Abstract
Mechanical loading on the skeleton stimulates bone formation. Although the exact mechanism underlying this process remains unknown, a growing body of evidence indicates that the Wnt signaling pathway is necessary for the skeletal response to loading. Recently, we showed that Wnts produced by osteoblast lineage cells mediate the osteo-anabolic response to tibial loading in adult mice. Here, we report that Wnt1 specifically plays a crucial role in mediating the mechano-adaptive response to loading. Independent of loading, short-term loss of Wnt1 in the Osx-lineage resulted in decreased cortical bone area in the tibias of 5-month old mice. In females, strain-matched loading enhanced periosteal bone formation in Wnt1F/F controls, but not in Wnt1F/F; OsxCreERT2 knockouts. In males, strain-matched loading increased periosteal bone formation in both control and knockout mice; however, the periosteal relative bone formation rate was 65% lower in Wnt1 knockouts versus controls. Together, these findings show that Wnt1 supports adult bone homeostasis and mediates the bone anabolic response to mechanical loading.
Keywords: Osteoblasts, Osteocytes, Bone, Osteogenesis, Mechanobiology, Wnt Signaling Pathway
Introduction
Mechanical loading on the skeleton stimulates bone formation. Although the mechanisms underlying this process are not fully defined, a growing body of evidence indicates that the Wnt signaling pathway plays a role in loading-induced bone formation. For example, ulnar loading suppresses expression of Wnt antagonist Sclerostin, and downregulation of this antagonist is in fact necessary for the bone anabolic response to loading1–2. Furthermore, mice lacking Wnt co-receptor Lrp5 exhibit a diminished bone anabolic response to loading, whereas mice with high-bone mass (HBM) mutations in Lrp5 have enhanced responses to loading3–5.
Wnt signaling initiates when one of 19 Wnt ligands (‘Wnts’) binds to Frizzled and Lrp5/6 co-receptors6. Recently, we reported that blocking Wnts secretion in the Osx-expressing cells of skeletally mature mice reduced the osteo-anabolic response to loading7. OsxCreERT2; WlsF/F; mice were tamoxifen-treated to inactivate Wntless – the Wnt-specific transport receptor responsible for shuttling Wnt ligands to the cell surface for exocytosis8 – thereby blocking the secretion of all Wnts from osteoblasts and osteocytes. We showed that the periosteal response to tibial loading was 65% lower in Wls knockouts relative to WlsF/F controls, indicating that Wnts produced by osteoblast-lineage cells are important mediators of loading-induced bone formation7.
Gene expression studies have aimed to identify the key factors that mediate loading-induced bone formation. In our lab and others, RNASeq analysis of cortical bone samples showed that Wnt1 and Wnt7b were differentially expressed in loaded vs non-loaded bones, suggesting that these Wnt ligands in particular may be necessary for the skeletal response to loading9–11. Furthermore, age-related declines in the skeletal response to loading are associated with dampened upregulation of Wnt1 and Wnt7b in the bone, while in vivo overexpression of Wnt1 and Wnt7b in the osteo-lineage causes increased bone mass12–14. Based on these studies, we hypothesized that loading-induced upregulation of Wnt1 and Wnt7b in osteoblasts is necessary for the bone anabolic response to loading. In this communication, we report that blocking Wnt1 upregulation in the osteoblast-lineage via Wnt1 deletion in Osx-expressing cells blunts the bone anabolic response to skeletal loading in mice.
Methods
Mice
Maintenance and genotyping.
All animal studies were approved by the Washington University IACUC. Mice were derived from breeders originally gifted by Drs. Henry Kronenberg (OsxCreERT215), Brendan Lee (Wnt1 floxed13), or obtained from Jackson Labs (Wnt7bC3 floxed; B6; 129X1-Wnt7btm2Amc/J; stock number 008467). Experimental mice were generated by breeding Wnt1F/F dams to OsxCreERT2; Wnt1F/F sires (for Wnt1 single conditional mutants), or Wnt1F/F; Wnt7bF/F dams to OsxCreERT2; Wnt1F/F; Wnt7F/F sires (for Wnt1/7b double conditional mutants). Mice were weaned at 21 days and housed up to five per cage with ad libitum access to normal chow and water. Mice were genotyped by Transnetyx (Transnetyx, USA) using tail snip DNA with probes for wild-type Wnt1 (Wnt1-1 WT), conditional Wnt1 (L1L2-Bact-P MD), wild-type Wnt7b (Wnt7b-2 WT), conditional Wnt7b (Wnt7b-2 FL), and OsxCreERT2 (Cre). Mice were euthanized by CO2 asphyxiation.
Tamoxifen induction.
Gene deletion was induced in 20 to 22 week old mice. Tamoxifen was dissolved in corn oil to a final concentration of 10mg/ml, and delivered by oral gavage for 5 consecutive days at a dose of 50mg/kg/day. The first day of dosing was “day 1” in the experimental timeline. Tamoxifen-treated Wnt1F/F and Wnt1F/F; Wnt7bF/F (cre-negative) mice served as genotype controls for Wnt1 and Wnt1/7b knockout experiments, respectively.
Knockdown validation
Tissue-specific gene deletion was confirmed by DNA recombination PCR, and RNA knockdown efficiency was evaluated by RT-qPCR (see Gene Expression below for RT-qPCR methods). Validation experiments were performed 3 weeks after the first tamoxifen dose, on day 22. Mice used for validation experiments were not subjected to tibial loading.
Recombination PCR.
In preparation for DNA recombination analysis, skeletal (cortical bone from tibia) and extra-skeletal tissues were collected from tamoxifen-treated control and knockout mice. Tissues were crushed manually with mortar and pestle, then digested overnight in Proteinase K. Genomic DNA was isolated using a commercially available kit from Qiagen (DNeasy). 5’-CTGCCCAGCTGGGTTTCTACTACG-3’ (FOR) and 5’-ACCAGCTGCAGACTCTTGGAATCCG-3’ (REV) were used to amplify wild-type Wnt1 DNA from the intact/conditional allele (800bp). 5’-AGTGAGCTAGTACGGGGTCC-3’ (FOR) and 5’-AGGACCATGAACTGATGGCG-3’ (REV) were used to amplify the modified/recombined Wnt1 locus, which produces a Wnt1 null allele (368bp). 5’-TGACAGAGGATGGGGAGAAG-3’ (FOR) and 5’-GGTCTTTCCAAGGGTGGTCT-3’ (REV) were used to amplify wild-type Wnt7b DNA from the intact locus. 5’-GAGGAAGTCAGGCAGGTGTC-3’ (FOR) and 5’-TATCCCACCGATACGCAAAC-3’ (REV) were used to amplify the modified/mutant Wnt7b allele as described previously16. PCR reactions were run separately, then combined and resolved by electrophoresis on a 2.5% agarose gel.
Knockdown efficiency qPCR.
Tibias were dissected and processed for knockdown efficiency testing as described under Gene Expression (RT-qPCR). PrimeTime assay Mm.PT.58.30187381 (Integrated DNA Technologies, USA) was used to assay Wnt1 expression in the cortical bone. These primers amplify a region spanning exons 2 and 3, which is deleted in knockout tissues. Deletion of exons 2-3 produces a Wnt1 null allele; thus, qPCR data reflects the abundance of wild-type Wnt1 mRNA in the bone. Wnt7b was assayed using primers spanning exons 2-3: FOR: 3’-CGGGCAAGAACTCCGAGTAG-5’; REV: 3’-GCGACGAGAAAAGTCGATGC-5’. Exon 3 is deleted in the Wnt7bC3 conditional mouse.
Tibial MicroCT
Sequential in vivo μCT scans of the tibia were used to evaluate the effects of short-term Wnt1 and Wnt1/7b deletion on cortical bone morphometry. Tibias were scanned at the mid-diaphysis 24-48 hours before the start of tamoxifen induction (day 0), followed by a second scan approximately 3 weeks later on day 22. Under anesthesia (1-3% isoflurane), the left tibia was positioned for scanning, and a 2.1 mm region centered 5 mm proximal to the distal TFJ was scanned on a vivaCT40 with settings optimized for bone (70 kVp, 115 μA, 300 ms integration, 0.4 sigma, 1 support, 1000 projections, 10.5 μm/voxel) (Scanco Medical, Switzerland). Post-scans were completed on mice in situ after sacrifice. For all scans a 1.05 mm region in the center of the scan was contoured in the Scanco software and evaluated for cortical bone area, total area, cortical thickness, polar moment of inertia, and tissue mineral density.
Tibial loading
Starting on day 22 after tamoxifen induction, mice were subjected to an in vivo tibial loading regimen to stimulate periosteal lamellar bone formation as previously described17–18. Briefly, the right tibias of isoflurane-anesthetized mice were positioned vertically between two fixtures and an ElectroPuls E1000 instrument (Instron, USA) was used to apply cyclic axial compression (60 cycles/day, 4Hz) on the right tibia for 5 consecutive days. Contralateral left tibias served as non-loaded controls.
Strain gage analysis was performed on mice that were sacrificed 3 weeks after tamoxifen induction (day 22). The right tibia was exposed and cleaned of soft tissue before a single-element strain gage was glued to the antero-medial surface 5 mm proximal to the distal tibiofibular junction (TFJ). The tibia was axially loaded on a material testing machine (Dynamite 8841) at a peak-to-peak force ranging from −2 N to −8 N for 12 cycles at 4 Hz. LabView data acquisition software was used to collect data, which was analyzed by linear regression to calculate the force-strain equation for each sex-genotype group (Fig S2).
Strain-matched and force-matched loading.
Force-strain equations were used to define the loading force necessary to engender a target strain of approximately −3000 μƐ in each group. In males, this required a loading force of −10.5 N for both genotypes. In females, strain-matched loading required −9.2 N and −5.4 N in control and knockout mice, respectively. These forces were found to induce a robust lamellar bone formation response in Wnt1F/F males and females (Fig S3). An additional cohort of Wnt1 knockout females were loaded to −9.2 N to provide a force-matched comparison with control females. In force-matched knockout females, −9.2 N was estimated to engender a peak strain of −5600 μƐ (Fig S2).
Dynamic histomorphometry
Bone formation was analyzed by dynamic histomorphometry. As outlined in Figures 4 and 5, mice were loaded for 5 days, then calcein (10 mg/kg; Sigma-Aldrich, St. Louis, MO, USA) and alizarin (30 mg/kg; Sigma-Aldrich) were given by IP injection on days 26 and 31, respectively, to label sites of active mineralization. Mice were sacrificed on day 33. Bilateral tibias (loaded and non-loaded) were de-hydrated through ethanol and embedded in MMA plastic as described previously19. Transverse sections from the mid-diaphysis (5mm proximal to the TFJ) were used to capture 20 μm z-stack images on a Leica confocal microscope (Leica DMi8) for analysis. Bioquant analysis software was used to calculate periosteal (Ps) and endocortical (Ec) bone formation indices, including % mineralizing surface (MS/BS), mineral apposition rate (MAR), and bone formation rate (BFR/BS) 20. Relative bone formation rates (rBFR/BS) were calculated as [loaded – non-loaded] in each animal. Thus, relative (r) values were used as a net measure of the effect of loading within each animal.
Figure 4. Loading-induced periosteal bone formation was blunted in Wnt1 knockout males.
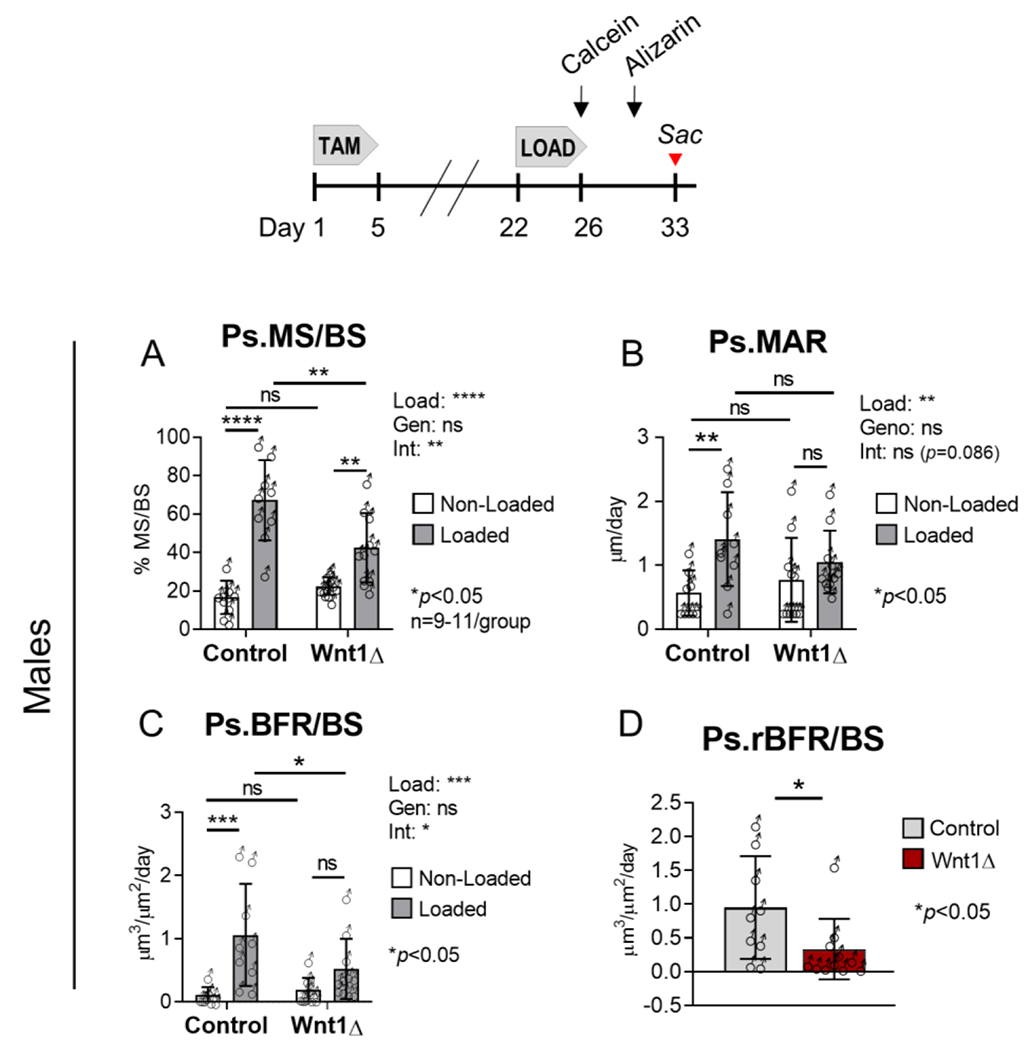
Dynamic histomorphometry was used to analyze bone formation after 5 days of strain-matched loading. (A-C) Periosteal bone formation indices in males subjected to strain-matched loading (approximately −3000 μƐ). Loading increased periosteal bone formation in both groups, albeit to a lesser degree in Wnt1 knockouts. (D) Relative bone formation rate (rBFR/BS), used as a net index of loading-induced bone formation, was 65% less in Wnt1 knockouts relative to controls. n=9-11/genotype.
Figure 5. Loading-induced periosteal bone formation was blunted in Wnt1 knockout females.
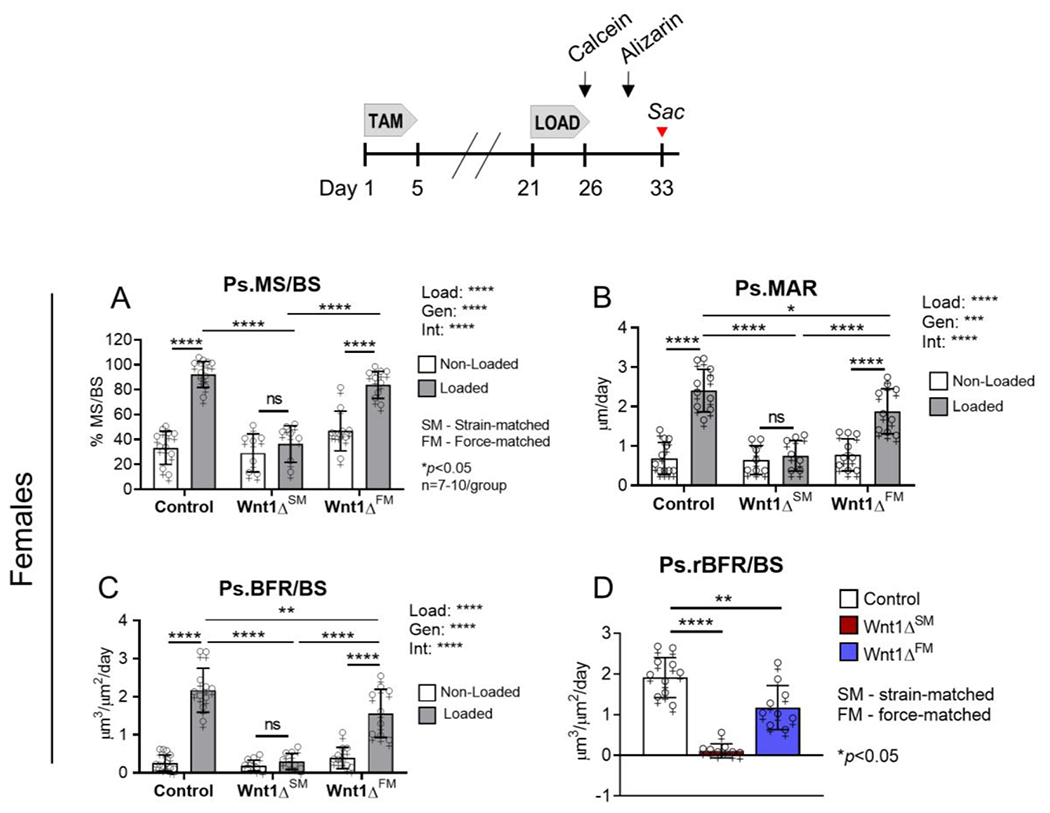
(A-D) Control females were loaded to −9.2 N to engender a peak target strain of −3040 μƐ. Strain-matched (SM) knockouts were loaded to −5.4 N to achieve a similar strain (−3200 μƐ), while force-matched (FM) knockouts were loaded to −9.2 N (approximately −5600 μƐ). (D) Comparison of the relative bone formation rates showed that the periosteal response to loading was 39% lower in force-matched knockouts relative to controls. n=7-10/genotype.
Gene Expression
Gene expression was analyzed by in situ hybridization and RT-qPCR on day 26, as outlined in Figure 6.
Figure 6. Loading-induced upregulation of bone formation genes was impaired in Wnt1 knockouts.
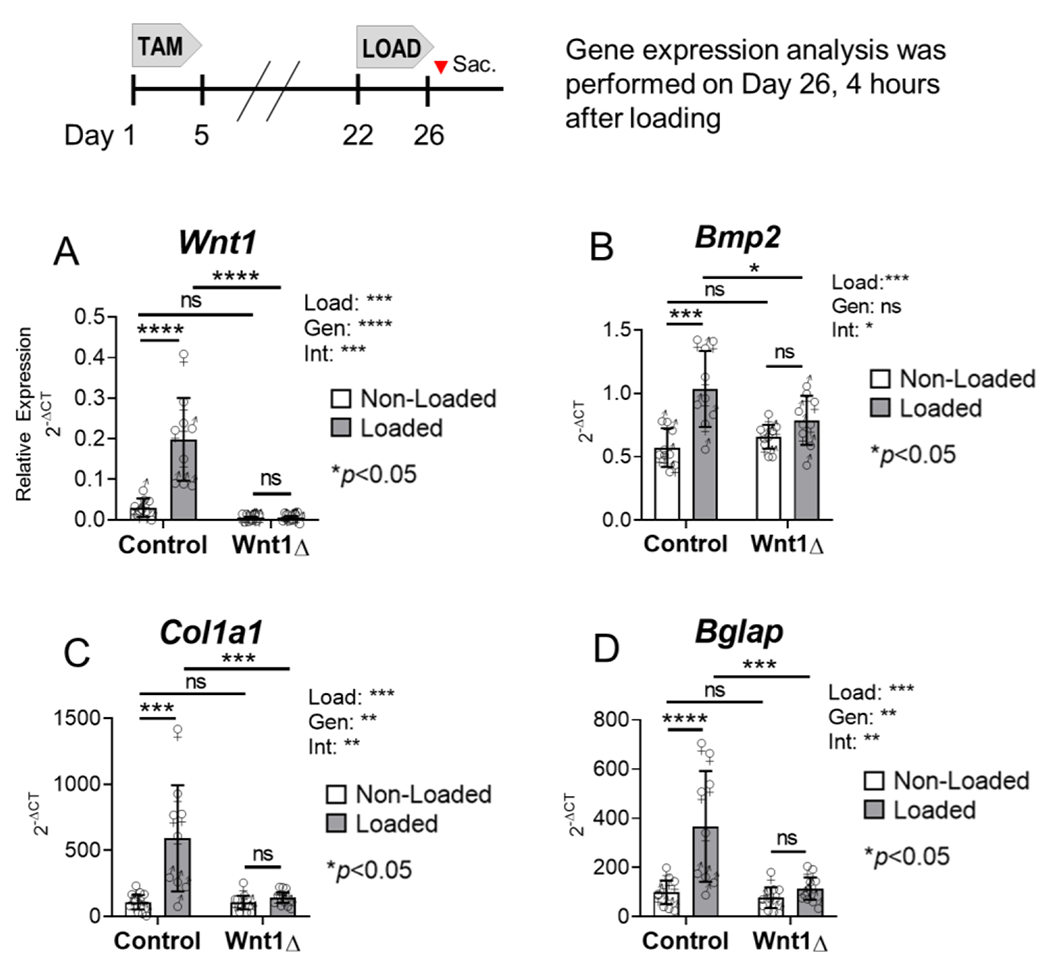
(A) Tibial loading potently induced Wnt1 expression in the bones of control but not knockout mice. (B-D) Loading-induced Bmp2, Col1a1, and Bglap upregulation was blunted in Wnt1 knockouts relative to strain-matched controls. n=4-5 per sex per genotype in each group for a total of n=9/genotype. Two-factor ANOVA was used to evaluate the effect of tibial loading (“Load)” and genotype (“Gen”), and their interaction (“Int”), on gene expression.
RNAScope in situ hybridization.
In situ hybridization was used to analyze gene expression in cre-negative Wnt1F/F and Wnt7bF/F male and female control mice. Tibias were fixed for 24 hours in 10% neutral buffered formalin (NBF), de-calcified in 14% EDTA, and submitted to the Musculoskeletal Histology and Morphometry Core (Washington University in St Louis, USA) for paraffin embedding and sectioning. Bones were sectioned to a thickness of 5 μm through the transverse plane approximately 5 mm distal to the tibial plateau. Hybridization was performed using the HybEZ II Hybridization System (ACDBio) in conjunction with the RNAScope 2.5HD Assay-BROWN kit. Probes for murine Wnt1, Bmp2, Bglap, Axin2, Opg, and Id1, and Wnt7b were purchased from ACDBio (Catalog No. 401091, 406661, 441211, 488961, 322335, and 401131). Sections were hematoxylin-counterstained and imaged on a Hamamatsu NanoZoomer slide scanning system at the Alafi Neuroimaging Laboratory (Washington University in St. Louis, USA). Wnt1 RNAScope was quantified by scoring cells at the site of peak compressive strain as negative, low, medium, or high for Wnt1 expression. Chi-square test was used for statistical analysis. Wnt1 RNAScope analysis was performed in n=4 mice, including 2 males and 2 females.
RT-qPCR.
Bilateral tibias were processed for RT-qPCR as described previously7, 12. Briefly, forceps were used to remove soft tissues from the bone surface, leaving the periosteum intact. Tibias were cut 2 mm below the tibial plateau and 1 mm below the tibio-fibrular junction to remove the epiphysis and distal tibia. The remainder – consisting largely of compact cortical bone – was spun at 13,000 rpm for 30 seconds to remove bone marrow. Bones were snap-frozen in liquid nitrogen, pulverized with a Mikro Dismembrator, lysed in Trizol, then phase separated with chloroform reagent. Total RNA was purified using RNeasy Total RNA kit (Qiagen), and cDNA was prepared from samples with RIN > 6.0 using the iScript cDNA Synthesis Kit (BioRad). Gene expression was analyzed with SYBR-based reagents on a StepOne Plus Machine (Applied Biosystems), and reported as a relative expression value (2−ΔCT), relative to reference gene Tbp. Gene expression analysis in males and females showed that loading had a significant or near-significant effect on osteogenic marker expression in Wnt1F/F control mice, irrespective of sex, but not in Wnt1F/F; OsxCreERT2 knockout males and females (Fig S10). Thus – because combining gene expression data from males and females did not impact how the results were interpreted – we chose to pool the data and present the findings as 4 panels instead of 8 in Figure 6.
Statistical Analysis
The main effects of genotype (knockout vs control) and loading (loaded vs non-loaded) and their interaction were analyzed by 2-factor ANOVA, and post hoc Sidak’s multiple comparisons test was used for pairwise comparisons (Prism 7.0, GraphPad Software, Inc., La Jolla, CA, USA). In this analysis the significance of the interaction (“Int”) term is crucial because it indicates whether the effect of loading differed between genotype groups. One-factor ANOVA was used for outcomes where loading was not a factor (eg knockdown efficiency, rBFR/BS). Significance was defined at p<0.05, with trends noted at 0.05< p <0.10. Individual data points and the mean ± standard deviation are plotted.
Results
Tibial loading induces Wnt1 expression in osteocytes
Tibial loading induces Wnt1 expression in bone9, 11–12. To identify the source of Wnt1 ligand expression in the bone, in situ hybridization was used to analyze Wnt1 RNA expression in the tibias of cre-negative Wnt1F/F and Wnt7bF/F mice after 5 days of tibial loading (Fig 1A). In loaded bones, Wnt1 RNA was detected in 39.3% (± 14.8%) of the osteocytes at the site of peak compressive strain, while in non-loaded bones Wnt1 expression was detected in only 3.8% (± 2.8%) of the osteocytes in the same region of interest (p<0.0001, chi-square test) (Fig 1B). Wnt1 expression was not detected by in situ hybridization on the bone surface (ie in osteoblasts) of either loaded or non-loaded bones (Fig S1).
Figure 1. Tibial loading increased Wnt1 expression in the bone.
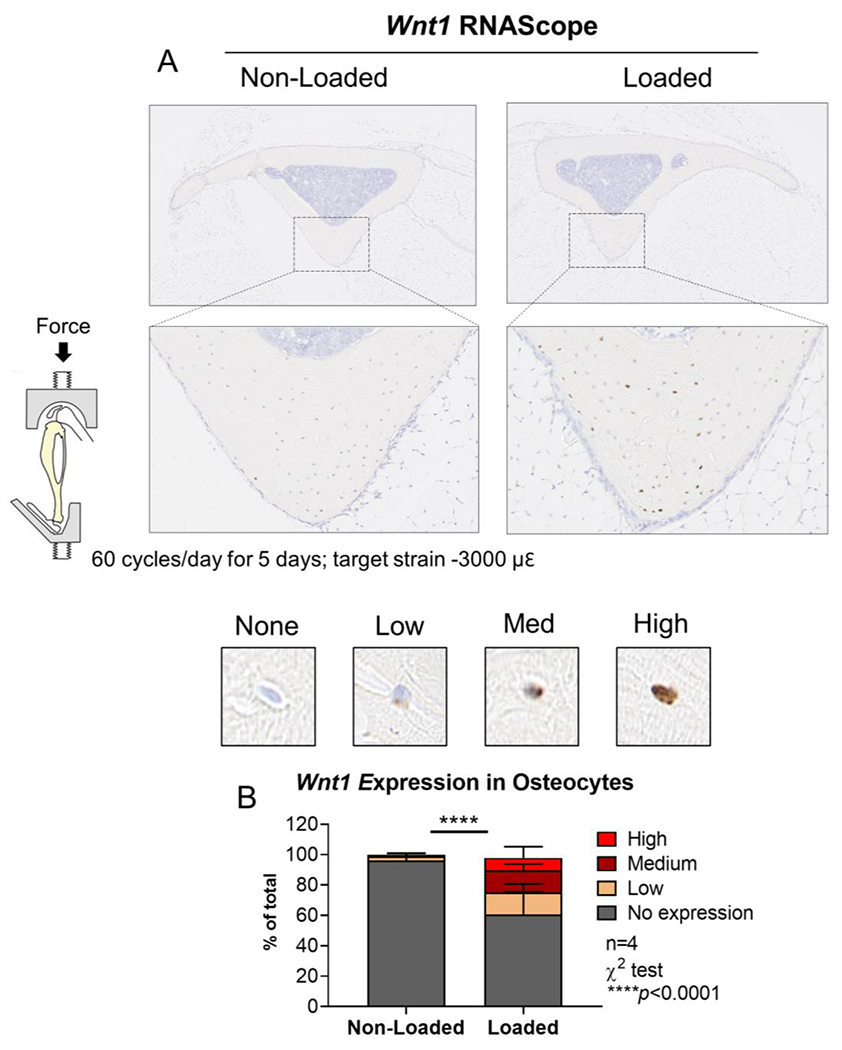
Wnt1 mRNA expression was analyzed by RNAScope in situ hybridization after 5 days of loading. (A) Tibial Wnt1 expression was induced by loading in a Wnt1F/F male. Results representative of n=4. (B) Osteocytes were scored for Wnt1 expression at the site of peak compressive strain (lower panels).
An inducible Cre/LoxP approach was used to delete Wnt1 in the Osx-lineage cells of 5-month old mice
To investigate the role of Wnt1 in the bone anabolic response to loading, an inducible Cre/LoxP approach was used to conditionally inactivate Wnt1 in the osteoblast-lineage cells of 5-month old mice. Gene deletion was confirmed on day 22 by DNA recombination PCR, and RT-qPCR was used to evaluate Wnt1 knockdown efficiency in the bone (Fig 2A). Recombination analysis using deletion-specific primers indicated that Wnt1 deletion was bone-specific; no off-target effects were observed in any of the extra-skeletal tissues surveyed (Fig 2B–C). Additionally, bone-specific deletion occurred in 5/5 knockout mice tested, but not in any (0/4) of the controls (Fig 2D). Finally, mRNA knockdown testing showed that tibial Wnt1 expression was 78% lower in Wnt1 knockouts compared to controls on day 22 (Fig 2E).
Figure 2. Wnt1 was conditionally deleted in the Osx-expressing cells of adult mice.
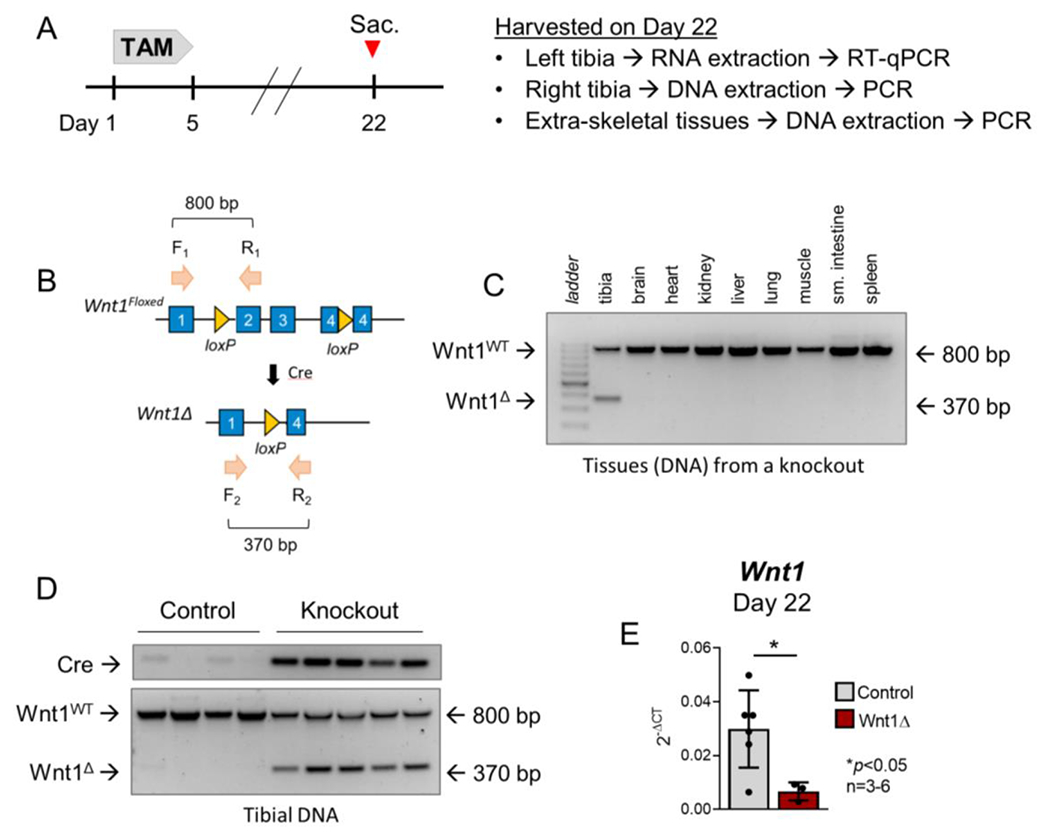
(A) Five-month old Wnt1F/F (control) and OsxCreERT2; Wnt1F/F (knockout) mice were dosed with tamoxifen to induce Wnt1 deletion in the osteoblast lineage. (B-D) DNA recombination analysis indicated that Wnt1 (exon 2-4) deletion was specific to the tibias of OsxCreERT2-positive Wnt1F/F mice. (E) Tibial Wnt1 RNA expression was 78% lower in Wnt1 knockouts compared to controls. Individual data points and the mean ± std deviation are shown. n=3-6/group.
Bone area decreased after short-term Wnt1 deletion
To evaluate the effects of short-term Wnt1 deletion on cortical bone morphometry, tibias were analyzed by in vivo μCT before (day 0) and after tamoxifen induction (day 22) (Fig 3, top panel). At baseline (day 0) no differences in cortical bone morphology were detected in control vs Wnt1 knockout mice, consistent with no phenotype prior to inducing gene deletion. However, between days 0 and 22, Ct.Ar decreased 11.6% and 8.6% in male and female knockouts, respectively (Fig 3B). Ct.Th also decreased between days 0 and 22 in male knockouts (−6.3%, p=0.055) while in female knockouts pMOI decreased 17.7%, consistent with a reduction in Tt.Ar and Ct.Ar between days 0 and 22 (Fig 3A, 3D, 3F). In contrast, an increase in Ct.Ar and Ct.Th was observed in Wnt1F/F controls – likely due to the anabolic effects of tamoxifen21.
Figure 3. Cortical bone area decreased after short-term Wnt1 deletion.
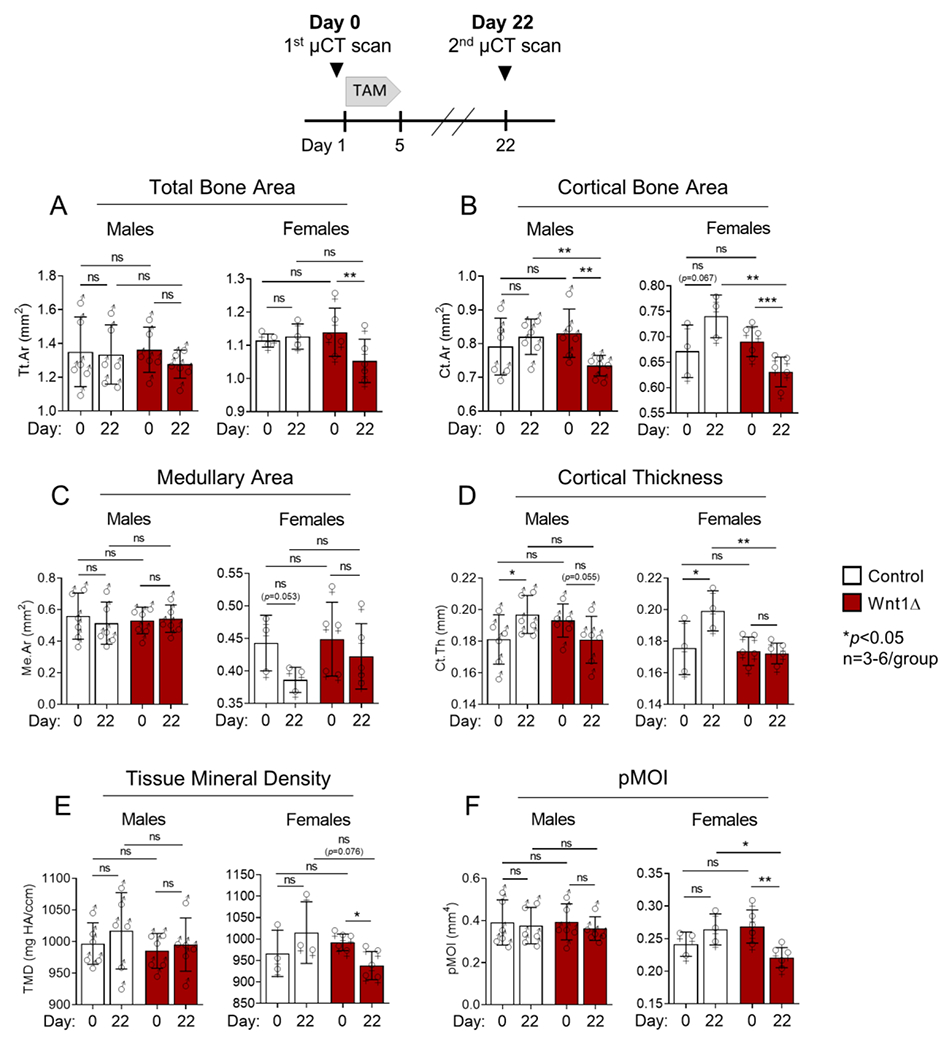
The tibias of naïve male and female mice were serially scanned to determine the effects of short-term Wnt1 deletion on cortical bone morphometry. Tibias were μCT scanned before tamoxifen induction on Day 0, and again on Day 22. Individual data points and the mean ± std deviation are shown. n=3-6/group.
In sum, Wnt1 deletion for only 3 weeks led to reduced cortical bone mass in adult mice, due either to loss of bone or reduced accrual of bone.
Loading-induced periosteal bone formation is impaired in Wnt1 knockouts
Next, to evaluate the role of Wnt1 in the loading response, in vivo tibial loading was used to induce cortical bone formation, starting on day 22. Based on a priori strain analysis, mice were subjected to strain-matched loading to a peak target strain of approximately −3000 μƐ for bone formation and gene expression analyses (Fig S2). Preliminary findings showed that this stimulus elicited a robust lamellar bone formation response in Wnt1F/F mice (Fig S3).
Ps bone formation, males
In males, tibial loading increased periosteal (Ps) bone formation in both groups, but the response to loading was blunted in Wnt1 knockouts. Two-factor ANOVA analysis indicated that the main effect of loading on Ps.MS/BS, Ps.MAR, and Ps.BFR/BS was significant (Fig 4A–C). Additionally, a significant (or near-significant) loading-genotype interaction was detected for Ps.MS/BS, Ps.MAR, and Ps.BFR/BS, indicating that the response to loading was different in control vs Wnt1 knockout mice.
In Wnt1F/F males, Ps.MS/BS and Ps.MAR were 4 and 2.5-fold higher in the loaded vs non-loaded limbs, resulting in an overall 9.6-fold increase in Ps.BFR/BS (Fig 4A–C). By comparison, in knockouts Ps.MS/BS and Ps.MAR were 1.9 and 1.4-fold higher in the loaded vs non-loaded limbs, resulting in a 2.8-fold increase in Ps.BFR/BS. A comparison of Ps.BFR/BS in the loaded tibias of control vs knockout mice showed that Ps.BFR/BS was statistically different in control vs knockout mice (p<0.05), indicating that Wnt1 was important for the skeletal response to loading (Fig 4C). Additionally, we calculated the relative bone formation rate (rBFR/BS), defined as BFR/BSLoaded minus BFR/BSNon-Loaded, as a measure of loading-induced bone formation. This analysis indicated that the periosteal response to loading was 65% lower in Wnt1 knockouts relative to controls (Fig 4D).
Ps bone formation, females
The bone anabolic response to loading was similarly blunted in Wnt1 knockout females. To achieve strain-matched loading in females, Wnt1F/F and Wnt1F/F; OsxCreERT2 mice were loaded to −9.2 N and −5.4 N, respectively (Fig S2). In Wnt1F/F females, Ps.MS/BS and Ps.MAR were 2.9 and 3.5-fold higher in the loaded vs non-loaded limbs, resulting in a 8.3-fold increase in Ps.BFR/BS (Fig 5A–C). In contrast, Ps.MS/BS, Ps.MAR, and Ps.BFR/BS were only negligibly affected by loading in strain-matched knockouts (Fig 5A–C). Ps.rBFR/BS indicated that in females, the periosteal response to loading was diminished 94% in Wnt1 knockouts relative to strain-matched controls (Fig 5D).
Force-matched loading in females
To achieve a strain-matched stimulus of −3000 μƐ in females, the force applied to Wnt1 knockouts (−5.4 N) was 40% less than the force required in controls (−9.2 N), consistent with the finding that pMOI was different in control vs knockout females on day 22 (Fig 3F). Because this level of stimulus did not induce a significant loading response in Wnt1F/F; OsxCreERT2 females (Wnt1ΔSM group), we asked if a higher magnitude stimulus might induce a response. To address this, a second cohort of knockout females was subjected to ‘force-matched’ loading (Wnt1ΔFM group, Figs 4A–D). In Wnt1F/F; OsxCreERT2 females, −9.2 N was estimated to engender a peak strain of −5600 μƐ (Fig S2), a level that typically induces a woven bone response12 (Fig S3); in knockout females −9.2 N induced lamellar bone formation.
In force-matched knockouts, Ps.MS/BS, Ps.MAR, and Ps.BFR/BS were 1.8, 2.4, and 4-fold higher in the loaded vs non-loaded limbs (p<0.0001), indicating that tibial loading stimulated Ps bone formation (Fig 5A–C). Notably, these values were nominally less than the respective fold-increases in control mice (2.9, 3.5 and 8.3), and a comparison of Ps.MAR and Ps.BFR/BS in the loaded limbs of control vs knockout mice showed that both indices were significantly lower in knockouts (Fig 5A–C). Finally, relative to the control group Ps.rBFR/BS was 39% lower in Wnt1 knockouts after force-matched loading (1.91 vs 1.17 μm3/μm2/day in control vs knockout mice; Fig 5D). Therefore, despite being loaded to 85% greater peak strain than control, Wnt1 knockout females still had a diminished response to loading.
Endocortical bone formation
In contrast to the strong anabolic response elicited on the periosteal surface in Wnt1F/F mice, tibial loading did not induce a strong endocortical response (Figs S4 and S5) – consistent with the lower values of strain engendered on this surface. In males, neither loading nor genotype had an effect on Ec.MS/BS, Ec.MAR, or Ec.BFR/BS, and Ec.rBFR/BS indicated that endocortical bone formation was not different in control vs knockout mice (Fig S4). In females, a significant or near-significant increase in Ec.MAR and Ec.BFR/BS was observed in controls and force-matched knockouts, but not in strain-matched knockouts (Fig S5). Loading-induced endocortical bone formation was not different in control vs force-matched knockouts (Fig S5D).
In sum, these findings show that Wnt1 was required for the bone anabolic response to skeletal loading, particularly for periosteal bone formation.
Loading-induced upregulation of bone formation genes was diminished in Wnt1 knockouts
To better understand the role of Wnt1 in loading-induced bone formation, gene expression analysis was performed after 5 days of loading, on day 26 (Fig 6, top panel). Gene expression was analyzed by RT-qPCR (control vs knockout mice) and by RNAScope in situ hybridization (control mice only).
Congruent with previous reports11–12, we found that loading potently induced Wnt1 mRNA expression in the bone (Fig 6A). RT-qPCR analysis indicated that Wnt1 expression was 6.6-fold higher in the loaded vs non-loaded tibias of Wnt1F/F controls (Fig 6A). In contrast, loading had no effect on the abundance of Wnt1 mRNA in the tibias of Wnt1F/F; OsxCreERT2 knockouts (loaded vs non-loaded, p=0.999), consistent with bone-targeted knockout.
Genes associated with osteo-induction, osteoblast differentiation, and matrix synthesis were also analyzed. RNAScope in situ hybridization showed that loading increased Bmp2 expression in osteocytes and Bglap expression in osteoblasts (Fig S6B, S6D). These findings were supported by qPCR, which showed that Bmp2 and Bglap were 1.8 and 3.7-fold higher, respectively, in the loaded vs non-loaded bones of Wnt1F/F mice (Fig 6B, 6D). In contrast, Bmp2 and Bglap were not significantly different in the loaded vs non-loaded bones of Wnt1 knockouts. Loading also significantly increased Col1a1 expression – a marker of bone matrix synthesis – in the tibias of control but not knockout mice (Fig 6C).
Wnt pathway-related genes were also analyzed by qPCR and in situ hybridization. Neither loading nor genotype were found to have a significant effect on the expression of Axin2 or Nkd2 (Figs S6E, S7A–B). In contrast, Opg was regulated by loading but not genotype (main effect of loading, p<0.05). Opg expression was 1.4 and 1.2-fold higher in the loaded vs non-loaded limbs of control and knockout mice, respectively (Fig S7C). In situ analysis showed that loading induced Opg expression in osteocytes (Fig S6F). Gene expression findings also showed that Sost and Dkk1 were not different in the loaded limbs of control vs knockout mice, indicating that the diminished loading response observed in knockouts was not due to elevated expression of these Wnt antagonists in the bone (Fig S7D–E). Finally, Wnt1 knockout mice had a marginally lower loading-induced induction of Wnt7b, while Wnt16 was unaffected by loading or genotype (Fig S7F–G). Analysis by in situ hybridization showed that Wnt7b mRNA was expressed in osteocytes (Fig S6A)
In sum, these findings show that Wnt1 plays a crucial role in mediating the bone anabolic response to mechanical loading.
The skeletal phenotype caused by Wnt1 deletion was recapitulated in Wnt1/7b knockout mice
Because Wnt7b is osteo-anabolic14 and is induced in the bone by loading12, we asked whether the phenotypes observed in Wnt1 knockouts would be exacerbated by also targeting Wnt7b. We confirmed DNA recombination of Wnt1 and Wnt7b genes in the tibias of Wnt1/7b knockout mice (tamoxifen-treated Wnt1F/F; Wnt7bF/F; OsxCreERT2) (Fig.S8A–B). We also confirmed knockdown of Wnt1 (−63% in the loaded limbs of Wnt1/7b knockouts vs controls, p=0.083) and Wnt7b (−87% in the loaded limbs of knockout vs control mice, p=0.194) at the mRNA level (Fig S9A–B).
The cortical bone phenotype observed in Wnt1 knockouts was recapitulated in Wnt1/7b knockouts. Before gene deletion (day 0) cortical bone morphology was not different between control vs Wnt1/7b knockout mice (Fig S8). However, Ct.Ar and pMOI decreased between days 0 and 22 in Wnt1/7b knockout mice, but increased in Wnt1F/F; Wnt7bF/F controls (Fig S8D, S8H). Ct.Th was not significantly different on days 0 and 22 in Wnt1/7b knockouts, but was significantly higher on day 22 relative to day 0 in controls (Fig S9F). We also found that Me.Ar decreased and TMD increased significantly between days 0 and 22 in control but not knockout mice (Fig S8E, S8G).
Finally, when Wnt1/7b knockouts and their controls were subjected to strain-matched loading (approximately −3000 μƐ), Wnt1/7b knockouts exhibited an impaired response to loading, similar to Wnt1 single knockouts. In male and female controls, loading increased Ps.MS/BS and Ps.MAR significantly (p<0.01), resulting in an overall 5.6 to 11-fold increase in Ps.BFR/BS (p<0.0001; Fig S9C–D). In contrast, loading generally did not have a significant effect on bone formation indices in Wnt1/7b knockouts. Analysis of relative bone formation rates showed that the periosteal response to loading was reduced 86-90% in Wnt1/7b knockouts compared to sex-matched controls.
Discussion
Mechanical loading on the skeleton stimulates bone formation. To investigate the role of Wnt1 in the skeletal response to loading, we tamoxifen-dosed 5-month old OsxCreERT2; Wnt1F/F mice to inactivate Wnt1 in the osteoblast lineage, then subjected mice to in vivo tibial loading 21 days later. Over 3 weeks, Ct.Ar decreased 9-12% in Wnt1 knockouts, concomitant with a 78% reduction in tibial Wnt1 expression (Figs 2E, 3B). In controls, loading increased Wnt1 expression in the bone (p<0.0001) and in situ hybridization showed that this induction occurred in osteocytes (Figs 1A, 6A). Loading-induced Wnt1 upregulation was associated with a 8 to 10-fold increase in Ps.BFR/BS in Wnt1F/F mice (Figs 4D, 5D). In knockouts, Osx-targeted Wnt1 deletion blocked Wnt1 upregulation (Fig 6A), leading to a 65 to 94% lower periosteal bone formation response (Figs 4E, 5D). Together, these findings show that Wnt1 is important for homeostasis of the adult skeleton, as well as for the bone anabolic response to mechanical loading.
Loading increases Wnt1 expression in osteocytes
Whole-bone transcriptional profiling studies have shown that mechanical loading increases the expression of Wnt family genes – including Wnt1 and Wnt7b – in the bone9–10. Subsequently, Harris and Silva used laser capture microdissection to isolate intracortical bone samples from loaded and non-loaded bones of C57Bl/6 mice, and showed that Wnt1 in these osteocyte-enriched samples was upregulated by loading11. Congruent with these results, our gene expression analyses showed that Wnt1 was 6.6-fold higher in the loaded vs non-loaded bones of Wnt1F/F mice after 5 days of loading (Fig 6A). Additionally, analysis by RNAscope® in situ hybridization showed that Wnt1 induction occurred primarily in osteocytes embedded in the intra-cortical matrix rather than in cells lining the bone surface (i.e., osteoblasts) (Figs 1A and S1). Analysis of in situ hybridization further revealed that loading increased not only the total percentage of Wnt1-positive cells in the bone, but also the magnitude of Wnt1 expression within those cells (Fig 1B). Together, these data show that mechanical loading stimulates Wnt1 expression in the bone, particularly in osteocytes.
Loading-induced bone formation is reduced in Wnt1 knockouts
OsxCreERT2 was used to inactivate Wnt1 in the Osx-lineage cells of 5-month old mice. Previously, we used a similar tamoxifen dosing strategy for Cre reporter analysis in 5-month old OsxCreERT2; Rosa-Ai9 mice, which showed robust TdTomato reporter expression on the bone surface (osteoblasts) and within the cortical bone (osteocytes) within days of tamoxifen treatment – indicating that OsxCreERT2 targets both osteoblasts and osteocytes in the bones of adult mice7.
OsxCreERT2-mediated Wnt1 deletion completely neutralized the effect of loading on Wnt1 expression in the bone (Figs 6A and S10A). While tibial Wnt1 expression increased 6.6-fold in loaded bones of Wnt1F/F controls, Wnt1 expression remained unchanged in the loaded vs non-loaded limbs of Wnt1 knockouts. qPCR analysis in male and female mice showed that although the gene expression response to loading was generally more robust in Wnt1F/F control females compared to Wnt1F/F males, the increase in Wnt1, Bmp2, Col1a1, and Bglap expression in response to loading was significant or near-significant in both sexes (Fig S10). These results were congruent with the finding that the bone formation response was more pronounced in Wnt1F/F females compared to Wnt1F/F males (Figs 4D and 5D). Meanwhile, the induction of Wnt1, Bmp2, Col1a1, and Bglap in response to loading was reduced in knockouts, irrespective of sex, indicating that loading-induced Wnt1 expression as required for the upregulation of Bmp2, Col1a1, and Bglap in the bone. In combination with our in situ hybridization findings, these results suggest that loading stimulates osteoblast differentiation/activity on the bone surface (Bglap, Fig S6D) by inducing Wnt1 ligand expression in osteocytes (Figs 1 and S1).
At the tissue level, Wnt1 inactivation in the Osx-lineage reduced the periosteal response to loading. In males, periosteal bone formation rate was higher in the loaded vs non-loaded bones of all mice, independent of genotype. However, the relative bone formation rate – which reflects the overall magnitude of the loading response – showed that the periosteal response to loading was 65% lower in knockouts (Fig 4D). In females, strain-matched loading increased all indices of periosteal bone formation in control but not knockout mice (Fig 5A–C). Compared to Wnt1F/F controls, the periosteal response to loading (i.e., Ps.rBFR/BS) was 94% lower in strain-matched Wnt1 knockouts (Fig 5D). Moreover, analysis of a force-matched cohort (−9.2 N, Wnt1 knockout females) showed that although a loading response could be elicited with greater strain magnitude (−5600 μƐ vs −3000 μƐ), the periosteal response to loading was still 39% less in knockouts vs controls (Fig 5D). Together, these results demonstrate that Wnt1 is required for loading-induced periosteal bone formation.
Bone area decreased in Wnt1 knockouts after 21 days deletion
Independent of loading, short-term Wnt1 deletion (3 weeks) in the Osx-lineage was associated with decreased bone area and cortical thickness (Fig 3B, 3D). These findings are congruent with a report by Wang, et al. which showed that deleting Wnt1 in the Osx-lineage impaired periosteal bone growth in young mice. Using a Dox-repressible system, Wang et al. showed that inactivating Wnt1 in the Osx-lineage of 4-week old mice produced measurable deficits in the skeleton 4 and 8 weeks later. Analysis of distal tibias from 12 week old mice revealed that these deficits were caused by a reduction in periosteal bone formation, while changes to endocortical bone formation were not significant22.
Similar findings were reported by Joeng, et al. and Luther, et al. Mice lacking Wnt1 in late osteoblasts (Dmp1Cre; Wnt1F/F) had skeletal deficits – including reduced Ct.Th, Tb.N, and Tb.Th – 2 months after birth13, while mice lacking Wnt1 in osteo-progenitors (Runx2Cre; Wnt1F/F) exhibited BV/TV and Ct.Th deficits in the femur at 24 weeks, leading to increased risk of spontaneous fractures23. Hypomorphic Wnt1 variants have also been linked to low bone mass and spontaneous fractures in mice24. Our study extends these findings by showing that short-term Wnt1 deletion in adults leads to bone loss. Thus, osteoblast/osteocyte Wnt1 is required for adult bone homeostasis.
The skeletal response to bone anabolic loading declines with age25–27. For example, tibial loading enhances periosteal bone formation in adult mice of different ages; however, the magnitude of the osteogenic response to loading is significantly lower in aged 22-month old vs. young adult 5-month old mice12. To understand the basis for these differences, Chermside-Scabbo, et al. used RNASeq to characterize the transcriptional responses to loading in 5 and 22-month old C57Bl/6 mice after 1, 3, and 5 days of loading10. In both age groups, Wnt1 emerged as a top 10 upregulated gene on days 1 and 3. The fold-change difference for Wnt1 in 5-month old mice was 5.8 and 6.7 on days 1 and 3, respectively, while in 22-month old mice the fold-change difference for Wnt1 was 1.4 and 2.3. Thus, loading increased Wnt1 in both age groups, albeit to a lesser degree in aged mice – concomitant with a diminished osteogenic response to loading. Taken together, these findings suggest that diminished induction of Wnt1 by loading may contribute to reduced loading-induced bone formation in the bones of aged mice.
The bone anabolic function of Wnt1 in the Osx-lineage is further supported by our findings in Wnt1/7b double conditional knockout mice, which exhibited cortical bone deficits 21 days post-deletion (Fig S8D). Moreover, like Wnt1 knockouts, Wnt1/7b knockouts did not exhibit a loading-induced increase in Wnt1 expression and had a blunted periosteal response to loading (Fig S9). In general, the magnitude of the deficits in Wnt1/7b knockouts were comparable to those in Wnt1 knockouts. Importantly, while we did see evidence of Wnt7b gene recombination in Wnt1/7b double knockout mice (Fig. S8B), we did not confirm significant knockdown of Wnt7b at the mRNA level (Fig. S9B). Thus, we are not able to infer whether Wnt7b has a functional role in loading-induced bone formation. Additional studies, perhaps using a different Wnt7b floxed mouse, are needed to address this question. Nonetheless, the consistent deficits in loading-induced bone formation in Wnt1 and Wnt1/7b knockout mice show that in two different mouse lines, Wnt1 in the Osx-lineage was essential for the response to loading.
In conclusion, our findings support a growing body of evidence that Wnt1 is essential for bone homeostasis in the adult skeleton. In particular, we showed that mechanical loading induced Wnt1 expression in osteocytes, and that loading-induced Wnt1 expression in the Osx-lineage was required for the periosteal bone formation response to tibial loading.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 AR047867, T32 AR060719 and the Washington University Musculoskeletal Research Center (P30 AR074992). We thank Crystal Idleburg and Samantha Coleman for histology support.
Conflict of Interest Statement
LYL, NM, CCS, JTS, KJ, and BL have no financial or non-financial competing interests to disclose. MJS is on the editorial board at Bone, Journal of Orthopaedic Research, and Calcified Tissue International, and served on the board of directors at the Orthopaedic Research Society (2018-2022). RC serves as Editor-in-Chief for Journal of Bone and Mineral Research and has received research support from Ultragenyx.
Data Availability Statement
The data that support the findings of this study are openly available in Biorxiv at https://doi.org/10.1101/2022.02.28.482178.
References
- 1.Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, Turner CH. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem. 2008. Feb 29;283(9):5866–75. doi: 10.1074/jbc.M705092200. Epub 2007 Dec 17 [DOI] [PubMed] [Google Scholar]
- 2.Tu X, Rhee Y, Condon KW, et al. Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone. 2012;50(1):209–217. doi: 10.1016/j.bone.2011.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawakami K, Robling AG, Ai M, Pitner ND, Liu D, Warden SJ, Li J, Maye P, Rowe DW, Duncan RL, Warman ML, Turner CH. The Wnt co-receptor LRP5 is essential for skeletal mechanotransduction but not for the anabolic bone response to parathyroid hormone treatment. J Biol Chem. 2006. Aug 18;281(33):23698–711. doi: 10.1074/jbc.M601000200. Epub 2006 Jun 20. [DOI] [PubMed] [Google Scholar]
- 4.Saxon LK, Jackson BF, Sugiyama T, Lanyon LE, Price JS. Analysis of multiple bone responses to graded strains above functional levels, and to disuse, in mice in vivo show that the human Lrp5 G171V High Bone Mass mutation increases the osteogenic response to loading but that lack of Lrp5 activity reduces it. Bone. 2011. Aug;49(2):184–93. doi: 10.1016/j.bone.2011.03.683. Epub 2011 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niziolek PJ, Farmer TL, Cui Y, Turner CH, Warman ML, Robling AG. High-bone-mass-producing mutations in the Wnt signaling pathway result in distinct skeletal phenotypes. Bone. 2011. Nov;49(5):1010–9. doi: 10.1016/j.bone.2011.07.034. Epub 2011 Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012. Jun 8;149(6):1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Lawson LY, Brodt MD, Migotsky N, Chermside-Scabbo CJ, Palaniappan R, Silva MJ. Osteoblast-Specific Wnt Secretion Is Required for Skeletal Homeostasis and Loading-Induced Bone Formation in Adult Mice. J Bone Miner Res. 2021. Sep 20. doi: 10.1002/jbmr.4445. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bänziger C, Soldini D, Schütt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006. May 5;125(3):509–22. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 9.Kelly NH, Schimenti JC, Ross FP, van der Meulen MC. Transcriptional profiling of cortical versus cancellous bone from mechanically-loaded murine tibiae reveals differential gene expression. Bone. 2016. May;86:22–9. doi: 10.1016/j.bone.2016.02.007. Epub 2016 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chermside-Scabbo CJ, Harris TL, Brodt MD, Braenne I, Zhang B, Farber CR, Silva MJ. Old Mice Have Less Transcriptional Activation But Similar Periosteal Cell Proliferation Compared to Young-Adult Mice in Response to in vivo Mechanical Loading. J Bone Miner Res. 2020. Sep;35(9):1751–1764. doi: 10.1002/jbmr.4031. Epub 2020 Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris TL, Silva MJ. Gene expression of intracortical bone demonstrates loading-induced increases in Wnt1 and Ngf and inhibition of bone remodeling processes. Bone. 2021. Sep;150:116019. doi: 10.1016/j.bone.2021.116019. Epub 2021 May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holguin N, Brodt MD, Silva MJ. Activation of Wnt Signaling by Mechanical Loading Is Impaired in the Bone of Old Mice. J Bone Miner Res. 2016. Dec;31(12):2215–2226. doi: 10.1002/jbmr.2900. Epub 2016 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joeng KS, Lee YC, Lim J, Chen Y, Jiang MM, Munivez E, Ambrose C, Lee BH. Osteocyte-specific WNT1 regulates osteoblast function during bone homeostasis. J Clin Invest. 2017. Jun 30;127(7):2678–2688. doi: 10.1172/JCI92617. Epub 2017 Jun 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song D, He G, Song F, Wang Z, Liu X, Liao L, Ni J, Silva MJ, Long F. Inducible expression of Wnt7b promotes bone formation in aged mice and enhances fracture healing. Bone Res. 2020. Feb 3;8:4. doi: 10.1038/s41413-019-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes C, Kobayashi T, Kronenberg HM. A novel transgenic mouse model to study the osteoblast lineage in vivo. Ann N Y Acad Sci. 2007. Nov;1116:149–64. doi: 10.1196/annals.1402.060. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopal J, Carroll TJ, Guseh JS, Bores SA, Blank LJ, Anderson WJ, Yu J, Zhou Q, McMahon AP, Melton DA. Wnt7b stimulates embryonic lung growth by coordinately increasing the replication of epithelium and mesenchyme. Development. 2008. May;135(9):1625–34. doi: 10.1242/dev.015495. Epub 2008 Mar 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Brodt MD, Zannit HM, Holguin N, Silva MJ. Evaluation of loading parameters for murine axial tibial loading: Stimulating cortical bone formation while reducing loading duration. J Orthop Res. 2018. Feb;36(2):682–691. doi: 10.1002/jor.23727. Epub 2017 Oct 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Main RP, Shefelbine SJ, Meakin LB, Silva MJ, van der Meulen MCH, Willie BM. Murine Axial Compression Tibial Loading Model to Study Bone Mechanobiology: Implementing the Model and Reporting Results. J Orthop Res. 2020. Feb;38(2):233–252. doi: 10.1002/jor.24466. Epub 2019 Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erben RG. Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J Histochem Cytochem. 1997. Feb;45(2):307–13. doi: 10.1177/002215549704500215. [DOI] [PubMed] [Google Scholar]
- 20.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013. Jan;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong ZA, Sun W, Chen H, Zhang H, Lay YE, Lane NE, Yao W. Optimizing tamoxifen-inducible Cre/loxp system to reduce tamoxifen effect on bone turnover in long bones of young mice. Bone. 2015. Dec;81:614–619. doi: 10.1016/j.bone.2015.07.034. Epub 2015 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Rummukainen P, Heino TJ, Kiviranta R. Osteoblastic Wnt1 regulates periosteal bone formation in adult mice. Bone. 2021. Feb;143:115754. doi: 10.1016/j.bone.2020.115754. Epub 2020 Nov 13. [DOI] [PubMed] [Google Scholar]
- 23.Luther J, Yorgan TA, Rolvien T, Ulsamer L, Koehne T, Liao N, Keller D, Vollersen N, Teufel S, Neven M, Peters S, Schweizer M, Trumpp A, Rosigkeit S, Bockamp E, Mundlos S, Kornak U, Oheim R, Amling M, Schinke T, David JP. Wnt1 is an Lrp5-independent bone-anabolic Wnt ligand. Sci Transl Med. 2018. Nov 7;10(466):eaau7137. doi: 10.1126/scitranslmed.aau7137. [DOI] [PubMed] [Google Scholar]
- 24.Joeng KS, Lee YC, Jiang MM, Bertin TK, Chen Y, Abraham AM, Ding H, Bi X, Ambrose CG, Lee BH. The swaying mouse as a model of osteogenesis imperfecta caused by WNT1 mutations. Hum Mol Genet. 2014. Aug 1;23(15):4035–42. doi: 10.1093/hmg/ddu117. Epub 2014 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner CH, Takano Y, Owan I. Aging changes mechanical loading thresholds for bone formation in rats. J Bone Miner Res. 1995. Oct;10(10):1544–9. doi: 10.1002/jbmr.5650101016. [DOI] [PubMed] [Google Scholar]
- 26.Birkhold AI, Razi H, Duda GN, Weinkamer R, Checa S, Willie BM. The influence of age on adaptive bone formation and bone resorption. Biomaterials. 2014. Nov;35(34):9290–301. doi: 10.1016/j.biomaterials.2014.07.051. Epub 2014 Aug 13. [DOI] [PubMed] [Google Scholar]
- 27.Meakin LB, Galea GL, Sugiyama T, Lanyon LE, Price JS. Age-related impairment of bones’ adaptive response to loading in mice is associated with sex-related deficiencies in osteoblasts but no change in osteocytes. J Bone Miner Res. 2014;29(8):1859–1871. doi: 10.1002/jbmr.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Biorxiv at https://doi.org/10.1101/2022.02.28.482178.


