ABSTRACT
Phytate is the main phosphorus storage molecule of plants and is therefore present in large amounts in the environment and in the diet of humans and animals. Its dephosphorylated form, the polyol myo-inositol (MI), can be used by bacteria as a sole carbon and energy source. The biochemistry and regulation of MI degradation were deciphered in Bacillus subtilis and Salmonella enterica, but a systematic survey of this catabolic pathway has been missing until now. For a comprehensive overview of the distribution of MI utilization, we analyzed 193,757 bacterial genomes, representing a total of 24,812 species, for the presence, organization, and taxonomic prevalence of inositol catabolic gene clusters (IolCatGCs). The genetic capacity for MI degradation was detected in 7,384 (29.8%) of all species for which genome sequences were available. IolCatGC-positive species were particularly found among Actinobacteria and Proteobacteria and to a much lesser extent in Bacteroidetes. IolCatGCs are very diverse in terms of gene number and functions, whereas the order of core genes is highly conserved on the phylum level. We predict that 111 animal pathogens, more than 200 commensals, and 430 plant pathogens or rhizosphere bacteria utilize MI, underscoring that IolCatGCs provide a growth benefit within distinct ecological niches.
IMPORTANCE This study reveals that the capacity to utilize inositol is unexpectedly widespread among soil, commensal, and pathogenic bacteria. We assume that this yet-neglected metabolism plays a pivotal role in the microbial turnover of phytate and inositols. The bioinformatic tool established here enables predicting to which extent and genetic variance a bacterial determinant is present in all genomes sequenced so far.
KEYWORDS: myo-inositol, phytate, catabolic pathway, prevalence, ecological niche, bacterial metabolism, virulence, genomics, large-scale approach
INTRODUCTION
Inositol is the structural basis for many biomolecules, such as phosphatidylinositol, that belong to the components of eukaryotic cell membranes. Their mono-, di-, or triphosphorylated derivatives, the phosphoinositides, are membrane constituents that act as cytosolic solutes and contribute to cell signaling, cell motility, membrane trafficking, and phagocytosis (1). Inositol phosphates (InsPs) are synthesized in animals and plants as secondary messengers. InsPs have been reported to represent 2% to 60% of the total organic phosphorus in animal waste (2), pointing to their role in eutrophication of the environment. InsP6, phytate, is the main storage molecule of phosphorus and minerals in plants. InsPs play multiple roles in eukaryotic cell functions, including growth, lipid metabolism, and insulin sensitivity (3). The most prominent example is the second messenger, inositol-1,4,5-triphosphate (InsP3). Several intestinal pathogens, such as Salmonella spp., Shigella spp., Escherichia coli serotypes, and Yersinia spp., modulate the InsP metabolism of the host in different ways (4). InsPs exist in various forms of phosphorylation with one to six phosphates and isomeric forms such as d-chiro, scyllo, and neo, with myo-inositol hexakiphosphate (InsP6), or phytate, the most abundant form in terrestrial and aquatic environments (5), cells (1), and in the diet as well as in the gut (6).
Phytate is present in plant tissues such as bran and seeds of legumes and cereals, including oil seeds and nuts. The dephosphorylated form of phytate is myo-inositol (MI), a polyol that is a readily available carbon and energy source for microorganisms. So far, the capacity to catabolize MI has been experimentally demonstrated for a few bacteria only, including Rhizobium leguminosarum (7), Sinorhizobium meliloti (8), Lactobacillus casei (9), Klebsiella aerogenes (10), Corynebacterium glutamicum (11), Legionella pneumophila (12), Yersinia mollaretii (13), and Citrobacter koseri (14). The transporters, repressor, and enzymes involved in MI utilization have been investigated in detail for Bacillus subtilis (15, 16), whereas the enteropathogen Salmonella enterica was used to elucidate the complex regulatory network controlling the MI pathway (17–19).
As MI is the most abundant carbon source detectable in the soil (20), we assumed that MI utilization is a widespread metabolic capability in microbial communities. However, it is largely unknown to which extent aquatic, soil, plant-associated, and gut bacteria are able to degrade MI that was released from phytate or cellular components. Here, we predict that MI is a substrate that can be utilized by an unexpectedly large number of bacteria as a source of carbon and energy. We determine the number and prevalence of bacterial species and genomes carrying the genetic determinant required for the degradation of dephosphorylated inositols and discuss the critical role of bacteria in the global turnover of these compounds.
RESULTS
Search for iol genes and inositol catabolic gene clusters.
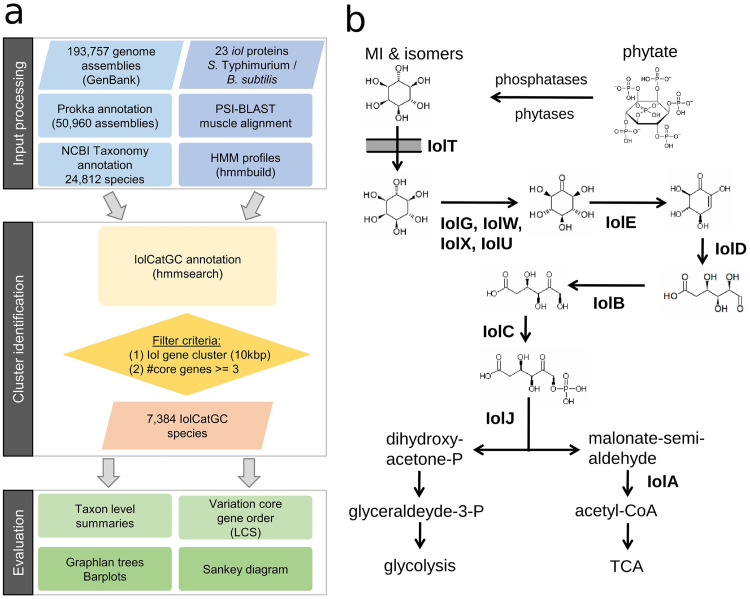
To obtain an overview of the distribution of inositol catabolic gene clusters (IolCatGCs), we performed a quality selection of bacterial genome sequences available in the GenBank database by applying RefSeq exclusion criteria, resulting in a total subset of 193,757 genomes that represent 24,812 species (Fig. 1a). In the case of 50,960 genome sequences, gene annotation files were not available in the database and were therefore generated using the annotation tool Prokka (21). Next, a basic set of 23 iol genes involved in the MI metabolism was taken from Salmonella enterica serovar Typhimurium and from B. subtilis, and its respective protein sequence was queried against the NCBI nonredundant (nr) database using PSI-BLAST. The resulting sequence hits were combined into hidden Markov models (HMMs) for each iol component. All 23 HMMs were searched against the total subset of genomes.
FIG 1.
Searching for IolCatGCs. (a) The workflow depicts the bioinformatic strategy applied here to perform a comprehensive prediction of all bacteria able to degrade MI. Starting with the processing of 193,757 input genome assemblies, filtering resulted in the identification of an iol gene cluster termed IolCatGC whose prevalence was then evaluated at all taxonomic levels of bacteria. (b) A schematic pathway of inositol catabolism in Gram-positive and negative bacteria is shown. Enzymes catalyzing the degradation of MI to glyceraldehyde and acetyl-CoA are indicated. IolT, transporter; IolG, IolW, IolX, IolU, IolJ, dehydrogenases of inositol and its stereoisomers; IolE, dehydratase; IolD, 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione hydrolase; IolB, isomerase; IolC, biphosphate aldolase; IolA, malonate-semialdehyde dehydrogenase; TCA, tricarboxylic acid cycle.
We defined a possibly functional IolCatGC to be present in a genome if at least three of the four core genes iolB, iolC, iolD, and iolE, which are essential for MI degradation (17) (Fig. 1b), were found in a distinct genetic determinant. Genes were assigned to an IolCatGC if they showed a maximum genetic distance of 10 kb. In 810 of 1,024 genomes with only 3 core genes, the 4th one was identified more than 10 kb apart from the IolCatGC elsewhere on the chromosome or within another contig, respectively. Possible reasons for a lack of the fourth gene are an incomplete genome sequence, assembly errors, or iol genes with frameshifts that were not considered to be functional. The genes iolG1, iolG2, iolU, and iolW encoding dehydrogenases of inositol isomers are expected to be present in genomes with a complete set of iol genes. Indeed, at least one of these dehydrogenase genes was identified in 6,502 IolCatGCs and outside the cluster in 881 species from our list. Genes encoding inositol transporters were not categorized as core genes due to their location apart from the IolCatGC in some bacteria (16) and not the genes encoding the malonate-semialdehyde dehydrogenase IolA and the biphosphate aldolase IolJ, which are also involved in the metabolism of valine or of fructose-1,6-biphosphate, respectively. The functions of genes associated with inositol catabolism are listed in Table S1 in the supplemental material. To not miss bacteria harboring an IolCatGC and to not erroneously state a species of not being capable of using MI, all genomes of strains belonging to the same species were analyzed for the presence and absence of iol genes. For each species with more than one genome available, we selected one IolCatGC that is representative in terms of cluster length and homology score (Table S2). We identified IolCatGCs within a total of 7,384 species corresponding to 29.8% of the 24,812 species’ genomes in the NCBI assembly database (Table S3). Among these are 3,651 Gram-negative and 3,733 Gram-positive species, corresponding to an almost equal distribution (14.7% versus 15%). Taken together, these findings highlight the relevance of MI degradation for many bacteria.
Length, composition, and genetic organization of the IolCatGCs.
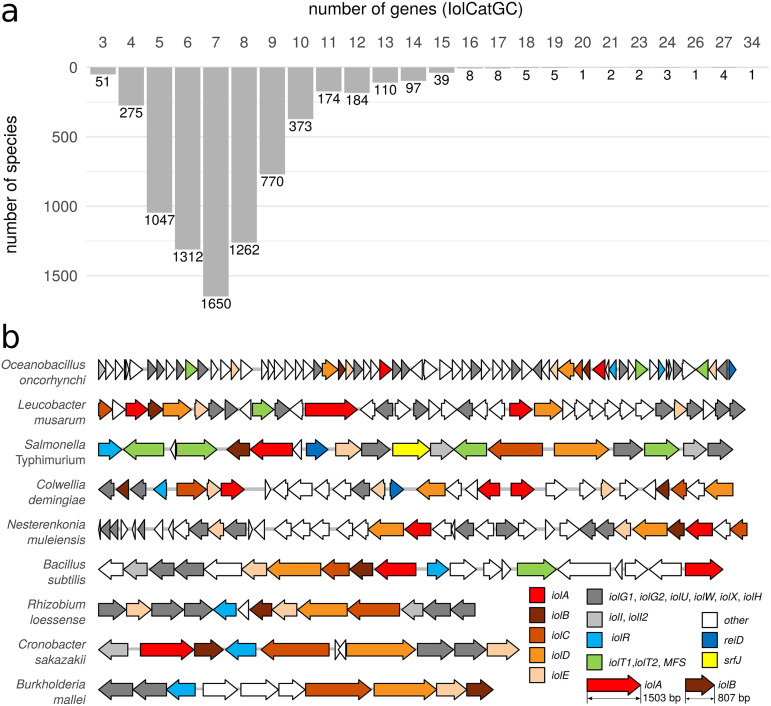
We observed a wide range of complexity of IolCatGCs with respect to the number of operons and genes associated with inositol degradation. The vast majority, namely, 6,689 of the 7,384 IolCatGCs investigated, here, comprise 4 to 10 genes, while only 644 IolCatGCs comprise 11 to 34 genes (Fig. 2a). In addition to the core gene set, larger clusters contain genes coding for regulators, transporters, dehydrogenases, hypothetical proteins with related metabolic functions, and duplications thereof. These clusters appear to be organized as a divergon comprising at least two operons and promoters.
FIG 2.
Complexity of the genetic IolCatGC organization. (a) The size of IolCatGCs in the genomes of bacterial species analyzed here is indicated by gene numbers. The species were considered that carry at least 3 and up to 34 genes within their IolCatGCs. The gene number was determined by counting genes encoding enzymes, regulators, transporters, and SrfJ, known or predicted to be involved in inositol utilization. Genes encoding transposases and hypothetical proteins or transporters without homology to putative Iol-degrading enzymes or characterized MI facilitators were not considered for defining the IolCatGC lengths. (b) Randomly selected IolCatGCs are depicted to illustrate their variability in length and composition. Regulators are stained blue; transporters, green; core genes, orange; and isomerases and dehydrogenases, gray. Numbers in brackets indicate the number of iol genes. White genes are genes of unknown function or not associated with inositol utilization.
The genome of Oceanobacillus oncorhynchi carries a 76-kb fragment with 34 genes predicted to be involved in inositol degradation. Many functions, including those of the two regulators ReiD and IolR, which play a pivotal role in the regulation of MI degradation (19, 22, 23), are encoded twice, and 12 putative dehydrogenases providing the substrate for IolE were identified (Fig. 2b). Further examples randomly chosen are the IolCatGC from Leucobacter musarum, which was isolated from a nematode; Colwellia demingiae, a psychrophilic Antarctic species; and Nesterenkonia muleiensis, a novel actinobacterium isolated from Populus euphratica. Species of the latter genus are found in soil and are characterized by a high metabolic versatility. A more compact IolCatGC organization was found in the genomes of Rhizobium loessense, a root nodule bacterium; Cronobacter sakazakii, a foodborne pathogen; and Burkholderia mallei, a pathogen causing glanders. The IolCatGC of Salmonella enterica is more complex with respect to operon organization and gene numbers than most others. Its canonical iol gene cluster (17), together with that of B. subtilis (24), is also shown in Fig. 2b.
Percentage of IolCatGC-positive genomes per species.
To exclude that a single genome erroneously represents a large number of IolCatGC-negative genomes, we analyzed the abundance of iol genes within all genomes available for each species. Out of the 7,384 species, 1,453 are represented by at least two genome sequences (Table S3a). In this group, we determined a high variability of a species pangenome with respect to the presence of iol genes. Regarding the 319 species for which at least 10 genome sequences met the quality criteria, 288 of them are represented by ≥0%, 246 by ≥50%, and 228 by ≥70% IolCatGC-positive genomes, respectively (Fig. S1; Table S3b). More than 1,000 genome sequences are available for 10 species (Table S3c). Among them are Listeria monocytogenes, Klebsiella pneumonia, Burkholderia pseudomallei, and Pseudomonas viridiflava, with at least 97% iol gene-positive sequences per species. Less than 2% of all 20,632 E. coli genome sequences comprise the IolCatGC.
IolCatGCs encoding the activator ReiD and the putative ceramidase SrfJ.
Of particular interest are IolCatGC genes that are absent in the majority of IolCatGCs and encode accessory functions not essential for MI degradation. The most interesting ones are reiD and srfJ, which code for a regulator and a putative ceramidase, respectively. ReiD belongs to the AraC family of transcriptional regulators and was experimentally determined to activate the transcription of iolE and iolG1, the genes that encode the enzymes responsible for the initial two steps in MI degradation (19). Gene srfJ is known to be activated by the two-component system SsrAB, which controls the expression of SPI-2 genes (25, 26). Its product shows a remarkable similarity to human lysosomal glycosylceramidases (E value of 4 × 10−62; 30.8% amino acid identity) and might be involved in the release of inositols from sphingolipids. Homologs of ReiD were found in 217 species and those of SrfJ in 10 species among the list of 7,384 genomes with IolCatGCs, indicating that both factors are not widespread among bacteria. Remarkably, 10 IolCatGCs harbor both genes (Fig. 3). The clusters of Citrobacter werkmanii and Mangrovibacter spp. are identical to that of Salmonella and nearly colinear in E. coli, Achromobacter spp., and Gibbsiella quercinecans. In comparison with S. Typhimurium, the genomes of Gibbsiella quercinecans, Achromobacter sp., and Escherichia coli ED1a are lacking the two genes iolI2 and iolH, and that of G. quercinecans a fourth transporter gene. The data suggest that additional regulatory, as well as enzymatic, determinants associated with MI degradation provide a benefit for some bacteria in their ecological niches.
FIG 3.
Complex IolCatGCs that harbor both the transcriptional activator gene reiD and the putative ceramidase gene srfJ. All IolCatGCs are shown that both carry the activator gene reiD and the putative ceramidase-encoding gene srfJ. The color code is the same as in Fig. 2. Genes encode transporters (green), core factors IolBCDE (orange), regulators (blue), the malonate-semialdehyde dehydrogenase IolA (red), the fructose-1,6-biphosphate aldolase IolJ (black), SrfJ (yellow), uncharacterized isomerases and dehydrogenases (gray), as well as transposases, insertion elements, genes not associated with MI utilization, and unknown genes (white). MFS, major facilitator superfamily; csbX, gene that encodes MFS efflux pump (69); xynC, srfJ homolog that encodes a glucuronoxylanase (70); rhaS, gene that codes for an activator of the l-rhamnose operon (71); glpR, gene coding for a repressor of sugar metabolism pathways (72); gntR, gene that encodes repressor of gluconate operon (73); hyp, hypothetical gene. Slashes indicate contig ends. Strain identification numbers or names are given. The classes the strains belong to are mentioned on the left.
IolCatGC gene order.
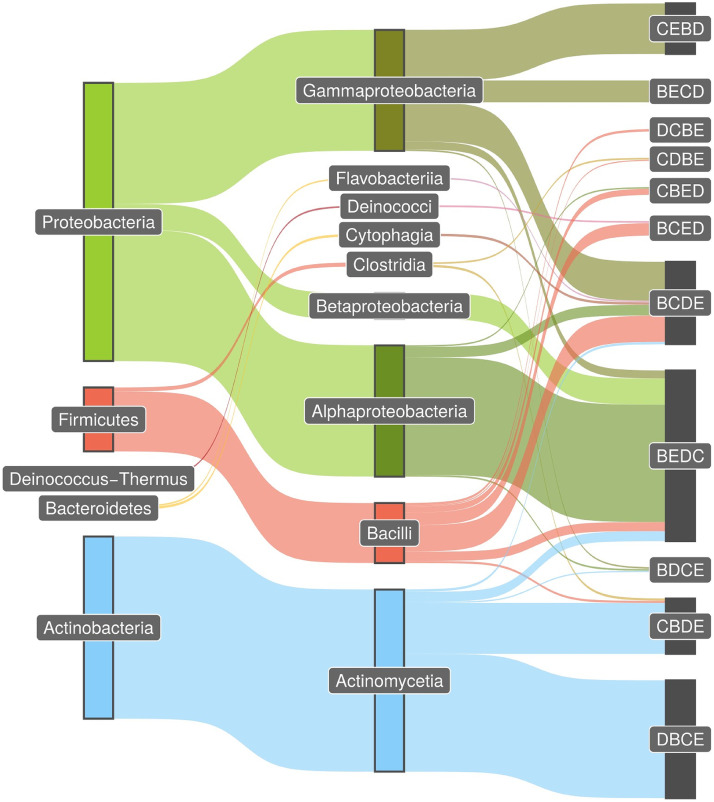
Next, we investigated whether there are distinct patterns of IolCatGCs with respect to the genetic organization of the four core genes iolB, iolC, iolD, and iolE. Within the genomes of species belonging to the same genus, we predominantly observed collinearity of the genes involved in MI metabolism, pointing to a high degree of conservation on this taxonomic level (data not shown). Performing a systematic phyla evaluation (Fig. 4), we found that 65% of all IolCatGC-positive species belonging to Actinobacteria harbor the pattern iolD-iolB-iolC-iolE (DBCE) on their genomes, and 28% have the pattern CBDE (Table S4). The most common iol gene orders were BEDC in Alphaproteobacteria (42%) and CEBD (18%) and BCDE (14%) in Gammaproteobacteria. In Firmicutes, we mainly identified the gene orders BCDE, BCED, and BEDC, corresponding to 41%, 20%, and 14%, respectively, of all species in this phylum. The pattern BCDE was present in all genomes belonging to the classes Cytophagia and Flavobacteria (each 44%) of Bacteroidetes and was also common in Chloroflexi (data not shown). Thus, the iol gene orders BCDE and BEDC are interphylum patterns present in Gram-negative, as well as Gram-positive, bacteria.
FIG 4.
Patterns of iol gene order. The Sankey diagram was employed with the R package ggsankey and shows the most common order pattern of the core genes iolB, iolC, iolD, and iolE in the phylum and class levels as taxonomic nodes. The thickness of the connections corresponds with the relative proportion of species genomes carrying the same pattern. Genomes from five phyla were analyzed. In total, we found IolCatGCs in 6,026 species with unique pattern hits (81.6%) and in 548 (7.4%) species with multiple matches. Multiple matches are mainly due to the occurrence of multiple core genes in the cluster and could not be unequivocally assigned to one permutation type. A minimum of 10 species per type were required to draw a connection between taxonomic class and cluster type. In summary, we show connections for 80% (5,879) of the species in our results table.
To summarize, we observed a close relationship between taxonomy and pattern type, and we identified the pattern BEDC as the most abundant and widespread one. These data point to a highly conserved, ancient pathway that further evolved by the acquisition of regulatory, transporter, and nonessential enzymatic genes.
Taxonomic prevalence of IolCatGC.
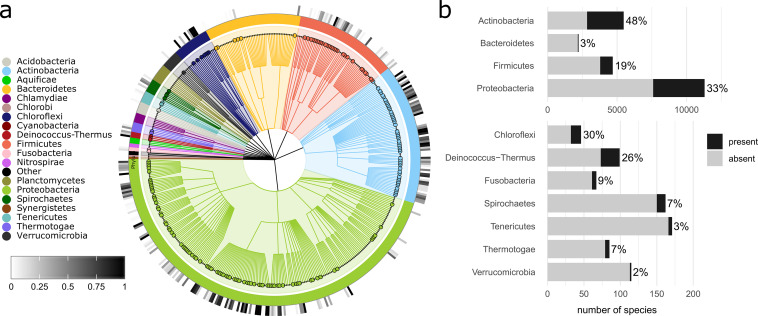
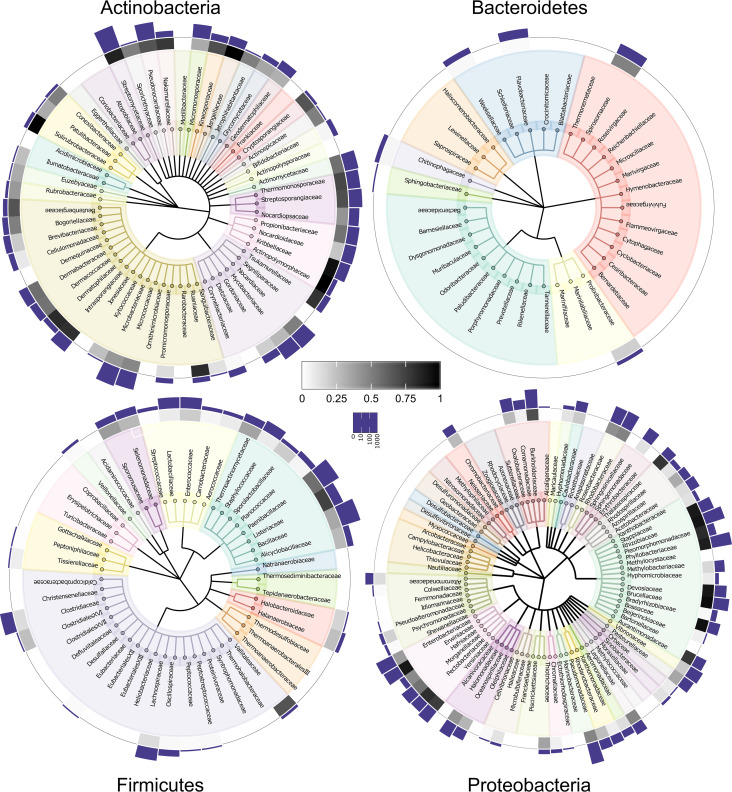
To examine the distribution of iol-positive species within different taxonomic levels of bacteria, we defined a species as positive with respect to inositol catabolism if at least one genome per species carried an IolCatGC. We found IolCatGCs in species belonging to 12 out of 21 phyla (Fig. 5a; Table S5), with substantial proportions in Actinobacteria (48% out of 5,473 species), Proteobacteria (33% of 11,297), and Firmicutes (19% of 4,686). In the next largest phylum, Bacteroidetes, IolCatGCs were found in the genomes of only 3% out of 2,227 species (Fig. 5b). These data indicate an uneven distribution of IolCatGCs at the phylum level.
FIG 5.
Prevalence of IolCatGCs among the bacterial kingdom. (a) Cladogram of analyzed bacterial species showing the prevalence of IolCatGCs at the family level. To filter out less relevant branches, only families with at least 10 species were considered. The taxonomic hierarchy includes phylum, class, order, and family (from inside to outside). Filled circles indicate families of which at least 10% of their species are IolCatGC positive. The outer circle heatmap indicates the percentage of species within a family that are IolCatGC positive. The figure was generated with GraPhlAn (version 1.1.4). (b) Bar plot indicating absolute and relative number of IolCatGC-positive species within selected bacterial phyla. The four largest phyla are shown in the upper part. The bar plot was generated with the help of R package ggplot2 (version 3.3.2).
On the family level, we identified IolCatGCs in 202 out of 473 bacterial families (Fig. 6). Analyzing the three largest families of the phylum Actinobacteria, we detected 87% IolCatGC positive out of 1,235 species of Streptomycetaceae dominated by the genus Streptomyces, 41% of 789 species belonging to Microbacteriaceae, and 28% of 395 species of Mycobacteriaceae. Fifty-six percent of the species belonging to Alphaproteobacteria and Gammaproteobacteria carry IolCatGCs. In particular, we identified iol genes in Pseudomonadaceae (48% of 1,221 species), Enterobacteriaceae (39% of 594 species), Phyllobacteriaceae (88% of 524 species), Burkholderiaceae (69% of 474 species), Rhizobiaceae (92% of 451 species), and Vibrionaceae (21% of 381 species). With respect to the phylum Firmicutes, we found IolCatGCs predominantly in Bacillaceae (34% of 1,078 species) and Paenibacillaceae (56% of 441 species) and, to lesser extent, in Lactobacillaceae (8% of 392 species), Clostridiaceae (14% of 352 species), Lachnospiraceae (14% of 324 species), and Staphylococcaceae (8% of 306 species). In Bacteroidetes, the capability of utilizing MI is less prevalent. IolCatGC-positive genomes were found in Flavobacteriaceae (2% of 810 species), Weeksellaceae (3% of 215 species), and Spirosomaceae (49% of 55 species), whereas no iol gene clusters were found in other 44 families, including the large families Bacteroidaceae and Prevotellaceae.
FIG 6.
Families predicted to degrade MI. Families comprising at least 2 (Actinobacteria, Bacteroidetes, and Firmicutes) or at least 10 (Proteobacteria) species are indicated. The bars in the logarithmic scale show the total number, and the heatmap shows the percentage of species within a family that are IolCatGC positive. Scales are indicated.
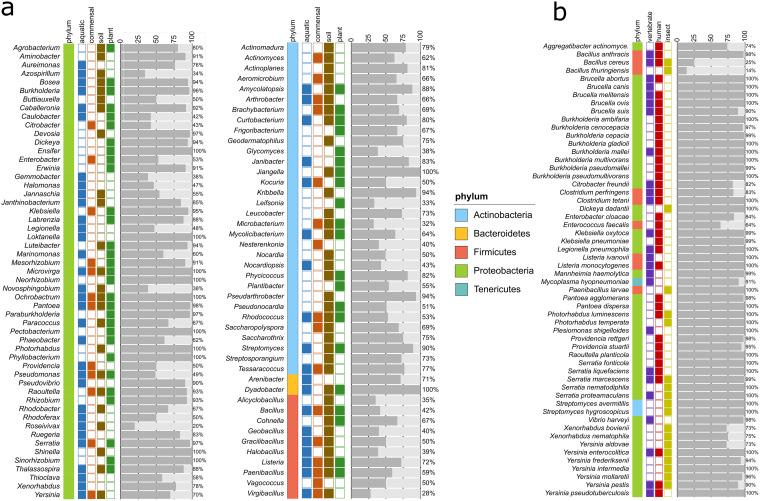
The 7,384 species identified above belong to 776 genera (Table S6a). To analyze the most relevant ones with respect to inositol metabolism, we selected all genera that comprise at least 10 species of which ≥30% carry the IolCatGCs (Table S6b). These criteria are fulfilled by 97 genera (Fig. 7a). For 86 of them, at least 40% of their species are positive with respect to the iol gene cluster. Examples from this group are genera with at least 200 species, such as Pseudomonas, Streptomyces, Bacillus, Mesorhizobium, Paenibacillus, Rhizobium, Burkholderia, and Rhodococcus.
FIG 7.
Distribution of the MI pathway at the genus level and in pathogenic bacteria. (a) Fraction of species within selected genera that carry an IolCatGC. Bacterial genera to which at least 10 species belong to, and of which ≥30% are IolCatGC positive, were selected. The number of species that carry (dark gray) or lack (light gray) the four core iol genes are shown. The site of sampling and/or typical ecological niche is indicated by colors. (b) Column plot showing the percentage of IolCatGC in all genomes of selected animal and human bacterial pathogens. Species for which at least 10 (insect pathogens, 5) genome sequences were available and of which at least 50% (with the exception of Bacillus cereus) were predicted as IolCatGC positive are shown.
Taken together, the high abundance of IolCatGCs within these species-rich and other genera shown in Table S3 points out the relevance of MI degradation in particular for bacteria that are found in soil, water, decaying vegetation, the rhizosphere, or in association with plants. In these environments, phytate and MI are present in large amounts as accessible carbon and energy sources for bacteria.
Plant pathogens and rhizosphere bacteria with iol genes.
From the list of IolCatGC-positive bacteria, we identified 82 species that are known as plant-pathogenic bacteria (27–29), most of them belonging to the genera Agrobacterium, Burkholderia, Dickeya, Dyadobacter, Erwinia, Pantoea, Pectobacterium, Pseudomonas, and Ralstonia (Table S7a). Among them are six of the top ten plant pathogens (30), namely A. tumefaciens, D. dadantii/solani, E. amylovora, P. carotovorum, P. syringae, and R. solanacearum (Fig. 7a). Another interesting group with respect to MI utilization are bacteria from the rhizosphere that promote plant growth by forming symbiotic root nodules. Six relevant genera from this group were therefore investigated for genomes with the capability of utilizing MI. Strikingly, IolCatGCs are highly prevalent in the genomes of 348 species (Table S7b) mainly belonging to Caballeronia (40 IolCatGC-positive genomes, corresponding to 93%), Ensifer (61 IolCatGC-positive genomes, corresponding to 100%), Mesorhizobium (459 IolCatGC-positive genomes, corresponding to 99%), Paraburkholderia (210 IolCatGC-positive genomes, corresponding to 100%), Rhizobium (829 IolCatGC-positive genomes, corresponding to 97%), and Sinorhizobium (305 IolCatGC-positive genomes, corresponding to 94%). To conclude, we hypothesize that these plant-associated bacteria gain a growth advantage by their inositol degradation capability in the phytate-rich plant environment.
Commensals capable of degrading MI.
Given that phytate and inositol derivatives contribute to the diet and are ubiquitously present in the gut, we determined which and how many members of the microbiota in the gut carry IolCatGCs. For this purpose, we compiled a list of intestinal and rumen bacteria from humans, pigs, and cattle (Table S8). Our human gut microbiota list comprises a total of 807 nonredundant bacterial species (31, 32) and 465 swine species, including data from a DSMZ list (pig intestinal bacterial collection [PiBAC]) (33–37). A cattle reference list of 485 nonredundant rumen species was composed of the Hungate collection and genome sequencing projects (38–40). Remarkably, a high percentage of these species were found to carry IolCatGCs, namely, 16.6% of the human gut species, 10.3% of those from the swine gut, and 10.9% of all ruminal bacterial species identified so far. Examples of genera with iol-positive species among these commensals are Bacillus, Blautia, Citrobacter, Clostridium, Corynebacterium, Enterobacter, Enterococcus, Klebsiella, and Paenibacillus (Fig. 7a). IolCatGCs are absent, however, in Faecalibacterium prausnitzii in all but one Bacteroides spp., all but three Bifidobacterium spp., Caprococcus spp., and Akkermansia municiphila. From these data, we conclude that a substantial percentage of gut commensals, namely, at least 10%, are capable to degrade inositols and use them as an alternative substrate in the gut. Moreover, it might be speculated that the fraction of IolCatGC-positive commensals in the gut directly depends on the diet and the metabolic state of the host.
Vertebrate pathogens harboring IolCatGC.
To identify bacterial pathogens equipped with iol genes, we compared Table S3 with a list of 636 species belonging to biosafety level 2 and 3 groups (Table S7c), resulting in a list of 87 species that are known as vertebrate pathogens, some of which are categorized as opportunistic, rare, or emerging pathogens (Table S7d). For an overview, we selected those 41 species for which ≥ 10 genomes were available, of which ≥ 50% carried IolCatGC (Fig. 7b; Table S7e and f). The genera Brucella, Burkholderia, Clostridium, Klebsiella, Listeria, Pantoea, Providencia, Serratia, and Yersinia are represented by two to eight species fulfilling these requirements. In addition, relevant pathogenic species such as Citrobacter freundii, Enterobacter cloacae, Enterococcus faecalis, Legionella pneumophila, Mannheimia haemolytica, S. enterica, and Vibrio harveyi were identified as encoding the capacity to utilize MI. Notably, most of those are enteropathogens. While 36% of all E. faecalis genomes do not carry iol genes, the genome sets of the other 41 pathogenic species shown in Fig. 7b with a proportion of ≥90% IolCatGC-positive sequences are much more coherent with respect to MI utilization. It is worth noting that the genes required for MI utilization are missing in the genera Bartonella, Bordetella, Borrelia, Campylobacter, Chlamydia, Mycobacterium, Rickettsia, and Streptococcus and in most Staphylococcus spp., including Staphylococcus aureus, all of which do not belong to Enterobacteriaceae and, with the exception of Campylobacter, do not proliferate in the gut.
To complement this survey on pathogens, we investigated the genomes of relevant bacterial insect pathogens (41–43) for the presence of IolCatGCs. We identified 20 entomopathogenic species without and 24 species with iol genes (Table S7g; Fig. 7b), among them, Bacillus thuringiensis, Photorhabdus luminescens, and Xenorhabdus nematophila, which play pivotal roles in pest control approaches. Paenibacillus larvae, the etiological agent of the American foul brood, colonizes the gut of honeybees and also carries an IolCatGC. Six insect-pathogenic Yersinia spp. (44) point to the fact that the interaction of species with invertebrates is, as of yet, underinvestigated.
Taken together, we identified a high number of pathogenic bacteria that are probably able to utilize MI and its derivatives. Given that the examples from above infect their hosts via the gut, lung, or bloodstream, we assume that the utilization of MI, which is present in food as well as in membrane compounds, provides a fitness advantage in different compartments of host organisms.
MI utilization by archaea or fungi.
Fungi, in particular, yeasts, have been reported to utilize MI (45). For example, Cryptococcus neoformans is known to grow with inositol as a sole carbon and energy source (46). In contrast to the enzymes encoded by IolCatGC, the environmental and pathogenic yeast relies on an inositol oxygenase activity that is responsible for the conversion of inositol to d-glucuronic acid (47). However, to the best of our knowledge, there is no literature that describes a fungal MI degradation pathway with enzymatic functions similar to those in bacteria. It was reported that inositol induces the sporulation response of Beauveria bassiana and Metarhizium anisopliae (48). The genome of B. bassiana encodes proteins with similarity to IolR, IolG1, IolG2, and IolE (~25% sequence identity each); IolC (31%); IolD (45%); and IolB (32%) of Actinomyces ruminicola, but a reiterated search of these fungi-specific protein sequences did not result in increasing similarity rates. By analyzing 1,511 genomes of Archaebacteria, homologs of enzymes involved in MI degradation were identified in two genomes each of Halobacteriales archaeon and Desulfurococcaceae archaeon and in Thermocladium modestius (Table S8). Some of these sequences were derived from metagenomics, and experimental evidence for a functionality of the respective IolCatGC was not found.
We therefore conclude that, so far, the capability of degrading MI via IolCatGCs is restricted to members of the kingdom Eubacteria.
DISCUSSION
The extensive genome sequences survey performed here required the public availability of hundreds of thousands of genome sequences, a well-adapted bioinformatics annotation pipeline, and the necessary parallel computing power. To our knowledge, this is one of the first comprehensive approaches that deciphers the prevalence and distribution of a single metabolic pathway across 24,812 bacterial species. A study addressing the highly complex production of the vitamin B12 family of cofactors investigated 11,000 bacterial species by comparative genomics (49). The annotation of IolCatGCs was based on stringent criteria to ensure high reliability of the results, including the definition of cluster core genes, a stringent cutoff for the HMM similarity score, and filtering for proximal localization of the annotated cluster components. The bioinformatics pipeline established here can be used to predict the prevalence of a selected bacterial pathway, genetic island, virulence factor, or any other genetic determinant in all taxonomic levels of bacteria. Moreover, the pipeline applied here and its data output can seamlessly be integrated into metagenomic studies.
Inositol phosphates accumulate in terrestrial environments where they constitute the major class of organic phosphorus compounds and are also present to great extent in aquatic environments (5). Thus, our finding that a huge number of bacterial genomes attributed to genera mainly found in the environment are potentially able to utilize MI is consistent with the presence of phytate and inositols in the environment (Fig. 6; see Table S6 in the supplemental material). Examples are Pseudomonas, Streptomyces, Bacillus, Paenibacillus, Clostridium, and Halomonas from saline environments and genera belonging to the ubiquitous Actinobacteria (Amycolatopsis, Actinomadura, Actinomyces, Curtobacterium, Gordonia, Mycobacterium, and Rathayibacter). Of particular interest is the microbiota of the rhizosphere for two reasons, namely, the availability of MI to promote bacterial growth and the dephosphorylation of phytate as organic phosphorus source (50). As expected, a majority of genomes belonging to main plant-associated bacterial genera carry the information to utilize MI. A benefit of inositol degradation was demonstrated for Rhizobium leguminosarum (7). This study supports the assumption that MI catabolism plays an important role for R. leguminosarum symbiosis with plants.
InsPs and phytates are widespread in organisms and in the diet, respectively, and MI is present in substantial amounts in the gut of humans and animals (51, 52). Indeed, we predicted more than 10% of commensal bacteria to be capable of utilizing MI for proliferation, indicating that IolCatGC is critical for some microbes to occupy microenvironments in the gut or to circumvent a depletion of other nutrients. In line with the substantial amount of IolCatGC-positive species frequently found in the gut, a metatranscriptome approach provided evidence that the capability of utilizing MI contributes to the bacterial fitness in a model of human gut microbial succession (53). When the metabolism of gut microbiota from mice was investigated by a metagenomic and metatranscriptomic approach, many iol genes were found to be differentially regulated in the presence of three antibiotics (54). These data corroborate iol gene activation in commensals and the functional role of the MI metabolism in the gut. Moreover, different amounts of mineral phosphorus and microbial phytases fed to chicken shaped the composition of their microbiota (55), and vegetarians’ microbiota revealed degradation of up to 100% phytate to MI-phosphate products lower than InsP3 (56). Therefore, it might be expected that nutrition affects the microbiological profile of gut microbiota in a IolCatGC-related manner. The two commensal species, Mitsuokella jalaludinii and Mitsuokella multacida, which are present in the porcine gastrointestinal tract as well as in the rumen of cattle (Tables S2 and S7), have been identified as antagonists of S. Typhimurium (57). As Mitsuokella spp. are able to degrade MI, it is tempting to speculate that the mechanism underlying this antagonism is based on a metabolic competition, which includes MI utilization.
Several pathogenic bacteria, mainly enteropathogens, are able to utilize MI and might thus gain an adaptive growth advantage during proliferation in gut niches in which other nutrients are not available or provide less energy than the polyol. Indeed, a transposon-directed insertion site sequencing (TraDIS)-based approach that systematically tested an S. Typhimurium transposon mutant library in chicken, pigs, and calves pointed to a strong attenuation of several iol gene mutants in enteritis models of these animals (58). Experimental data demonstrate that Legionella pneumophila utilizes MI to promote its infection of amoebae and macrophages (12).
Conclusion.
Our comprehensive search for iol gene clusters exploited nearly 200,000 bacterial genomes and revealed that this metabolic capacity is more widely distributed among the bacterial kingdom than thought so far. Analysis of all bacterial taxonomic levels revealed an uneven distribution of the MI degradation pathway. Many soil and rhizosphere bacteria carry IolCatGCs in their genomes and benefit from this pathway due to the high concentration of phytate and inositol isomers in the environment. Remarkably, 10% to 16% of the human and animal microbiota members were identified as being capable of degrading MI, pointing to a metabolic niche that provides a growth advantage for gut bacteria. The presence of conserved iol gene clusters in one-quarter of all bacterial species sequenced so far strongly suggests that the MI degradation pathway plays a yet underestimated role in the metabolism and ecology of bacteria.
MATERIALS AND METHODS
Selection of genome assemblies.
Bacterial genome assemblies were downloaded from GenBank (accessed August 2020) using the public NCBI FTP server (ftp://ftp.ncbi.nlm.nih.gov/genomes/), including the nucleotide sequence fasta (fna), the annotated feature file (gff), and the translated protein sequences (faa) for each accession number. Assemblies were discarded if they matched a RefSeq exclusion criterion (https://www.ncbi.nlm.nih.gov/assembly/help/anomnotrefseq/). Genome assemblies with missing gene annotation files (50,960) were further analyzed for protein-coding sequences using Prokka (version 1.14.5) with default parameters (21).
Genome data analysis.
The genome sequence of S. Typhimurium strain 14028 (GenBank accession no. NC_003197) was used to select the iol genes (iolA, iolB, iolC, iolD, iolE, iolG1, iolG2, iolH, iolI, iolI2, srfJ, reiD, iolR, iolT1, and iolT2) as an input (17). The genes iolX, iolW, iolU, iolJ, and iolS were taken from B. subtilis (GenBank accession no. NC_000964) (59, 60). For each gene associated with MI utilization (see Table S1 in the supplemental material), the following steps were applied. Translated protein sequences were queried against the NCBI nonredundant (nr) database using PSI-BLAST (61) with a total of 5 iterations; a cutoff of 70% percentage identity and 70% sequence coverage was chosen to select for highly similar sequences. The resulting list of orthologous protein sequences was aligned with the tool MUSCLE (62, 63), and the resulting multiple alignment was loaded into hmmbuild of package hmmer (version 3.3.2) to generate an iol gene-specific hidden Markov model (HMM) (64). In total, we obtained 23 HMMs for the gene cluster identification (Table S9).
IolCatGC screening.
To implement a large-scale high-throughput search for IolCatGCs, we developed a custom R annotation pipeline which is based on a series of processing steps. For each genome, this included an import of the genome assembly file (gff) and translated protein sequences (faa) into R using packages rtracklayer and Biostrings (65). Next, hmmsearch applied the precomputed HMMs on the imported protein sequences with default parameters. Gram-positive strains were additionally queried with models iolX, iolW, iolU, iolJ, and iolS. All resulting hits were filtered by a stringent E value cutoff of 10−10. Next, all iol gene hits were assigned to gene clusters, which were identified by a minimum distance of 10 kbp between two consecutive hit genes on the same contig sequence. One representative cluster with the largest number of iol core genes (iolC, iolB, IolD, and iolE) and the largest number of total unique iol genes was selected for the assembly. This annotation procedure was applied to all input genome assemblies by using multicore functions of R package parallel.
IolCatGC selection at the species level.
In order to select the most representative gene cluster of a species, we implemented a selection method which ranks the identified IolCatGC according to the following criteria: the most frequent IolCatGC size across all genomes per species, average HMM score, and a minimum number of three iol core genes. The top-ranked genome is considered the candidate IolCatGC representative genome for each species. The entire summary table, including information about all species, candidate IolCatGC size, and frequency of occurrence, is available as Table S2. Gene maps were generated with R package gggenes to display the composition and structure of selected clusters (66).
Taxonomic analysis.
We used the taxonomic data supplied by the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy). The two central database files, nodes.dmp and names.dmp, were downloaded (on 25 February 2022) and processed by the R package taxonomizr (version 0.5.3; https://github.com/sherrillmix/taxonomizr). These files provide hierarchical relationships between the taxonomic identity of species and strains and the respective taxonomic levels, including genus, family, order, class, and phylum. Mapping functions of the package provided the assignment of genome species IDs to their respective taxonomy. Only genomes with taxonomic assignments on all five levels were retained. Candidate species were not considered. All phylogenetic figures were generated with GraPhlAn (version 1.1.4) (67), requiring the generation of tree and annotation files as described (https://github.com/biobakery/graphlan).
Cluster analysis.
The cluster analysis was done by comparing the core gene order in all IolCatGCs. To implement this comparison, we generated all possible permutations of the four core genes (iolB, iolC, iolD, and iolE [BCDE]) and removed reverse duplicates. As the gene sequence can be interrupted by other noncore genes, we used the LCS algorithm from R package qualV in forward and reverse directions to screen for clusters which are structured in the same order (68).
Data availability.
All data are available in the tables in the supplemental material.
ACKNOWLEDGMENT
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael Weber, Email: michael.weber@fli.de.
Thilo M. Fuchs, Email: thilom.fuchs@fli.de.
Yanbin Yin, University of Nebraska—Lincoln.
REFERENCES
- 1.Michell RH. 2008. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol 9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 2.Peperzak P, Caldwell AG, Hunziker RR, Black CA. 1959. Phosphorus fractions in manures. Soil Science 87:293–302. doi: 10.1097/00010694-195905000-00010. [DOI] [Google Scholar]
- 3.Huber K. 2016. Cellular myo-inositol metabolism, p 53–60. In Walk CL, Kühn I, Stein HH, Kidd MT, Rodehutscord M (ed), Phytate destruction: consequences for precision animal nutrition. Wageningen Academic Publishers, Wageningen, Netherlands. [Google Scholar]
- 4.Heyer CM, Weiss E, Schmucker S, Rodehutscord M, Hoelzle LE, Mosenthin R, Stefanski V. 2015. The impact of phosphorus on the immune system and the intestinal microbiota with special focus on the pig. Nutr Res Rev 28:67–82. doi: 10.1017/S0954422415000049. [DOI] [PubMed] [Google Scholar]
- 5.Turner BL, Paphazy MJ, Haygarth PM, McKelvie ID. 2002. Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci 357:449–469. doi: 10.1098/rstb.2001.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staib L, Fuchs TM. 2014. From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology (Reading) 160:1020–1039. doi: 10.1099/mic.0.078105-0. [DOI] [PubMed] [Google Scholar]
- 7.Fry J, Wood M, Poole PS. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol Plant Microbe Interact 14:1016–1025. doi: 10.1094/MPMI.2001.14.8.1016. [DOI] [PubMed] [Google Scholar]
- 8.Galbraith MP, Feng SF, Borneman J, Triplett EW, de Bruijn FJ, Rossbachl S. 1998. A functional myo-inositol catabolism pathway is essential for rhizopine utilization by Sinorhizobium meliloti. Microbiology 144:2915–2924. doi: 10.1099/00221287-144-10-2915. [DOI] [PubMed] [Google Scholar]
- 9.Yebra MJ, Zuniga M, Beaufils S, Perez-Martinez G, Deutscher J, Monedero V. 2007. Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl Environ Microbiol 73:3850–3858. doi: 10.1128/AEM.00243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman T, Magasanik B. 1966. The pathway of myo-inositol degradation in Aerobacter aerogenes. Ring scission. J Biol Chem 241:807–813. doi: 10.1016/S0021-9258(18)96837-7. [DOI] [PubMed] [Google Scholar]
- 11.Krings E, Krumbach K, Bathe B, Kelle R, Wendisch VF, Sahm H, Eggeling L. 2006. Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on L-lysine formation. J Bacteriol 188:8054–8061. doi: 10.1128/JB.00935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manske C, Schell U, Hilbi H. 2016. Metabolism of myo-inositol by Legionella pneumophila promotes infection of amoebae and macrophages. Appl Environ Microbiol 82:5000–5014. doi: 10.1128/AEM.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wauters G, Janssens M, Steigerwalt AG, Brenner DJ. 1988. Yersinia mollaretii sp. nov. and Yersinia bercovieri sp. nov., formerly called Yersinia enterocolitica biogroups 3A and 3B. Int J Syst Evol Microbiol 38:424–429. doi: 10.1099/00207713-38-4-424. [DOI] [Google Scholar]
- 14.Yuan C, Yang P, Wang J, Jiang L. 2019. Myo-inositol utilization by Citrobacter koseri promotes brain infection. Biochem Biophys Res Commun 517:427–432. doi: 10.1016/j.bbrc.2019.07.112. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida KI, Aoyama D, Ishio I, Shibayama T, Fujita Y. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J Bacteriol 179:4591–4598. doi: 10.1128/jb.179.14.4591-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida K, Yamamoto Y, Omae K, Yamamoto M, Fujita Y. 2002. Identification of two myo-inositol transporter genes of Bacillus subtilis. J Bacteriol 184:983–991. doi: 10.1128/jb.184.4.983-991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröger C, Fuchs TM. 2009. Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. J Bacteriol 191:545–554. doi: 10.1128/JB.01253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kröger C, Rothhardt JE, Brokatzky D, Felsl A, Kary SC, Heermann R, Fuchs TM. 2018. The small RNA RssR regulates myo-inositol degradation by Salmonella enterica. Sci Rep 8:17739. doi: 10.1038/s41598-018-35784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothhardt JE, Kröger C, Broadley SP, Fuchs TM. 2014. The orphan regulator ReiD of Salmonella enterica is essential for myo-inositol utilization. Mol Microbiol 94:700–712. doi: 10.1111/mmi.12788. [DOI] [PubMed] [Google Scholar]
- 20.Fry J, Poole PS, Wood M. 1998. myo-inositol utilisation by Rhizobium leguminosarum. In Elmerich C, Kondorosi A, Newton WE (ed), Biological nitrogen fixation for the 21st century. Current plant science and biotechnology in agriculture, vol. 31. Springer Dordrecht, Dordrecht, Netherlands. [Google Scholar]
- 21.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Hellinckx J, Heermann R, Felsl A, Fuchs TM. 2017. High binding activity of repressor IolR avoids costs of untimely induction of myo-inositol utilization by Salmonella Typhimurium. Sci Rep 7. doi: 10.1038/srep44362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kröger C, Srikumar S, Ellwart J, Fuchs TM. 2011. Bistability in myo-inositol utilization by Salmonella enterica serovar Typhimurium. J Bacteriol 193:1427–1435. doi: 10.1128/JB.00043-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida K, Yamaguchi M, Morinaga T, Kinehara M, Ikeuchi M, Ashida H, Fujita Y. 2008. myo-Inositol catabolism in Bacillus subtilis. J Biol Chem 283:10415–10424. doi: 10.1074/jbc.M708043200. [DOI] [PubMed] [Google Scholar]
- 25.Worley MJ, Ching KH, Heffron F. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol 36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 26.Cordero-Alba M, Bernal-Bayard J, Ramos-Morales F. 2012. SrfJ, a Salmonella type III secretion system effector regulated by PhoP, RcsB, and IolR. J Bacteriol 194:4226–4236. doi: 10.1128/JB.00173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrios GN. 2005. Plant pathology, 5th ed. Elsevier Academic Press, New York. [Google Scholar]
- 28.Kim JS, Yoon SJ, Park YJ, Kim SY, Ryu CM. 2020. Crossing the kingdom border: human diseases caused by plant pathogens. Environ Microbiol 22:2485–2495. doi: 10.1111/1462-2920.15028. [DOI] [PubMed] [Google Scholar]
- 29.Pscheidt JW. 2005. Pacific Northwest plant disease management handbook. Oregon State University, Corvallis, OR. [Google Scholar]
- 30.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, Sun H, Xia Y, Liang S, Dai Y, Wan D, Jiang R, Su L, Feng Q, Jie Z, Guo T, Xia Z, Liu C, Yu J, Lin Y, Tang S, Huo G, Xu X, Hou Y, Liu X, Wang J, Yang H, Kristiansen K, Li J, Jia H, Xiao L. 2019. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 37:179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. 2021. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 39:105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao L, Estelle J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AO, Kjeldsen NJ, Liu C, Maguin E, Dore J, Pons N, Le Chatelier E, Prifti E, Li J, Jia H, Liu X, Xu X, Ehrlich SD, Madsen L, Kristiansen K, Rogel-Gaillard C, Wang J. 2016. A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 34.Wylensek D, Hitch TCA, Riedel T, Afrizal A, Kumar N, Wortmann E, Liu T, Devendran S, Lesker TR, Hernandez SB, Heine V, Buhl EM, M D'Agostino P, Cumbo F, Fischoder T, Wyschkon M, Looft T, Parreira VR, Abt B, Doden HL, Ly L, Alves JMP, Reichlin M, Flisikowski K, Suarez LN, Neumann AP, Suen G, de Wouters T, Rohn S, Lagkouvardos I, Allen-Vercoe E, Sproer C, Bunk B, Taverne-Thiele AJ, Giesbers M, Wells JM, Neuhaus K, Schnieke A, Cava F, Segata N, Elling L, Strowig T, Ridlon JM, Gulder TAM, Overmann J, Clavel T. 2020. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat Commun 11:6389. doi: 10.1038/s41467-020-19929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenske GJ, Ghimire S, Antony L, Christopher-Hennings J, Scaria J. 2020. Integration of culture-dependent and independent methods provides a more coherent picture of the pig gut microbiome. FEMS Microbiol Ecol 96:fiaa022. doi: 10.1093/femsec/fiaa022. [DOI] [PubMed] [Google Scholar]
- 36.Jeong Y, Park J, Kim EB. 2020. Changes in gut microbial community of pig feces in response to different dietary animal protein media. J Microbiol Biotechnol 30:1321–1334. doi: 10.4014/jmb.2003.03021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Zhou Y, Fu H, Xiong X, Fang S, Jiang H, Wu J, Yang H, Gao J, Huang L. 2021. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat Commun 12:1106. doi: 10.1038/s41467-021-21295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshadri R, Leahy SC, Attwood GT, Teh KH, Lambie SC, Cookson AL, Eloe-Fadrosh EA, Pavlopoulos GA, Hadjithomas M, Varghese NJ, Paez-Espino D, Hungate project c, Perry R, Henderson G, Creevey CJ, Terrapon N, Lapebie P, Drula E, Lombard V, Rubin E, Kyrpides NC, Henrissat B, Woyke T, Ivanova NN, Kelly WJ, Hungate1000 Project Collaborators . 2018. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat Biotechnol 36:359–367. doi: 10.1038/nbt.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, Liachko I, Snelling TJ, Dewhurst RJ, Walker AW, Roehe R, Watson M. 2018. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun 9:870. doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart RD, Auffret MD, Warr A, Walker AW, Roehe R, Watson M. 2019. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol 37:953–961. doi: 10.1038/s41587-019-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallet-Gely I, Lemaitre B, Boccard F. 2008. Bacterial strategies to overcome insect defences. Nat Rev Microbiol 6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 42.Ruiu L. 2015. Insect pathogenic bacteria in integrated pest management. Insects 6:352–367. doi: 10.3390/insects6020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galac MR, Lazzaro BP. 2011. Comparative pathology of bacteria in the genus Providencia to a natural host, Drosophila melanogaster. Microbes Infect 13:673–683. doi: 10.1016/j.micinf.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchs TM, Bresolin G, Marcinowski L, Schachtner J, Scherer S. 2008. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol 8:214. doi: 10.1186/1471-2180-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnett JA. 1976. The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem 32:125–234. doi: 10.1016/s0065-2318(08)60337-6. [DOI] [PubMed] [Google Scholar]
- 46.Healy ME, Dillavou CL, Taylor GE. 1977. Diagnostic medium containing inositol, urea, and caffeic acid for selective growth of Cryptococcus neoformans. J Clin Microbiol 6:387–391. doi: 10.1128/jcm.6.4.387-391.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Wear M, Kohli G, Vij R, Giamberardino C, Shah A, Toffaletti DL, Yu CA, Perfect JR, Casadevall A, Xue C. 2021. Inositol metabolism regulates capsule structure and virulence in the human pathogen Cryptococcus neoformans. mBio 12:e0279021. doi: 10.1128/mBio.02790-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell RK, Barnes GL, Cartwright BO, Eikenbary RD. 1983. Growth and sporulation of Beauveria bassiana and Metarhizium anisopliae in a basal medium containing various carbohydrate sources. J Invert Pathol 41:117–121. doi: 10.1016/0022-2011(83)90242-2. [DOI] [Google Scholar]
- 49.Shelton AN, Seth EC, Mok KC, Han AW, Jackson SN, Haft DR, Taga ME. 2019. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J 13:789–804. doi: 10.1038/s41396-018-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unno Y, Shinano T. 2013. Metagenomic analysis of the rhizosphere soil microbiome with respect to phytic acid utilization. Microbes Environ 28:120–127. doi: 10.1264/jsme2.me12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. 2012. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep 2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommerfeld V, Kunzel S, Schollenberger M, Kuhn I, Rodehutscord M. 2018. Influence of phytase or myo-inositol supplements on performance and phytate degradation products in the crop, ileum, and blood of broiler chickens. Poult Sci 97:920–929. doi: 10.3382/ps/pex390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng L, Raman AS, Hibberd MC, Cheng J, Griffin NW, Peng Y, Leyn SA, Rodionov DA, Osterman AL, Gordon JI. 2020. Identifying determinants of bacterial fitness in a model of human gut microbial succession. Proc Natl Acad Sci USA 117:2622–2633. doi: 10.1073/pnas.1918951117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabral DJ, Penumutchu S, Reinhart EM, Zhang C, Korry BJ, Wurster JI, Nilson R, Guang A, Sano WH, Rowan-Nash AD, Li H, Belenky P. 2019. Microbial metabolism modulates antibiotic susceptibility within the murine gut microbiome. Cell Metab 30:800–823.e7. doi: 10.1016/j.cmet.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tilocca B, Witzig M, Rodehutscord M, Seifert J. 2016. Variations of phosphorous accessibility causing changes in microbiome functions in the gastrointestinal tract of chickens. PLoS One 11:e0164735. doi: 10.1371/journal.pone.0164735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markiewicz LH, Honke J, Haros M, Świątecka D, Wróblewska B. 2013. Diet shapes the ability of human intestinal microbiota to degrade phytate–in vitro studies. J Appl Microbiol 115:247–259. doi: 10.1111/jam.12204. [DOI] [PubMed] [Google Scholar]
- 57.Levine UY, Bearson SM, Stanton TB. 2012. Mitsuokella jalaludinii inhibits growth of Salmonella enterica serovar Typhimurium. Vet Microbiol 159:115–122. doi: 10.1016/j.vetmic.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morinaga T, Ashida H, Yoshida KI. 2010. Identification of two scyllo-inositol dehydrogenases in Bacillus subtilis. Microbiology (Reading) 156:1538–1546. doi: 10.1099/mic.0.037499-0. [DOI] [PubMed] [Google Scholar]
- 60.Kang DM, Tanaka K, Takenaka S, Ishikawa S, Yoshida KI. 2017. Bacillus subtilis iolU encodes an additional NADP(+)-dependent scyllo-inositol dehydrogenase. Biosci Biotechnol Biochem 81:1026–1032. doi: 10.1080/09168451.2016.1268043. [DOI] [PubMed] [Google Scholar]
- 61.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23:205–211. doi: 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- 65.Pagès H, Aboyoun P, Gentleman R, DebRoy S. 2020. Biostrings: efficient manipulation of biological strings. R package version 2.58.0.
- 66.Wilkins D. 2020. Gggenes: drawgene arrowmaps in 'ggplot2'. R package version 041.
- 67.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jachner S, Boogaart G, Petzoldt T. 2007. Statistical methods for the qualitative assessment of dynamic models with time delay (R Package qualV). J Stat Soft 22:1–30. doi: 10.18637/jss.v022.i08. [DOI] [Google Scholar]
- 69.Page WJ, Kwon E, Cornish AS, Tindale AE. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol Lett 228:211–216. doi: 10.1016/S0378-1097(03)00753-5. [DOI] [PubMed] [Google Scholar]
- 70.St John FJ, Rice JD, Preston JF. 2006. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J Bacteriol 188:8617–8626. doi: 10.1128/JB.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tobin JF, Schleif RF. 1987. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol 196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- 72.Schweizer HP, Po C. 1996. Regulation of glycerol metabolism in Pseudomonas aeruginosa: characterization of the glpR repressor gene. J Bacteriol 178:5215–5221. doi: 10.1128/jb.178.17.5215-5221.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoskisson PA, Rigali S. 2009. Chapter 1: variation in form and function of the helix-turn-helix regulators of the GntR superfamily. Adv Appl Microbiol 69:1–22. doi: 10.1016/S0065-2164(09)69001-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1, Table S1, and list of additional files. Download spectrum.02013-22-s0001.pdf, PDF file, 0.9 MB (920.8KB, pdf)
Table S2. Download spectrum.02013-22-s0002.xlsx, XLSX file, 0.8 MB (809KB, xlsx)
Table S3. Download spectrum.02013-22-s0003.xlsx, XLSX file, 0.5 MB (479.1KB, xlsx)
Table S4. Download spectrum.02013-22-s0004.xlsx, XLSX file, 0.01 MB (11.3KB, xlsx)
Table S5. Download spectrum.02013-22-s0005.xlsx, XLSX file, 0.01 MB (8.8KB, xlsx)
Table S6. Download spectrum.02013-22-s0006.xlsx, XLSX file, 0.06 MB (57.6KB, xlsx)
Table S7. Download spectrum.02013-22-s0007.xlsx, XLSX file, 0.07 MB (77KB, xlsx)
Table S8. Download spectrum.02013-22-s0008.xlsx, XLSX file, 0.3 MB (355.9KB, xlsx)
Table S9. Download spectrum.02013-22-s0009.xlsx, XLSX file, 0.8 MB (782.2KB, xlsx)
Data Availability Statement
All data are available in the tables in the supplemental material.