vgrG islands. Using T6SS core component-null mutant strains, Western blotting, fluorescence microscopy, and competition assays, we showed that the C. violaceum T6SS is active and required for competition against Gram-negative bacteria such as Pseudomonas aeruginosa but dispensable for C. violaceum infection in mice. Characterization of single and multiple vgrG mutants revealed that, despite having high sequence similarity, the six VgrGs show little functional redundancy, with VgrG3 showing a major role in T6SS function. Our coimmunoprecipitation data support a model of VgrG3 interacting directly with the other VgrGs. Moreover, we determined that the promoter activities of T6SS genes increased at high cell density, but the produced Hcp protein was not secreted under such condition. This T6SS growth phase-dependent regulation was dependent on CviR but not on CviI, the components of a C. violaceum quorum sensing (QS) system. Indeed, a ΔcviR but not a ΔcviI mutant was completely defective in Hcp secretion, T6SS activity, and interbacterial competition. Overall, our data reveal that C. violaceum relies on a QS-regulated T6SS to outcompete other bacteria and expand our knowledge about the redundancy of multiple VgrGs.
IMPORTANCE The type VI secretion system (T6SS) is a contractile nanomachine used by many Gram-negative bacteria to inject toxic effectors into adjacent cells. The delivered effectors are bound to the components of a puncturing apparatus containing the protein VgrG. The T6SS has been implicated in pathogenesis and, more commonly, in competition among bacteria. Chromobacterium violaceum is an environmental bacterium that causes deadly infections in humans. In this work, we characterized the single T6SS of C. violaceum ATCC 12472, including its six VgrG proteins, regarding its function and regulation. This previously undescribed C. violaceum T6SS is active, regulated by QS, and required for interbacterial competition instead of acute infection in mice. Among the VgrGs, VgrG3, encoded outside the main T6SS cluster, showed a major contribution to T6SS function. These results shed light on a key contact-dependent killing mechanism used by C. violaceum to antagonize other bacteria.
KEYWORDS: type VI secretion system, VgrG proteins, interbacterial competition, Chromobacterium violaceum, quorum sensing
INTRODUCTION
Bacteria engage in complex ecological relationships with other organisms and very often compete with other bacteria. Interbacterial competition can occur at a distance through diffusible secreted inhibitory factors or may be mediated by contact (1–4). Contact-mediated competition involves the injection of toxins directly into a target cell through complex protein machineries called secretion systems, such as the type IV, type V, and type VI secretion systems (3, 5). The type VI secretion system (T6SS), found in many Gram-negative bacteria, delivers toxic effectors into both bacterial and host eukaryotic cells (6–8). However, its main function is to mediate interbacterial competition, affecting the dynamics of bacterial communities in multiple ecological contexts, such as during interactions between bacterial pathogens and the microbiota (4, 9). In addition to hijacking host cellular processes and intoxicating competing bacteria, novel roles have been described for the T6SS and its effectors, including antifungal activity, metal acquisition, resistance against amoeba predation, and several intraspecies social interactions (9–12).
T6SS is a complex nanomachine resembling the contractile tail of bacteriophages. It is classified into four subtypes (T6SSi to T6SSiv) that differ in structure and number of core components (9, 13). The canonical T6SSi commonly found in Proteobacteria is composed of at least 14 core components that are assembled in subcomplexes: a cytoplasmic baseplate, an intermembrane complex, a contractile sheath-like structure (VipAB), and an inner tube of polymerized Hcp (hemolysin-coregulated protein) sharpened at its tip by VgrG (valine-glycine repeat protein G) trimers and PAAR (proline-alanine-alanine-arginine) proteins (14–17). The translocation of toxic protein effectors by the T6SS directly inside a target cell involves the firing of a puncturing apparatus (Hcp/VgrG/PAAR) by contraction of the VipAB sheath (18). The ATPase ClpV recycles the sheath components, which are then reused for new T6SS firing events (14, 19). The effectors can be “specialized” effectors, when they are covalently fused to the expelled components VgrG, PAAR, or Hcp, or “cargo” effectors, which comprise independent proteins interacting directly or via adaptor proteins with VgrG, PAAR, or Hcp (16, 20–22). VgrG proteins have a modular architecture containing a core gp27-gp5 portion with puncturing and structural functions and sometimes an additional C-terminal region serving as an effector or adaptor domain (6, 16, 23, 24). Even in bacteria harboring a single T6SS, the presence of multiple VgrG proteins encoded by genes located in the main T6SS cluster and vgrG islands is common (25). However, little is known about the redundancy of multiple VgrGs for the function of the T6SS (25–28).
The expression and activity of T6SSs can be tightly regulated at several levels in response to different environmental cues, such as reactive oxygen species, metal scarcity, temperature, pH, and membrane damage caused by the attack of competitor bacteria (17, 29, 30). For bacteria having more than one T6SS, each of them can be differentially regulated to exert a particular function (12, 29). Among the multitude of regulatory mechanisms controlling T6SS expression, regulation by quorum sensing (QS) systems is a recurrent theme with very distinct outcomes (29). For instance, the QS regulation of T6SSs has been described in several bacteria, such as Pseudomonas aeruginosa (31), Vibrio cholerae (32), Burkholderia thailandensis (33), Aeromonas hydrophila (34), and Vibrio fluvialis (35).
Chromobacterium violaceum is a free-living Gram-negative bacterium found in the soil and water of tropical and subtropical regions. Despite its free-living lifestyle, C. violaceum causes systemic infections in humans and animals by employing several virulence factors, such as a Cpi-1/1a T3SS, which injects effectors into host eukaryotic cells (36–39). C. violaceum produces violacein, a violet hydrophobic compound with broad-spectrum biocide activity that targets the cytoplasmic membranes of Gram-positive bacteria (40, 41). By delivering violacein packed into outer membrane vesicles (OMVs), C. violaceum outcompetes Gram-positive but not Gram-negative bacteria in a contact-independent manner (42, 43). In addition to producing violacein, bacteria of the genus Chromobacterium antagonize other organisms, such as pathogenic fungi, parasites, and larvae of disease-transmitting insects, by using extracellular secreted molecules, such as chitinases, proteases, hydrogen cyanide, and romidepsin (44–47). Most of these compounds are specifically produced at high cell density via activation by the QS system CviI/CviR, a circuit composed of the regulator CviR and CviI, an enzyme that produces homoserine lactone (HSL)-type autoinducers (48–51). However, the role of contact-dependent mechanisms for C. violaceum competition remains largely unknown.
In this study, we used interbacterial competition assays to reveal that C. violaceum employs its single T6SS to overcome competitor Gram-negative bacteria, such as P. aeruginosa. Virulence assays with a murine infection model indicated that C. violaceum T6SS mutants were as virulent as the wild-type strain. These data indicate that the C. violaceum T6SS is more relevant to interbacterial competition than to bacterial pathogenesis. Furthermore, we demonstrate that the C. violaceum T6SS is active, expressed under laboratory conditions, and regulated by cell density and QS. The study of the six vgrG genes found in the C. violaceum genome revealed little functional redundancy, given that VgrG3 showed a preponderant role in relation to the other five VgrGs in the assembly of a functional T6SS with antibacterial activity. Coimmunoprecipitation assays indicated that VgrG3 interacts directly with other VgrGs in vivo, suggesting that VgrG3 could facilitate the secretion of effectors bound to the other VgrGs.
RESULTS
C. violaceum has a single T6SS containing multiple VgrG proteins.
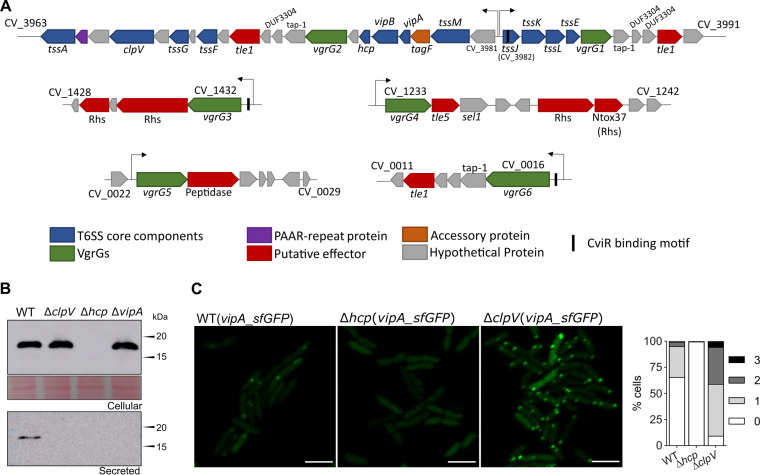
We searched for T6SS genes in the genome of C. violaceum ATCC 12472 (52) using BLAST searches with conserved T6SS core components (VipA and Hcp) and the TXSScan tool (53). These in silico analyses retrieved a single T6SS whose genes are located in a main cluster containing 31 open reading frames (from CV_3963 to CV_3991) organized as two divergently transcribed large putative operons (Fig. 1A). This T6SS main cluster contains all genes encoding the T6SS core components (tssA to tssM and PAAR) found in a single copy, except for vgrG (tssI), which was found in two copies (vgrG1 and vgrG2). There is also a tagF gene, described as a posttranslational inhibitor of T6SS activity in other bacteria (17). Downstream of both vgrG1 and vgrG2, there are genes encoding putative effectors (the phospholipases of the Tle1 family with the DUF2235 domain, CV_3990 and CV_3971) (54), chaperone/adaptor proteins (TEC/Tap-1, DUF4123) (55, 56), and putative immunity proteins (DUF3304) (Fig. 1A). Using vgrG1 and vgrG2 for BLAST searches, we detected four additional vgrG genes (vgrG3, vgrG4, vgrG5, and vgrG6) dispersed in other regions of the C. violaceum genome. Each such vgrG gene is located in small orphan VgrG clusters that contain genes encoding putative effectors, such as Rhs repeat-containing proteins (CV_1429, CV_1431, CV_1238, and CV_1239), Tle5 (CV_1234) and Tle1 (CV_0012) phospholipases, and a protein with lysozyme-like and peptidase M23 domains (CV_0024) (Fig. 1A). These analyses indicate that C. violaceum has a single T6SS and a repertoire of potential T6SS-delivered effectors encoded by genes located near those encoding six VgrG proteins.
FIG 1.
The single T6SS of C. violaceum is active and contains multiple VgrG proteins. (A) The genes encoding the T6SS components are located in a main T6SS cluster and four minor orphan VgrG clusters in the genome of C. violaceum ATCC 12472. Conserved T6SS components and effectors are colored according to their functions. The six VgrGs are in green. The arrows above the genes indicate promoter regions. Genes of the main T6SS are organized into two putative large operons transcribed from divergent promoters (regions between CV_3981 and tssJ). (B) Secretion of Hcp depends on T6SS core components. Western blot of Hcp with an anti-Hcp antibody from the C. violaceum WT and the indicated T6SS-null mutant strains using whole cellular extracts or cell-free supernatants. Protein loading was confirmed by membrane Ponceau staining. (C) Activity of the T6SS visualized by fluorescence microscopy of VipA_sfGFP. Assembled and contracted sheaths appear as GFP foci. The foci were counted manually, and the percentages indicate the proportions of bacterial cells with one, two, or three GFP foci at the same time. Bars = 3 μm.
The C. violaceum T6SS is functional and active under laboratory growth conditions.
To verify the functionality of the C. violaceum T6SS, we analyzed the expression and secretion of Hcp by immunoblotting assays with an anti-Hcp serum developed against the purified His-Hcp protein. In the cellular fraction, Hcp was detected from Luria-Bertani (LB)-grown log-phase cultures of wild-type (WT), ΔclpV, and ΔvipA strains (Fig. 1B). However, Hcp was detected in the cell-free supernatants of the WT but not the ΔclpV and ΔvipA strains, indicating that the T6SS core components VipA (sheath structure) and ClpV (sheath depolymerization) are required for Hcp secretion. As expected, Hcp was not detected in either the cellular or secreted fraction of the Δhcp mutant strain (Fig. 1B). To assess the dynamics of the T6SS sheath in vivo, we constructed a plasmid-borne carboxy-terminal fusion of the VipA protein with the superfolded green fluorescent protein (sfGFP) (18), and the resulting construct was transferred to the WT and T6SS mutants. Visualization by fluorescence microscopy in the WT strain showed a high activity of the T6SS sheath in log-phase cultures, as shown by the high number of sfGFP foci (Fig. 1C). As expected, no sfGFP focus was observed in the Δhcp strain (Fig. 1C), since Hcp polymerization is necessary for sheath assembly. Furthermore, the images suggest that nondynamic foci accumulated in ΔclpV, possibly because, in the absence of ClpV, the VipAB sheath structure remained assembled after the firing event (Fig. 1C). These results indicate that the C. violaceum T6SS is functionally active and requires the T6SS core components Hcp, VipA, and ClpV for its proper function.
The T6SS contributes to interbacterial competition but not to virulence in C. violaceum.
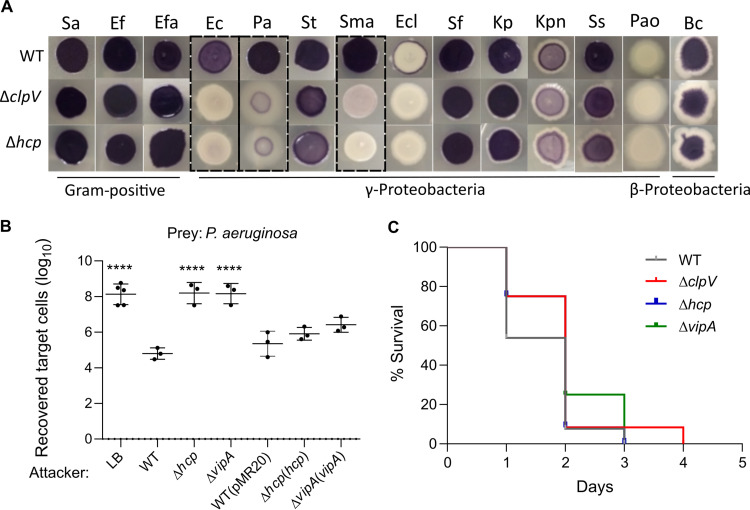
Considering that T6SSs mediate bacterial competition and are involved in bacterial pathogenesis (4, 8, 57, 58), we tested whether the T6SS is involved in such processes in C. violaceum (Fig. 2). To verify the competitive capacity of C. violaceum against a broad range of bacteria, we performed a visual screening that consisted of the coculture of C. violaceum WT or T6SS mutants (purple due to violacein) with phylogenetically distinct bacteria (Fig. 2A). C. violaceum WT was able to overcome almost all targeting bacteria (purple output), except for Enterobacter cloacae ATCC 13047 and P. aeruginosa PAO1 (Fig. 2A). However, the C. violaceum advantage was lost for the ΔclpV and Δhcp mutant strains in cocultures against Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, and Stenotrophomonas maltophilia and to a lesser extent against Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae ATCC 13883, and Shigella sonnei (Fig. 2A). The T6SS mutant strains grew similarly to the WT strain in LB medium (see Fig. S1A in the supplemental material), suggesting that their decreased competitive fitness is due to the loss of the T6SS functionality itself instead of a general fitness impairment.
FIG 2.
The T6SS contributes to interbacterial competition but not to virulence in C. violaceum. (A) C. violaceum outcompetes many Gram-negative bacteria via the T6SS. Qualitative growth competition assay of C. violaceum WT or T6SS mutants against Staphylococcus aureus (Sa), Enterococcus faecium (Ef), Enterococcus faecalis (Efa), Escherichia coli (Ec), Pseudomonas aeruginosa (Pa), Salmonella Typhimurium (St), Stenotrophomonas maltophilia (Sma), Enterobacter cloacae (Ecl), Shigella flexneri (Sf), Klebsiella pneumoniae (Kp), Klebsiella pneumoniae ATCC BAA-1705 (Kpn), Shigella sonnei (Ss), Pseudomonas aeruginosa PAO1 (Pao), and Burkholderia cepacia (Bc) (details in Table S3). Predominantly purple spots (due to violacein production by C. violaceum) indicate the cocultures in which C. violaceum was able to outcompete the target bacteria. The C. violaceum T6SS showed a major role in competition against E. coli, P. aeruginosa ATCC 27853, and S. maltophilia. (B) C. violaceum employs its T6SS to kill P. aeruginosa. A quantitative antibacterial assay was performed by counting the CFU of P. aeruginosa ATCC 27853 without (LB) or after coculture with C. violaceum WT, T6SS mutants (Δhcp and ΔvipA strains), or complemented strains. Data are from at least three independent biological assays. Bars show means and standard deviations (SD). Statistical analyses were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Asterisks indicate variations in relation to the WT. ****, P < 0.0001. (C) The T6SS does not contribute to C. violaceum virulence. BALB/c mice were intraperitoneally infected with 106 CFU of the indicated C. violaceum strains. The animals were monitored for 5 days (n = 13 for WT; n = 12 for the ΔclpV strain; n = 11 for the Δhcp strain; and n = 4 for the ΔvipA strain). Statistical analysis by the log-rank (Mantel-Cox) test showed no significant difference among the strains.
To further confirm the role of the T6SS as an antibacterial weapon, we performed a quantitative antibacterial assay using cocultures of C. violaceum (attacker) with P. aeruginosa ATCC 27853 (prey) at a ratio of 5:1 (Fig. 2B). The recovery of P. aeruginosa was at least three logs higher in competition against the T6SS mutants than against the WT strain, indicating C. violaceum T6SS-mediated killing activity. In fact, the prey recovery competing against the T6SS mutants was at the same levels as the negative control using LB medium instead of C. violaceum. The complemented strains killed P. aeruginosa similarly to the WT strain (Fig. 2B). Together, these results indicate that the T6SS is a killing machinery that provides a competitive advantage for C. violaceum against other bacteria, likely helping it persist in mixed bacterial populations.
The role of the T6SS in virulence was assessed using a mouse model of acute infection (59). After intraperitoneal injection, animals inoculated with three distinct T6SS mutants succumbed as fast as those inoculated with the WT strain (Fig. 2C), indicating that the T6SS does not contribute to C. violaceum virulence, at least under our tested conditions.
Among the six VgrGs, VgrG3 has major roles in T6SS assembly and function.
The protein VgrG is the sole T6SS core component whose genes were found in multiple copies in the C. violaceum genome (Fig. 1A). The six VgrG proteins from C. violaceum showed high sequence identity (70% to 93%) and a similar domain organization (Fig. S2), raising the question of whether they would have redundant roles. To address this question, we constructed null mutant strains with individual or sequential deletions of the vgrG genes and a sextuple mutant (ΔvgrG1-6) with each single vgrG gene reintroduced using a plasmid. The vgrG mutant strains showed no growth impairment, as assessed by growth curves (Fig. S1B and C). These strains were used to investigate the roles of the six VgrGs in the activity/assembly (Fig. 3) and antibacterial function (Fig. 4) of the T6SS.
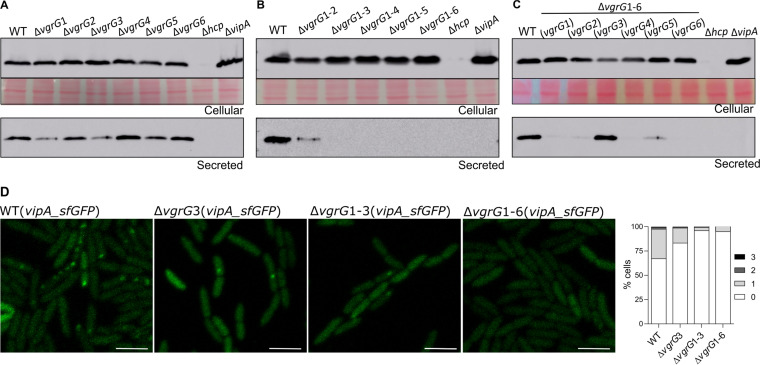
FIG 3.
Each VgrG distinctly affects T6SS activity, but VgrG3 has a preponderant role. (A to C) Analysis of VgrG-dependent activity of the T6SS evaluating Hcp secretion. Western blots with anti-Hcp to evaluate Hcp expression (cellular) and secretion (secreted) in (A) individual vgrG mutants, (B) sequential vgrG mutants, or (C) the sextuple ΔvgrG1-6 mutant harboring each single vgrG gene. Protein loading was confirmed by membrane Ponceau staining. (D) Effects of VgrGs on the assembly and activity of the T6SS visualized by the formation of VipA_sfGFP foci. The GFP foci were observed by fluorescence microscopy and counted in the indicated vgrG mutant strains. Bars = 3 μm.
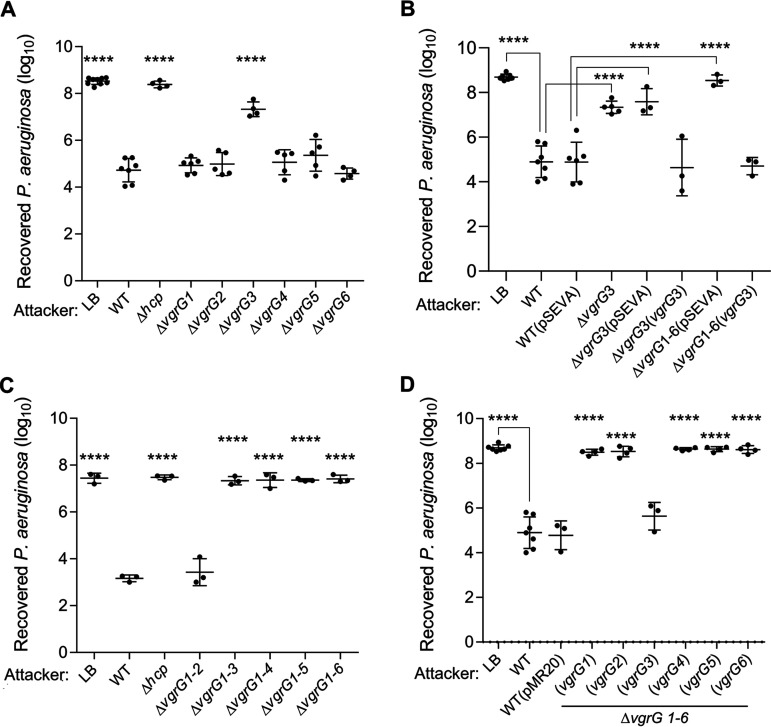
FIG 4.
VgrG3 is the most important VgrG for C. violaceum antibacterial activity. A quantitative antibacterial assay was performed by counting the CFU of P. aeruginosa ATCC 27853 without (LB) or after coculture with C. violaceum. Competitions were performed using (A) the individual vgrG mutants, (B) the complemented ΔvgrG3(vgrG3) strain, (C) the sequential vgrG mutants, and (D) the sextuple ΔvgrG1-6 mutant harboring each single vgrG gene. Each point represents biological replicates of distinct assays. Bars show means and SD. Statistical analyses were performed using one-way ANOVA with Tukey’s post hoc test. ****, P < 0.0001 in relation to the WT, WT(pMR20) or WT(pSEVA), as indicated.
The immunoblotting of secreted Hcp from log-phase cultures revealed that the individual deletion of each vgrG had a distinct impact on the T6SS functionality, with marked decreases in Hcp secretion in the ΔvgrG1 and ΔvgrG3 mutants and slight or no effects in the other vgrG mutants (Fig. 3A). The ΔvgrG1-2 double mutant showed residual secretion of Hcp, but no Hcp secretion was detected from the triple to the sextuple vgrG mutants, indicating that the combined deletion of vgrG1, vgrG2, and vgrG3 resulted in the complete loss of T6SS functionality (Fig. 3B). Immunoblotting assays using the sextuple mutant with the reintroduction of each single vgrG gene indicated that the presence of vgrG3 fully restored the ability of the ΔvgrG1-6 mutant to secrete Hcp, while the presence of the other five individual vgrG genes resulted in very weak Hcp secretion (Fig. 3C). As expected, the Hcp protein was no longer detected in the Δhcp mutant (cellular or supernatant fractions) or in the supernatant of the ΔvipA mutant (Fig. 3A to C). These results indicate that VgrG3 has a preponderant role over the other VgrGs in Hcp secretion. The role of VgrG3 in T6SS functionality was further confirmed by fluorescence microscopy using VipA_sfGFP. The number of sfGFP foci decreased in the ΔvgrG3 mutant compared to that in the WT strain and was almost abolished in the ΔvgrG1-3 and ΔvgrG1-6 mutant strains (Fig. 3D).
To verify the contribution of each VgrG to interbacterial competition, we performed competition assays against P. aeruginosa ATCC 27853 using our panel of C. violaceum vgrG mutants (Fig. 4). Among the single vgrG mutants, only the ΔvgrG3 strain showed a decreased competitive fitness compared to that of the WT strain, although at levels lower than the control (LB) and the Δhcp mutant (Fig. 4A). This phenotype was fully reverted in the ΔvgrG3(vgrG3) complemented strain and was unaffected in the strains with the empty vector (Fig. 4B). Regarding the sequential vgrG mutants, the ΔvgrG1-2 strain displayed no change in competitive capacity, but the ΔvgrG1-3 strain and all other multiple vgrG mutants showed strong impairments in competition, similar to that found for the Δhcp mutant (Fig. 4C). The complete loss of competitiveness of the sextuple mutant ΔvgrG1-6 was fully reversed when this strain was provided with the vgrG3 gene, but no effect was observed using the other vgrG genes (Fig. 4D). Altogether, these data demonstrate that VgrG3 is the most important VgrG for the antibacterial activity of the C. violaceum T6SS, probably owing to its important structural role for the overall T6SS function.
VgrG3 interacts directly with other VgrGs in vivo.
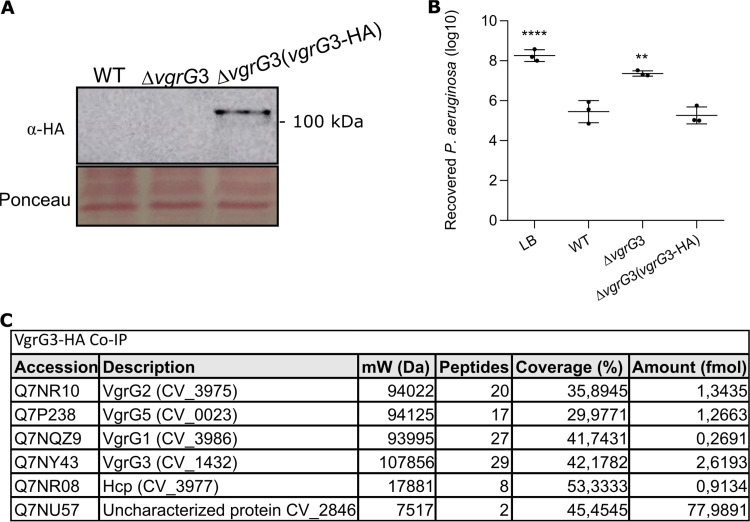
Given the main role of VgrG3 in T6SS assembly and function, we hypothesize that VgrG3 could interact with other VgrGs to allow the delivery of their cognate effectors. To test this hypothesis, we developed a construct in the pMR20 vector (vgrG3-HA) to express VgrG3 fused to a hemagglutinin epitope at its C terminus (VgrG3-HA) for coimmunoprecipitation assays (Co-IP) using a monoclonal anti-HA antibody (Fig. 5). After introduction of this construct into the ΔvgrG3 mutant, we confirmed by immunoblotting that the VgrG3-HA protein was expressed (Fig. 5A) and able to complement the competitive deficiency of this mutant strain against P. aeruginosa ATCC 27853 (Fig. 5B). Co-IP assays were performed from the ΔvgrG3(vgrG3-HA) strain and the WT(pMR20) strain (a negative control), and the immunoprecipitated proteins were identified by mass spectrometry (Table S4). Excluding many abundant cellular proteins detected in the Co-IPs from both strains, such as ribosomal proteins (Table S4), six proteins were detected specifically from the ΔvgrG3(vgrG3-HA) strain (Hcp, VgrG1, VgrG2, VgrG3, VgrG5, and CV_2846) (Fig. 5C). Therefore, the Co-IP data showed that VgrG3 interacts with VgrG1, VgrG2, and VgrG5 in vivo.
FIG 5.
VgrG3 interacts directly with other VgrGs in vivo. (A) Confirmation of VgrG3-HA expression. Western blotting with a monoclonal anti-HA antibody revealed VgrG3-HA expression (predicted molecular weight of 110 kDa) in the ΔvgrG3 strain harboring a vector expressing the construct vgrG3-HA. (B) VgrG3-HA functionally complements the ΔvgrG3 mutant strain. Interbacterial competition of the C. violaceum indicated strains against P. aeruginosa ATCC 27853. Competitive fitness was rescued in the ΔvgrG3(vgrG3-HA) strain. Bars indicate means and SD. Statistical analyses were performed using one-way ANOVA with Tukey’s post hoc test. Asterisks indicate variations in relation to the WT. ****, P < 0.0001; **, P < 0.01. (C) Several VgrGs coimmunoprecipitated with VgrG3-HA. Proteins identified after mass spectrometry of coimmunoprecipitation with VgrG3-HA. As a control, Co-IP was also performed with the WT(pMR20) strain. Many abundant cellular proteins detected in the Co-IP from both strains (Table S4) may represent contaminants.
Regulation of the C. violaceum T6SS is growth phase dependent and affected by CviR.
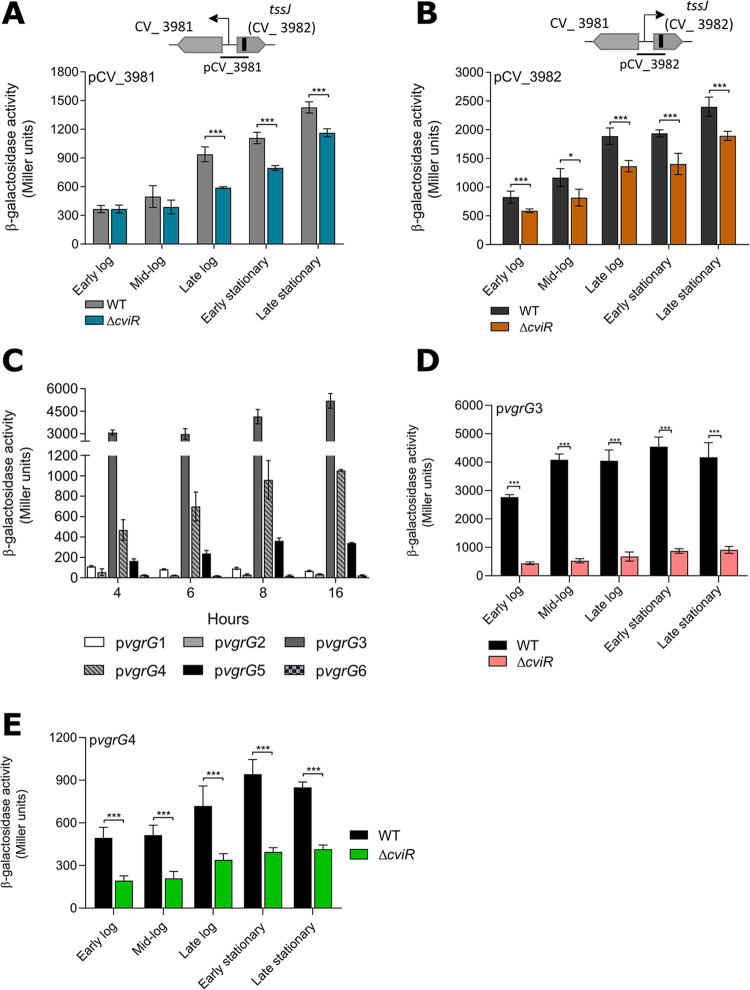
It has been demonstrated in other bacteria that the T6SS is regulated by QS (29, 32, 34, 35). The HSL-based QS circuit CviI/CviR controls many processes in C. violaceum (48–51). Considering the presence of three predicted CviR binding sites associated with T6SS genes in C. violaceum (49) (Fig. 1A), we hypothesize that T6SS expression is under QS regulation. To test this hypothesis, we measured the activity of T6SS promoters along growth curves by β-galactosidase activity assays using transcriptional reporter fusions (Fig. 6). The expression levels of the two divergent promoters that drive transcription of the two operons in the main T6SS cluster (Fig. 1A) increased throughout the growth curve in the WT strain but to a lesser extent in the ΔcviR strain, indicating that CviR activates these promoters mainly at high cell density (Fig. 6A and B). With respect to the six vgrG genes, the regions upstream of vgrG1, vgrG2, and vgrG6 showed no promoter activity regardless of the growth phase, while the promoters of vgrG3, vgrG4, and, at lower levels, vgrG5 were active and showed a slight tendency to be more expressed at high cell density than at low cell density (Fig. 6C). The absence of activity for the vgrG1 and vgrG2 fusions and the intraoperonic locations of vgrG1 and vgrG2 suggest that these genes lack their own promoters and are transcribed from the promoters of the two divergent operons. Detailed analysis of the two more active vgrG promoters, vgrG3 and vgrG4, revealed that their expression levels were markedly decreased in the ΔcviR mutant strain at all points along the growth curve (Fig. 6D and E), indicating a full dependence on CviR. Overall, these data show that the QS regulator CviR exerts transcriptional activation on many T6SS genes in C. violaceum.
FIG 6.
The promoter activity of T6SS genes increases at high cell density and is activated by the QS regulator CviR in C. violaceum. β-Galactosidase activity assays of the indicated promoters were performed along the indicated time points of the growth curve. The strains (WT or ΔcviR) harboring the lacZ fusions were cultivated in liquid LB from an OD600 of 0.01 until the indicated growth phases: early log phase, OD600 ~ 0.5; mid-log phase, OD600 ~ 1.0; late log phase, OD600 ~ 3.0; early stationary phase, OD600 ~ 5.0 to 6.0 (an 8-h culture); and late stationary phase, an overnight culture. (A and B) Promoters pCV_3981 and pCV_3982, driving the expression of the two divergent operons of the main T6SS cluster. (C) Promoters of the six vgrG genes along the growth curve. (D and E) Effects of CviR on the promoter activity levels of vgrG3 and vgrG4. Bars show means and SD. *, P < 0.05; ***, P < 0.001 (unpaired Student’s t test).
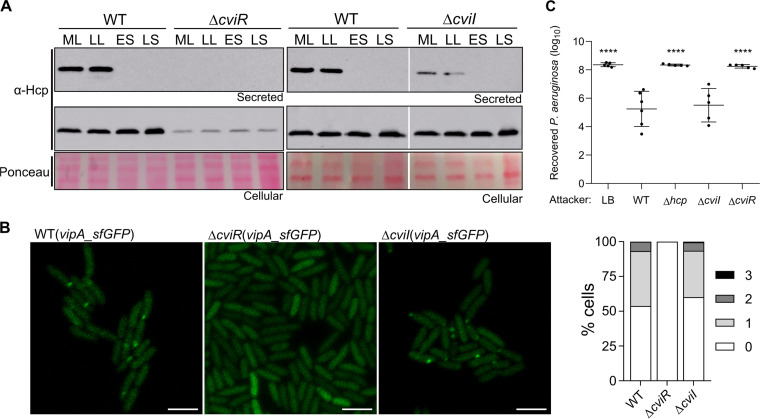
Considering the CviR-dependent transcriptional regulation of T6SS genes, we tested the effects of CviR and CviI on T6SS functionality (Fig. 7). To determine whether T6SS activity was dependent on cell density, Hcp expression and secretion throughout the growth curve were verified by immunoblotting (Fig. 7A). In the WT strain, although the Hcp protein was highly expressed throughout the growth curve (cellular fraction), its secretion occurred at low but not at high cell density (Fig. 7A), suggesting that C. violaceum inhibits T6SS activity at high cell density by a posttranslational mechanism. Surprisingly, Hcp expression and secretion were markedly impaired in the ΔcviR strain, independent of the growth phase, while for the ΔcviI strain, Hcp expression was unaffected but its secretion was decreased (Fig. 7A). Furthermore, in fluorescence microscopy assays with VipA_sfGFP, the GFP foci were abolished in the ΔcviR strain but almost unaffected in the ΔcviI strain (Fig. 7B). Similarly, in competition assays against P. aeruginosa ATCC 27853, the ΔcviR mutant showed a complete loss of competitive capacity, as observed for the Δhcp mutant, while the ΔcviI mutant killed P. aeruginosa as efficiently as the WT strain (Fig. 7C). Altogether, these results indicate that C. violaceum depends on CviR but not on CviI for the assembly of a functional T6SS, suggesting that the QS system CviI/CviR regulates the T6SS by a noncanonical mechanism.
FIG 7.
C. violaceum depends on CviR but not on CviI for the assembly of a functional T6SS. (A) Effects of cell density, CviR, and CviI on Hcp production and secretion. Western blot of Hcp in cellular and secreted fractions of WT, ΔcviR, and ΔcviI strains at different points of the growth curve: ML, mid-log phase (OD600 ~ 1.0); LL, late log phase (OD600 ~ 3.0); ES, early stationary phase (OD600 ~ 5.0); LS, late stationary phase (an overnight culture with OD600 ranging from 7.0 to 8.0). (B) Fluorescence microscopy showing T6SS sheath assembly through the formation of VipA_sfGFP foci. Bars = 3 μm. The graph shows the average number of foci per cell. (C) Recovery of P. aeruginosa ATCC 27853 after 4 h in coculture with the indicated C. violaceum strains. Bars indicate means and SD. Statistical analyses were performed using one-way ANOVA with Tukey’s post hoc test. ****, P < 0.0001 relative to the WT.
DISCUSSION
The T6SS has been characterized in detail in some model bacteria. Studying this system in several different bacterial species allows us to gain novel insights regarding its function and regulation and find new families of antibacterial and anti-eukaryotic T6SS effectors (8, 17, 21). In this work, we characterized the function and regulation of the T6SS of C. violaceum, an environmental pathogenic bacterium. Our results show that the single T6SS of C. violaceum is active, is regulated by QS, and has a major role in contact-dependent interbacterial competition. These findings add the T6SS to the already-known armamentarium used by C. violaceum to antagonize other bacteria, which includes diffusible molecules and violacein delivered into OMVs (38, 40, 42, 43, 51). Some of the putative effectors identified in T6SS-associated clusters are members of Tle1 and Tle2 phospholipases, which can act as anti-eukaryotic or antibacterial effectors (8, 54). However, given that the T6SS mutants showed no virulence attenuation and the other predicted T6SS effectors seem to be antibacterial toxins, it is tempting to speculate that C. violaceum relies on its Cpi-1/1a T3SS but not on its T6SS to inject effectors that affect key functions in host mammalian cells (36, 39).
The presence of six VgrG proteins in C. violaceum showing high sequence identity and no toxic domains gave us the opportunity to study the functional redundancy of VgrGs for T6SS function. These proteins are encoded by vgrG genes located in the main T6SS cluster and four vgrG islands and surrounded by putative T6SS effector genes (encoding phospholipases, Rhs-repeat-containing proteins, and a putative peptidase/lysozyme-like enzyme), a genomic organization commonly found in other bacteria (25, 28, 54, 60). Moreover, all six VgrGs presented a similar domain organization: a core gp27-gp5 portion followed by an additional DUF2345 C-terminal domain, which has been found in many VgrGs (27) and contributes to the VgrG/effector interaction (23). VgrG3 and VgrG4 also presented an additional C-terminal unstructured region after the DUF2345 domain. However, we provide experimental evidence, using single and multiple vgrG mutant strains, that in C. violaceum the VgrGs showed little functional redundancy, with the VgrG3 protein playing a more important role in relation to the other five VgrGs to the T6SS function: (i) among the single vgrG mutants, the ΔvgrG3 strain showed a marked decrease in Hcp secretion and loss of competitive fitness against P. aeruginosa; (ii) only vgrG3 among the six vgrG genes fully restored the ability of the ΔvgrG1-6 mutant to secrete Hcp and to kill P. aeruginosa; (iii) the vgrG3 promoter was the most active among the vgrG promoters and showed full dependence on the QS regulator CviR; and (iv) VgrG3 interacted in vivo with other VgrGs (VgrG1, VgrG2, and VgrG5), most likely favoring the assembly of VgrG heterotrimers to allow the secretion of effectors associated with each VgrG, as described for the three VgrGs of V. cholerae (61). Different results, indicative of greater functional redundancy, have been described in other bacteria harboring multiple VgrGs. For instance, in Burkholderia cenocepacia (ten VgrGs), Serratia marcescens (two VgrGs), and Agrobacterium tumefaciens (four VgrGs), the individual deletion of any vgrG had no impact on Hcp secretion (25, 27, 60). Conversely, in A. hydrophila and V. cholerae (each with three VgrGs), the individual deletion of specific vgrG genes abolished Hcp secretion (61, 62). As expected, single-copy T6SS core components such as Hcp, VipA, and ClpV were absolutely required for the T6SS functionality in C. violaceum.
Our data indicate that the C. violaceum T6SS is an offensive system because it is active even in laboratory conditions (as indicated by the high number of sfGFP foci and the easy detection of Hcp in supernatants of log-phase liquid cultures) and efficiently killed many Gram-negative bacteria regardless of whether they had a T6SS (such as E. coli ATCC 25922) for a counterattack. This offensive pattern, described in other bacteria such as S. marcescens (63), differs from the defensive strategy employed by P. aeruginosa, where the T6SS assembly and firing occur only in response to an incoming attack and involves complex posttranslational regulation by the threonine phosphorylation pathway (TPP) (17, 64). Although the C. violaceum T6SS can be considered an offensive system, our results indicate that Hcp secretion ceased at high cell density despite the normal production of Hcp and the maximum expression of the T6SS promoters under this condition via activation by the QS regulator CviR. Similarly, a T6SS2 of V. fluvialis was regulated by QS (35), and Hcp secretion was repressed at high cell density (65). C. violaceum seems to lack the TPP pathway but harbors TagF encoded in the main T6SS cluster; TagF is a protein that can block T6SS activity independent of TPP in other bacteria (17, 63, 66). We speculate that C. violaceum posttranslationally blocks T6SS firing events in nonpolymicrobial cultures at high cell density by activating tagF together with the other T6SS genes from a CviR-dependent promoter. This strategy ensures that the T6SS components will be produced but that the T6SS firing will be blocked, avoiding physical damage and activation of stress responses in nearby kin cells inhabiting densely populated cultures. These adventitious events have been described in recipient cells targeted by T6SSs at least during interspecies competition (67, 68).
Many processes regulated by QS in C. violaceum depend on both CviI and CviR, the sole QS circuit described in this bacterium (48, 49, 51). Our results indicate a clear role of CviR but not CviI in the T6SS function, suggesting that CviR could act independently of the HSL autoinducers produced by CviI. Indeed, a recent work found six new HSL molecules in C. violaceum ATCC 12472 in addition to the already known C10-HSL and 3-OH-C10-HSL, but it remains unknown how they are synthesized (51). There is a possibility that one of these newly identified HSLs may interact with CviR, activating this regulator and its target genes, which would explain our distinct regulatory and phenotypic outcomes observed for CviR and CviI. Future works investigating the global impact of CviI and CviR on the C. violaceum transcriptome will contribute to revealing whether this QS system operates together with other QS circuits to control the T6SS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table S1. E. coli and C. violaceum strains were grown with agitation at 37°C in LB medium or on LB agar plates (LB supplemented with 15 g/L agar). When necessary, media were supplemented with antibiotics at the following concentrations: for C. violaceum, gentamicin at 40 μg/mL, tetracycline 5 to 10 μg/mL, and kanamycin at 50 μg/mL, and for E. coli, gentamicin at 20 μg/mL, tetracycline at 12 μg/mL, kanamycin at 50 μg/mL, and ampicillin at 100 μg/mL.
Construction of the C. violaceum mutant and complemented strains.
Strains harboring in-frame deletions of the selected T6SS genes were constructed by an allelic exchange protocol, as described previously (42, 69, 70). Briefly, the flanking regions of the selected genes were amplified by PCR with specific primers (Table S2) and cloned into the suicide vector pNPTS138. After electroporation into E. coli S17-1, the constructs were transferred by conjugation to C. violaceum ATCC 12472. Transconjugants were selected on LB agar with kanamycin at 50 μg/mL and ampicillin at 100 μg/mL. Isolated colonies were grown in liquid LB followed by counterselection on LB plates with 16% sucrose. For the multiple vgrG mutants, the procedure was repeated sequentially, from the ΔvgrG1 mutant to the final ΔvgrG1-6 sextuple mutant. The null mutant strains were confirmed by PCR using specific primers (Table S2). To complement the T6SS mutants, the respective T6SS genes were amplified by PCR with specific primers (Table S2) and cloned into the replicative noninducible plasmid pMR20 or pSEVA. Genes were cloned with their native promoter sequences or with approximately 50 bp upstream of ATG in the case of genes without intergenic regions. The resulting constructs were transferred to the respective C. violaceum mutant strain by conjugation. Similarly, each vgrG gene cloned into pMR20 was also introduced into the ΔvgrG1-6 sextuple mutant.
Interbacterial competition.
For an initial screening, attacker C. violaceum strains were competed against several prey bacteria (Table S3) and analyzed for a purple output by visual inspection (indicating C. violaceum overgrowth). Attacker and prey bacteria were grown overnight on LB agar plates, resuspended in LB at an optical density at 600 nm (OD600) of 1, and mixed at a ratio of 1:1. The mixtures (5 μL) were spotted on LB agar plates and incubated at 37°C overnight prior to image acquisition. For the quantitative competition assay of C. violaceum strains against P. aeruginosa ATCC 27853, bacterial strains were grown overnight on LB agar, with antibiotics added for the C. violaceum strains harboring plasmids. Bacteria were resuspended in liquid LB and normalized to an OD600 of 5. C. violaceum WT and T6SS mutant strains (attacker) or LB (control) were mixed with the target bacterium at a proportion of 5:1. The mixtures (10 μL) were spotted on LB agar to allow contact-dependent competition. After 4 h at 37°C, the bacterial spots were recovered and resuspended in 1 mL of LB. Serial dilutions were plated on LB agar with kanamycin to allow P. aeruginosa CFU counting.
Mouse virulence assay.
C. violaceum virulence in mice was assayed using an intraperitoneal (i.p.) infection model, as previously described (59). Briefly, 6-week-old BALB/c female mice were challenged with i.p. injections of 106 CFU of C. violaceum WT and mutant strains. To check the injected dose, serially diluted bacterial samples were plated on LB agar for CFU counting. The survival of infected mice was monitored for 5 days. All experiments using mice were performed in the Animal Facility of the Faculdade de Medicina de Ribeirão Preto (FMRP-USP) according to the ARRIVE guidelines, and protocol number 037/2018 was approved by the local animal ethics committee (CEUA, FMRP-USP). The CEUA follows the Ethical Principles in Animal Research adopted by the National Council for the Control of Animal Experimentation (CONCEA).
β-Galactosidase activity assay.
The promoters of the two large operons located in the main T6SS cluster (divergent regions between CV_3981 and CV_3982, tssJ) and the promoter regions of each vgrG were amplified by PCR (Table S2), cloned into the pGEM-T Easy plasmid (Promega), and subcloned into the pRKlacZ290 vector to generate transcriptional fusions to the promoterless lacZ gene. The resulting constructs were introduced into C. violaceum wild-type and ΔcviR strains. To assess the expression of these promoters according to the cell density, the strains were diluted to an OD600 of 0.01 and grown in LB, and samples were harvested through the growth curve. The β-galactosidase assay was performed with ONPG (o-nitrophenyl-β-d-galactoside) as the substrate, and the results were calculated as Miller units normalized by OD600 (71).
Fluorescence microscopy.
T6SS firing events were detected by fluorescence microscopy using a VipA_sfGFP fusion separated by a 3XAla 3XGly linker, as previously described (18). Briefly, the vipA (without its stop codon) and sfGFP genes were amplified by PCR and cloned into the pJN105 vector to express a VipA_sfGFP fusion protein from an arabinose-inducible promoter. This construct was introduced by conjugation into the C. violaceum strains. The strains were grown under shaking in liquid LB at 37°C until mid-log phase, when l-arabinose was added at a final concentration of 0.05% for 30 min prior to image acquisition. Cells were spotted in a thin pad of 1% agarose in phosphate-buffered saline (PBS) and imaged in a Carl Zeiss LSM 780 microscope.
Hcp cloning, expression, and purification.
The hcp (CV_3977) coding region was amplified by PCR (Table S2) and cloned into the expression vector pET-15b. The construct was transformed into E. coli BL21(DE3). Recombinant His-tagged Hcp was induced by the addition of 1 mM isopropyl-d-thiogalactopyranoside (IPTG) to E. coli log-phase cultures. After 2 h, the cells were harvested and lysed by sonication. His-Hcp was purified from the soluble fraction by nitrilotriacetic acid (NTA)-resin chromatography (Qiagen). Purified protein was desalted with a PD-10 desalting column (GE Healthcare) in storage buffer (20 mM Tris-Cl [pH 7.4], 100 mM NaCl, 0.1 mM EDTA, and 5% [vol/vol] glycerol). Samples from the induction and purification steps were evaluated using 15% SDS-PAGE.
Hcp detection in cellular and secreted fractions by Western blotting.
C. violaceum strains were diluted to an OD600 of 0.01 and grown in liquid LB at 37°C until mid-log phase or the indicated time points throughout the growth curve. Two samples of each culture were harvested to obtain cellular and secreted fractions. For the cellular fractions, the samples were pelleted by centrifugation at 4,000 × g for 5 min. The secreted fractions were obtained by centrifugation at 4,000 × g for 15 min followed by filtration with 0.22-μm membranes to obtain cell-free supernatants. The pellets and supernatants were resuspended in SDS sample buffer to adjust the volumes according to the OD600 values of the cultures. The proteins were resolved by 15% SDS-PAGE and transferred to a nitrocellulose membrane. Western blotting was carried out with a Protein Detector LumiGLO Western blotting (KPL) kit according to the manufacturer’s instructions. The polyclonal anti-Hcp antibody was used at a dilution of 1:10,000 for the cellular fractions and 1:2,000 for the supernatant fractions. Sample loading was assessed by membrane staining with 0.1% Ponceau S. The polyclonal anti-Hcp antibody was developed after immunization of 6-week-old female BALB/c mice with three subcutaneous injections of the purified His-Hcp protein emulsified in Freund adjuvant (Sigma).
VgrG3-HA Co-IP and MS.
To generate a VgrG3 protein fused at its C terminus to a hemagglutinin tag (VgrG3-HA), the vgrG3 gene containing its promoter and coding region but without its stop codon was amplified by PCR with specific primers. The long reverse primer contained the sequences for a duplicated HA tag and a stop codon (Table S2). The PCR-amplified fragment was digested with specific restriction enzymes and cloned into pMR20 (vgrG3-HA). This construct was introduced into the ΔvgrG3 mutant, generating the ΔvgrG3(vgrG3-HA) strain. For the Co-IP assays, the WT(pMR20) strain as a control and the ΔvgrG3(vgrG3-HA) strain were diluted in LB to an OD600 of 0.01 and cultured until an OD600 of 0.8 was reached. The cultures (80 mL) were harvested by centrifugation, and the cells were resuspended in 7 mL of lysis buffer (150 mM NaCl, 10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 1% Triton X-100, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) prior to sonication. Lysed cells were treated with 1 mL 10% streptomycin for 20 min under agitation prior to centrifugation and filtration with a 0.45-μm filter. Proteins from the supernatant at 2 mg/mL were used for the Co-IP assay using protein G magnetic beads (New England Biolabs) and a mouse monoclonal anti-HA antibody (Sigma). The anti-HA antibody was cross-linked to the protein G magnetic beads using triethanolamine and ethanolamine. The protein samples were precleaned by incubation with protein G magnetic beads. For Co-IP, cleaned samples (400 μg) were incubated with protein G magnetic beads containing cross-linked anti-HA antibody for 1 h at 4°C. The beads were washed three times with lysis buffer, and the proteins were eluted with elution buffer (0.1 M glycine, pH 2.5). For mass spectrometry (MS), the eluted proteins were trypsinized, and the resulting peptides were analyzed by liquid chromatography-tandem MS (LC-MS/MS) at the Life Sciences Core Facility (LaCTAD) of the State University of Campinas (UNICAMP).
ACKNOWLEDGMENTS
This research was supported by grants from the São Paulo Research Foundation (FAPESP; grants 2020/00259-8 and 2021/06894-0) and Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da FMRP-USP (FAEPA). During the course of this work, J.A.A. (grant 2018/03979-1) and M.P.-M. (grant 2018/12461-6) were supported by FAPESP fellowships, and F.C.L. was granted a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
We thank the staff of the Life Sciences Core Facility (LaCTAD) of the State University of Campinas (UNICAMP) for the proteomics analysis and Roberta Rosales for technical support in microscopy.
J.A.A., F.C.L., and J.F.D.S.N. planned the experiments; J.F.D.S.N. and J.A.A. wrote the manuscript; J.A.A., F.C.L., and M.P.-.M. performed the experimental work; J.A.A., F.C.L., and J.F.D.S.N. analyzed and interpreted the data; J.F.D.S.N. participated in study coordination and funding acquisition. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
José F. da Silva Neto, Email: jfsneto@usp.br.
Eric Cascales, Centre national de la recherche scientifique, Aix-Marseille Université.
REFERENCES
- 1.Ghoul M, Mitri S. 2016. The ecology and evolution of microbial competition. Trends Microbiol 24:833–845. doi: 10.1016/j.tim.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Stubbendieck RM, Straight PD. 2016. Multifaceted interfaces of bacterial competition. J Bacteriol 198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 4.Raffatellu M. 2018. Learning from bacterial competition in the host to develop antimicrobials. Nat Med 24:1097–1103. doi: 10.1038/s41591-018-0145-0. [DOI] [PubMed] [Google Scholar]
- 5.Chassaing B, Cascales E. 2018. Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol 26:329–338. doi: 10.1016/j.tim.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Alcoforado Diniz J, Liu YC, Coulthurst SJ. 2015. Molecular weaponry: diverse effectors delivered by the type VI secretion system. Cell Microbiol 17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hachani A, Wood TE, Filloux A. 2016. Type VI secretion and anti-host effectors. Curr Opin Microbiol 29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Monjarás Feria J, Valvano MA. 2020. An overview of anti-eukaryotic T6SS effectors. Front Cell Infect Microbiol 10:584751. doi: 10.3389/fcimb.2020.584751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallegos-Monterrosa R, Coulthurst SJ. 2021. The ecological impact of a bacterial weapon: microbial interactions and the type VI secretion system. FEMS Microbiol Rev 45:fuab033. doi: 10.1093/femsre/fuab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer-Santos E, Lima LDP, Ceseti LM, Ratagami CY, de Santana ES, da Silva AM, Farah CS, Alvarez-Martinez CE. 2018. Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF σ factor and cognate Ser/Thr kinase. Environ Microbiol 20:1562–1575. doi: 10.1111/1462-2920.14085. [DOI] [PubMed] [Google Scholar]
- 11.Trunk K, Peltier J, Liu YC, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ. 2018. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3:920–931. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Liu H, Zhang Y, Shen X. 2021. Roles of type VI secretion system in transport of metal ions. Front Microbiol 12:756136. doi: 10.3389/fmicb.2021.756136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unni R, Pintor KL, Diepold A, Unterweger D. 2022. Presence and absence of type VI secretion systems in bacteria. Microbiology (Reading) 168. doi: 10.1099/mic.0.001151. [DOI] [PubMed] [Google Scholar]
- 14.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen VS, Douzi B, Durand E, Roussel A, Cascales E, Cambillau C. 2018. Towards a complete structural deciphering of type VI secretion system. Curr Opin Struct Biol 49:77–84. doi: 10.1016/j.sbi.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Cherrak Y, Flaugnatti N, Durand E, Journet L, Cascales E. 2019. Structure and activity of the type VI secretion system. Microbiol Spectr 7:PSIB-0031-2019. doi: 10.1128/microbiolspec.PSIB-0031-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Brodmann M, Basler M. 2019. Assembly and subcellular localization of bacterial type VI secretion systems. Annu Rev Microbiol 73:621–638. doi: 10.1146/annurev-micro-020518-115420. [DOI] [PubMed] [Google Scholar]
- 18.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brackmann M, Nazarov S, Wang J, Basler M. 2017. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol 27:623–632. doi: 10.1016/j.tcb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez RE, Gallegos-Monterrosa R, Coulthurst SJ. 2020. Type VI secretion system effector proteins: effective weapons for bacterial competitiveness. Cell Microbiol 22:e13241. doi: 10.1111/cmi.13241. [DOI] [PubMed] [Google Scholar]
- 22.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaugnatti N, Le TTH, Canaan S, Aschtgen MS, Nguyen VS, Blangy S, Kellenberger C, Roussel A, Cambillau C, Cascales E, Journet L. 2016. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol 99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 24.Renault MG, Zamarreno Beas J, Douzi B, Chabalier M, Zoued A, Brunet YR, Cambillau C, Journet L, Cascales E. 2018. The gp27-like hub of VgrG serves as adaptor to promote Hcp tube assembly. J Mol Biol 430:3143–3156. doi: 10.1016/j.jmb.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Santos MNM, Cho ST, Wu CF, Chang CJ, Kuo CH, Lai EM. 2019. Redundancy and specificity of type VI secretion vgrG loci in antibacterial activity of Agrobacterium tumefaciens 1D1609 strain. Front Microbiol 10:3004. doi: 10.3389/fmicb.2019.03004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jové D, Filloux A. 2011. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem 286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aubert DF, Hu S, Valvano MA. 2015. Quantification of type VI secretion system activity in macrophages infected with Burkholderia cenocepacia. Microbiology (Reading) 161:2161–2173. doi: 10.1099/mic.0.000174. [DOI] [PubMed] [Google Scholar]
- 28.Zong B, Zhang Y, Wang X, Liu M, Zhang T, Zhu Y, Zheng Y, Hu L, Li P, Chen H, Tan C. 2019. Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 10:118–132. doi: 10.1080/21505594.2019.1573491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata ST, Bachmann V, Pukatzki S. 2013. Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol 62:663–676. doi: 10.1099/jmm.0.053983-0. [DOI] [PubMed] [Google Scholar]
- 30.Yu KW, Xue P, Fu Y, Yang L. 2021. T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int J Mol Sci 22:478. doi: 10.3390/ijms22020478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. 2012. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci USA 107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majerczyk C, Schneider E, Greenberg EP. 2016. Quorum sensing control of type VI secretion factors restricts the proliferation of quorum-sensing mutants. Elife 5:e14712. doi: 10.7554/eLife.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khajanchi BK, Sha J, Kozlova EV, Erova TE, Suarez G, Sierra JC, Popov VL, Horneman AJ, Chopra AK. 2009. N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology (Reading) 155:3518–3531. doi: 10.1099/mic.0.031575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Pan J, Gao H, Han Y, Zhang A, Huang Y, Liu P, Kan B, Liang W. 2021. CqsA/LuxS-HapR quorum sensing circuit modulates type VI secretion system VflT6SS2 in Vibrio fluvialis. Emerg Microbes Infect 10:589–601. doi: 10.1080/22221751.2021.1902244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miki T, Akiba K, Iguchi M, Danbara H, Okada N. 2011. The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol Microbiol 80:1186–1203. doi: 10.1111/j.1365-2958.2011.07637.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang CH, Li YH. 2011. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J Chin Med Assoc 74:435–441. doi: 10.1016/j.jcma.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Batista JH, da Silva Neto JF. 2017. Chromobacterium violaceum pathogenicity: updates and insights from genome sequencing of novel Chromobacterium species. Front Microbiol 8:2213. doi: 10.3389/fmicb.2017.02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan F, Huang C, Wang X, Tan J, Cheng S, Wan M, Wang Z, Wang S, Luo S, Li A, Guo X, Feng M, Liu X, Zhu Y, Zhou Y. 2020. Threonine ADP-ribosylation of ubiquitin by a bacterial effector family blocks host ubiquitination. Mol Cell 78:641–652.E9. doi: 10.1016/j.molcel.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Durán N, Justo GZ, Durán M, Brocchi M, Cordi L, Tasic L, Castro GR, Nakazato G. 2016. Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite: a review. Biotechnol Adv 34:1030–1045. doi: 10.1016/j.biotechadv.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Cauz ACG, Carretero GPB, Saraiva GKV, Park P, Mortara L, Cuccovia IM, Brocchi M, Gueiros-Filho FJ. 2019. Violacein targets the cytoplasmic membrane of bacteria. ACS Infect Dis 5:539–549. doi: 10.1021/acsinfecdis.8b00245. [DOI] [PubMed] [Google Scholar]
- 42.Batista JH, Leal FC, Fukuda TTH, Alcoforado Diniz J, Almeida F, Pupo MT, da Silva Neto JF. 2020. Interplay between two quorum sensing-regulated pathways, violacein biosynthesis and VacJ/Yrb, dictates outer membrane vesicle biogenesis in Chromobacterium violaceum. Environ Microbiol 22:2432–2442. doi: 10.1111/1462-2920.15033. [DOI] [PubMed] [Google Scholar]
- 43.Choi SY, Lim S, Cho G, Kwon J, Mun W, Im H, Mitchell RJ. 2020. Chromobacterium violaceum delivers violacein, a hydrophobic antibiotic, to other microbes in membrane vesicles. Environ Microbiol 22:705–713. doi: 10.1111/1462-2920.14888. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ, Choi HS, Yang SY, Kim IS, Yamaguchi T, Sohng JK, Park SK, Kim JC, Lee CH, Gardener BM, Kim YC. 2014. Both extracellular chitinase and a new cyclic lipopeptide, chromobactomycin, contribute to the biocontrol activity of Chromobacterium sp. C61. Mol Plant Pathol 15:122–132. doi: 10.1111/mpp.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, Tripathi A, Mlambo G, Dimopoulos G. 2014. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog 10:e1004398. doi: 10.1371/journal.ppat.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraiva RG, Huitt-Roehl CR, Tripathi A, Cheng YQ, Bosch J, Townsend CA, Dimopoulos G. 2018. Chromobacterium spp. mediate their anti-Plasmodium activity through secretion of the histone deacetylase inhibitor romidepsin. Sci Rep 8:6176. doi: 10.1038/s41598-018-24296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Short SM, van Tol S, MacLeod HJ, Dimopoulos G. 2018. Hydrogen cyanide produced by the soil bacterium Chromobacterium sp. Panama contributes to mortality in Anopheles gambiae mosquito larvae. Sci Rep 8:8358. doi: 10.1038/s41598-018-26680-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GSAB, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology (Reading) 143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 49.Stauff DL, Bassler BL. 2011. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol 193:3871–3878. doi: 10.1128/JB.05125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans KC, Benomar S, Camuy-Vélez LA, Nasseri EB, Wang X, Neuenswander B, Chandler JR. 2018. Quorum-sensing control of antibiotic resistance stabilizes cooperation in Chromobacterium violaceum. ISME J 12:1263–1272. doi: 10.1038/s41396-018-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mion S, Carriot N, Lopez J, Plener L, Ortalo-Magné A, Chabrière E, Culioli G, Daudé D. 2021. Disrupting quorum sensing alters social interactions in Chromobacterium violaceum. NPJ Biofilms Microbiomes 7:40. doi: 10.1038/s41522-021-00211-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci USA 100:11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha EP. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unterweger D, Kostiuk B, Ötjengerdes R, Wilton A, Diaz-Satizabal L, Pukatzki S. 2015. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG. 2015. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci USA 112:9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alcoforado Diniz J, Coulthurst SJ. 2015. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Previato-Mello M, Meireles DA, Netto LES, da Silva Neto JF. 2017. Global transcriptional response to organic hydroperoxide and the role of OhrR in the control of virulence traits in Chromobacterium violaceum. Infect Immun 85:e00017-17. doi: 10.1128/IAI.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. 2016. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog 12:e1005735. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sha J, Rosenzweig JA, Kozlova EV, Wang S, Erova TE, Kirtley ML, van Lier CJ, Chopra AK. 2013. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology (Reading) 159:1120–1135. doi: 10.1099/mic.0.063495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ostrowski A, Cianfanelli FR, Porter M, Mariano G, Peltier J, Wong JJ, Swedlow JR, Trost M, Coulthurst SJ. 2018. Killing with proficiency: integrated post-translational regulation of an offensive type VI secretion system. PLoS Pathog 14:e1007230. doi: 10.1371/journal.ppat.1007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Y, Du P, Zhao M, Liu W, Du Y, Diao B, Li J, Kan B, Liang W. 2017. Functional characterization and conditional regulation of the type VI secretion system in Vibrio fluvialis. Front Microbiol 8:528. doi: 10.3389/fmicb.2017.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. 2011. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol 82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong TG, Dong S, Catalano C, Moore R, Liang X, Mekalanos JJ. 2015. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci USA 112:2181–2186. doi: 10.1073/pnas.1425007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamal F, Liang X, Manera K, Pei TT, Kim H, Lam LG, Pun A, Hersch SJ, Dong TG. 2020. Differential cellular response to translocated toxic effectors and physical penetration by the type VI secretion system. Cell Rep 31:107766. doi: 10.1016/j.celrep.2020.107766. [DOI] [PubMed] [Google Scholar]
- 69.da Silva Neto JF, Negretto CC, Netto LES. 2012. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a Cys-based redox sensor regulated by thioredoxin. PLoS One 7:e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barroso KCM, Previato-Mello M, Batista BB, Batista JH, da Silva Neto JF. 2018. EmrR-dependent upregulation of the efflux pump EmrCAB contributes to antibiotic resistance in Chromobacterium violaceum. Front Microbiol 9:2756. doi: 10.3389/fmicb.2018.02756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01576-22-s0001.pdf, PDF file, 1.0 MB (1.1MB, pdf)