Abstract
The motility of Halobacillus halophilus as observed on swarm agar plates was strictly dependent on the chloride concentration. Cl− was apparently not used as the coupling ion for flagellar rotation. Cells grown in the absence of chloride were devoid of flagella, but flagellation was restored upon the addition of chloride. These experiments indicate that chloride is involved in synthesis of flagella in H. halophilus.
Halobacillus halophilus (formerly Sporosarcina halophila) is an obligate, moderately halophilic bacterium which was isolated from salt marsh at the North Sea coast of Germany (1). Most interestingly, growth of vegetative cells as well as germination of endospores of H. halophilus is strictly chloride dependent (1a, 4). H. halophilus is the first obligate chloride-dependent bacterium known to date, and projects to study the physiological basis of the chloride dependence have just begun. In one of these projects, we noted that H. halophilus creates an electrochemical chloride potential (ΔμCl−) of about −190 to −230 mV across the membrane (M. Roeßler and V. Müller, unpublished data). The ΔμCl− is, in principal, sufficient to drive energy-dependent reactions. Flagellar rotation in bacteria is such an energy-dependent process, driven by the electrochemical ion gradient (Na+, H+) across the membrane (3). Since H. halophilus is motile by means of flagella, we asked whether ΔμCl− is used as a driving force for flagellar rotation.
Motile cells were selected by several passages over swarm agar plates, which contained 2.96 g of NH4NO3, 1.21 g of Tris-HCl, 1 ml of vitamin solution (8), 2.1 mg of FeSO4, 1 g of glucose, and 0.25 g of yeast extract per liter of artificial seawater (2). MgCl2 was replaced by MgSO4; NaCl and NaNO3 were added separately in concentrations as indicated. The pH was adjusted to 7.5, and the incubation temperature was 30°C.
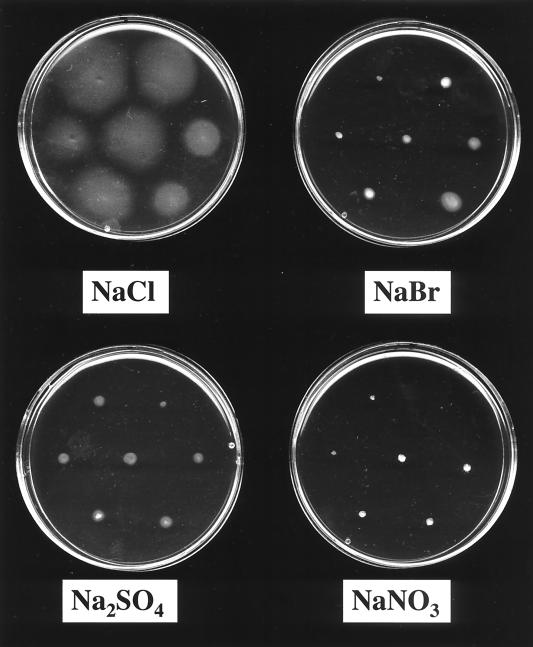
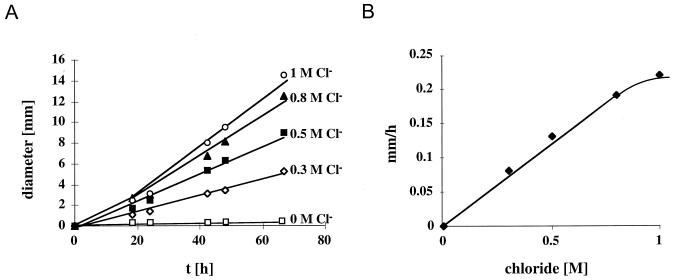
As can be seen in Fig. 1, H. halophilus was motile on swarm agar plates containing 1 M NaCl, as visible by the diffuse zone of bacteria around the colonies. It should be noted that H. halophilus is much more slowly moving than other bacteria, and, therefore, true chemotactic rings were not observed. To analyze the effect of Cl−, NaCl was replaced by other sodium ion salts. It is evident from Fig. 1 that there is a specific requirement for the anion. Neither NaBr nor NaNO3 or Na2SO4 could promote motility. Since at least NaBr and NaNO3 have been shown to promote growth of H. halophilus in the absence of NaCl (4), the apparent nonmotility in the presence of NaNO3 or NaBr is not due to a general inhibition of metabolism by these salts. These experiments demonstrate that motility of H. halophilus is strictly chloride dependent. To further characterize the chloride dependence, quantitative studies with swarm agar plates containing increasing Cl− concentrations were performed. The salt concentration was always kept constant at 1 M by appropriate addition of NaNO3. An increase in the Cl− concentration led to a concomitant increase in motility of H. halophilus, as determined by the time-dependent increase of the swarm size (Fig. 2A). The optimum was reached at around 1.0 M Cl− (Fig. 2B). This value is well within the range of the Cl− concentration required for optimal growth, which was determined to be 0.5 to 2 M (4).
FIG. 1.
Chloride-dependent motility of H. halophilus. Swarm agar plates containing 1 M NaCl, 1 M NaBr, 0.66 M Na2SO4, or 1 NaNO3 were inoculated with H. halophilus and incubated for 48 h at 30°C.
FIG. 2.
Kinetic analyses of chloride-dependent motility of H. halophilus. Colonies were spotted on swarm agar plates with increasing NaCl concentrations. The salt concentration was kept constant at 1 M by appropriate addition of NaNO3. (A) The swarm sizes were measured at the time points indicated and plotted against the incubation time. (B) Migration rates were calculated and plotted against the chloride concentration.
To determine whether ΔμCl− is the driving force for flagellar rotation, free-swimming cells suspended in complex medium (8 g of nutrient broth per liter, 50 mM MgSO4, 1 M NaCl or NaNO3) were observed under the phase-contrast light microscope. Cells grown overnight in chloride-containing medium were motile, whereas cells grown in chloride-free but nitrate-containing medium were not. Addition of chloride to NaNO3-grown cultures did restore motility only after a further incubation for 10 to 12 h. These experiments are not in favor of Cl− as the coupling ion for flagellar rotation.
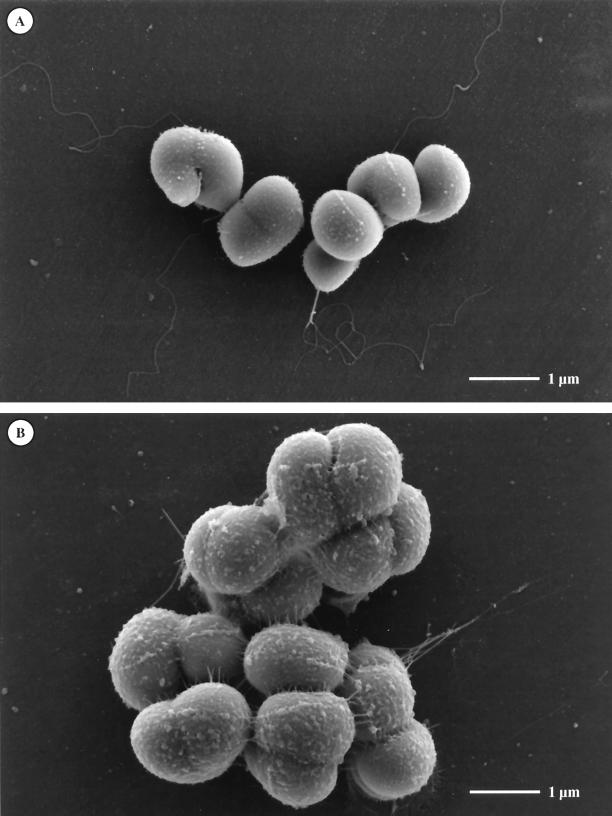
Scanning electron microscopic investigations of cells grown in chloride- or nitrate-containing media were performed to analyze whether chloride might affect the synthesis of flagella. Therefore, cells were grown in complex medium supplemented with either 1 M NaCl or 1 M NaNO3. Cells were harvested by centrifugation and fixed with 2.5% glutaraldehyde in fixative buffer essentially as described previously (7). In addition, the buffer contained 1 M NaCl or 1 M NaNO3 to avoid effects caused by hypoosmosis. Samples were analyzed in a Hitachi S-4100 field emission scanning electron microscope, and representative electron micrographs are depicted in Fig. 3. As can be seen in Fig. 3A, cells grown in chloride-containing medium have one flagellum per cell. In cells grown in chloride-free, nitrate-containing medium, the cell surface had a rough appearance (Fig. 3B). In addition, fibrillar structures extruding into the medium were sometimes visible. These structures have a significantly smaller diameter than flagella and, therefore, most likely are not flagella which are deformed due to lack of chloride. In the presence of NaNO3, cell size was increased and cells tended to aggregate. Most interestingly, we could never observe any flagella in cells grown in nitrate-containing media. A possible inhibition of flagellar synthesis by nitrate could be excluded since flagellation of cells grown in the presence of both 1 M NaCl and 1 M NaNO3 was indistinguishable from cells grown in 1 M NaCl. (data not shown).
FIG. 3.
Chloride-dependent flagellar synthesis in H. halophilus. Cells were grown in complex medium supplemented with 1 M NaCl (A) or 1 M NaNO3 (B) and analyzed by scanning electron microscopy as described in the text.
This is the first demonstration of chloride-dependent flagellation and motility in any living cell. The finding of chloride-dependent synthesis of flagella adds another function to chloride in H. halophilus. The role of chloride could be to serve as an intracellular signal for transcription of flagellar genes or it could be involved in export and assembly of subunits and/or stability of the flagella. A molecular study addressing these questions has to await the generation of protein- and/or RNA-directed probes. However, since germination of endospores as well as growth of the moderate halophile H. halophilus is strictly chloride dependent, it is tempting to speculate that H. halophilus uses Cl− as a signal to measure the salinity of its environment. Cl− could trigger expression of genes, directly or indirectly, involved in flagellar synthesis. Cl−-induced gene expression was recently observed in Lactococcus lactis (5, 6). The elucidation of the role of chloride in flagellar synthesis and assembly in H. halophilus is a challenging task for future experiments, which have just started in our laboratory.
Acknowledgments
This work was supported by a grant from the Fonds der Chemischen Industrie.
The excellent technical assistance of S. Dobler and R. Ulbrich is gratefully acknowledged.
REFERENCES
- 1.Claus D, Fahmy F, Rolf H J, Tosunoglu N. Sporosarcina halophila sp. nov., an obligate, slightly halophilic bacterium from salt marsh soils. Syst Appl Microbiol. 1983;4:496–506. doi: 10.1016/S0723-2020(83)80007-1. [DOI] [PubMed] [Google Scholar]
- 1a.Dohrmann A B, Müller V. Chloride dependence of endospore germination in Halobacillus halophilus. Arch Microbiol. 1999;172:264–267. doi: 10.1007/s002030050769. [DOI] [PubMed] [Google Scholar]
- 2.Fahmy F, Mayer F, Claus D. Endospores of Sporosarcina halophila: characteristics and ultrastructure. Arch Microbiol. 1985;140:338–342. [Google Scholar]
- 3.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 4.Roeßler M, Müller V. Quantitative and physiological analyses of chloride dependence of growth of Halobacillus halophilus. Appl Environ Microbiol. 1998;64:3813–3817. doi: 10.1128/aem.64.10.3813-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders J W, Leenhouts K, Burghoorn J, Brands J R, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 6.Sanders J W, Venema G, Kok J. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl Environ Microbiol. 1997;63:4877–4882. doi: 10.1128/aem.63.12.4877-4882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanner G, Formanek F, Galli D, Wirth R. Localization of aggregation substances of Enterococcus faecalis after induction by sex pheromones. An ultrastructural comparison using immuno labelling, transmission and high resolution scanning electron microscopic techniques. Arch Microbiol. 1989;151:491–497. doi: 10.1007/BF00454864. [DOI] [PubMed] [Google Scholar]
- 8.Wolin E A, Wolin M J, Wolfe R S. Methane formation from bacterial extracts. J Biol Chem. 1963;238:2883–2886. [PubMed] [Google Scholar]