Abstract
Growth rate-independent rrn P1 promoter mutants were tested for their ability to respond to changes in rrn gene dosage. Most were found to be normal for the feedback response. In addition, cellular levels of the initiating nucleoside triphosphates remained unchanged when the rrn gene dosage was altered. These results suggest that the feedback response cannot be the mechanism for growth rate-dependent control of rRNA synthesis and that the relationship between these two processes may be more complicated than is currently understood.
In rapidly growing Escherichia coli cultures, the level of rRNA synthesis controls the protein biosynthetic capacity of the cells (for reviews, see references 5 and 11). The synthesis of rRNA per unit amount of protein increases with the square of the growth rate (14); this phenomenon is called growth rate-dependent control. However, when rrn gene dosage is increased (by the addition of an rrn operon on a multicopy plasmid) or decreased (by deletion of rrn operons from the chromosome), expression from the individual rrn operons changes so that the total rRNA content stays the same (3, 4, 12, 17). This effect is called the feedback response. The rrn P1 promoter is both the site of growth rate-dependent control and the target site of feedback control, suggesting that the feedback response might be the mechanism of growth rate-dependent control (2, 6, 7, 10).
A model correlating initiating nucleoside triphosphate (NTP) concentration with growth rate-dependent control has been proposed (9). In vitro, open complexes of the rrn P1 promoters require high concentrations of either GTP (the initiating nucleotide for rrnD) or ATP (the initiating nucleotide for the remaining six rrn operons). Correspondingly, in vivo, NTP concentrations rise in the cell as the growth rate increases. Also, one growth rate-independent P1 mutant loses the ability to respond to changes in the initiating nucleotide. To explain these results, Gaal et al. propose a model in which an increase in nutrient availability (and hence an increase in NTP concentration) leads to increased rRNA synthesis until a level is reached in which the availability of NTPs is in equilibrium with the consumption of ATP and GTP by the protein synthetic apparatus. These authors suggest that this model can also explain the feedback response: a change in the number of ribosomes as the result of altered gene dosage can lead to a change in ATP and GTP consumption, thus affecting the level of rrn transcription.
In this study, we have sought to determine if the feedback response and growth rate-dependent control are analogous processes. We have approached this question by asking if promoter elements that are required for growth rate-dependent control are the same as those required for the feedback response. Specifically, we tested a representative sample of rrn P1 promoters, which are mutated so that they are now growth rate independent, for a response to changes in rrn gene dosage. One of the mutations, a single base substitution at −1 (C-1T; JV2979) does not raise the level of the promoter above the normal level at high growth rates (2). The other mutations are promoter-up mutations; the activity of promoters with these mutations is not only growth rate independent but also highly elevated over the normal P1 levels, even at high growth rates. One of these mutations is a single base insertion in the region between the −10 and −35 motifs, increasing the spacing from 16-bp to the 17-bp spacing of the consensus E. coli non-rrn promoter (Ains-22; JV1063). One is a single base substitution within the −35 region, also bringing this region to the E. coli consensus (T-33A; JV2935), and one contains double alterations at positions −1 and −15 (C-1T,C-15G; JV901). The mutations in this double mutant have been tested separately (2); the C-1T promoter is growth rate independent as indicated above, and the C-15G promoter is wild type (wt) in its growth rate dependence, although it retains higher-than-wt promoter activity. Most of the promoter fragments contain only the core rrn promoter region plus a small section of the UP element, a 20-bp region just upstream of the P1 promoter-35 sequence which interacts with the α subunit of RNA polymerase and increases the activity of P1 by as much as 30-fold (11). The C-1T,C-15G double mutant (JV901) and its wt cognate (JV1100), however, contain the entire UP element and the promoter-proximal FisI site as well.
The wt and growth rate-independent P1 promoters were fused to lacZ and were present on the chromosome in single copies as lambda prophage. The feedback response was elicited by changing the rrn gene dosage: the number of rrn operons was increased by the addition of rrn operon-containing plasmids to the lysogenic strain or decreased by using lysogenic strains in which rrn operons had been deleted. Measurements of β-galactosidase levels obtained from the growth rate-independent mutant promoters were used to assess the response of these promoters to the feedback control normally seen with altered rrn gene dosage.
Response of rrn P1 growth rate-independent mutants to changes in rrn gene dosage.
A decrease in the wt complement of rrn operons leads to a feedback response in which the remaining operons produce more rRNA to sustain the same level of ribosomes as the normal number of operons would provide (4). An increase in β-galactosidase activity in ΔBAGH, indicative of a normal feedback response, was observed in all but one of the mutant growth rate-independent promoters (Table 1). The rise in expression was similar to the increase in expression observed from the wt promoters.
TABLE 1.
Expression of growth rate-independent rrn promoters in strains with altered rrn gene dosage
| Lysogena | rrnB P1 type | Fragment end points | β-Gal activityb
|
ΔBAGH/W1485d activity ratio | β-Gal activityb
|
pNO1301/pBR322d activity ratio | ||
|---|---|---|---|---|---|---|---|---|
| ΔBAGHc | W1485 | W1485(pNO1301)e | W1485(pBR322) | |||||
| JV1100 | wt | −88, +1 | 3,960 | 2,690 | 1.47 | 1,430 | 1,930 | 0.741 |
| JV783 | wt | −50, +2 | 557 | 364 | 1.53 | 243 | 371 | 0.655 |
| JV2978 | wt | −46, +1 | 3,480 | 2,490 | 1.40 | 725 | 901 | 0.805 |
| JV901 | C-1T,C-15G | −88, +1 | 5,630 | 4,480 | 1.26 | 2,970 | 4,230 | 0.702 |
| JV1063 | Ains-22 | −50, +2 | 2,160 | 2,150 | 1.00 | 1,480 | 2,140 | 0.692 |
| JV2935 | T-33A | −48, +1 | 5,100 | 3,740 | 1.39 | 3,050 | 4,260 | 0.716 |
| JV2979 | C-1T | −46, +1 | 2,670 | 2,110 | 1.27 | 926 | 939 | 0.986 |
Lysogenic strains containing various rrnB P1-lacZ fusions on lambda prophages were the gift of Richard L. Gourse. We have maintained his numbering for identifying the prophages but we have changed the preceding initials to indicate that these prophages are in a different host background (e.g., JV1063 contains the prophage obtained from strain RLG1063; see references 2 and 6 for descriptions of the RLG strains). Lambda phages containing the rrnB P1-lacZ fusions were purified and used to infect both W1485 and ΔBAGH, and the presence of a single prophage was confirmed by PCR (16).
β-Galactosidase (β-Gal) assays were performed as described by Miller (15) except that extracts of sonicated cells were used instead of permeabilized cells. Ampicillin (final concentration, 200 μg/ml) was added to the media for assays of the plasmid-containing strains. At an OD600 of 0.6 to 0.8, each culture was divided to create triplicates that were then sonicated and measured separately for β-galactosidase. The measurements are reported as specific activity, in units per milligram of protein. Because of changes in the protein standard, values for JV2978 and JV2979 differ from the values in the plasmid-containing strains and in Table 2. The triplicates were averaged to obtain the day's value. Experiments on each strain were performed on at least two different days.
In strain W1485ΔBAGH (referred to as ΔBAGH) rrnB, rrnA, rrnG, and rrnH are inactivated by antibiotic cassette insertion and deletion) as described in reference 4.
Standard errors of ratios were less than 10%.
Plasmid pNO1301, a pBR322 derivative that contains the entire rrnB operon, was the gift of Richard L. Gourse (12).
Addition of an rrn-containing plasmid to a cell causes a drop in expression from the individual rrn operons so that the overall amount of rRNA synthesized remains the same (10, 12). All but one of the mutant promoters tested in the pNO1301-containing strain retained the ability to respond to an increase in rrn gene dosage, as indicated by lowered expression relative to that in the strain containing pBR322 (Table 1).
Growth rate-dependent control in strains with decreased rrn gene dosage.
Baracchini and Bremer (1) observed that growth rate-dependent control still operates in strains in which the rrn gene dosage has been increased. We have verified here that a decrease in rrn gene dosage also does not alter the rrn promoter's response to changes in growth rate. We measured β-galactosidase activity in cell extracts of JV2978 (wt P1) and JV2979 (C-1T) after growth of cultures at low and high growth rates. In both the ΔBAGH and W1485 strains, β-galactosidase expression from the wt promoter increased with increased growth rate, while expression from the mutant promoter remained unchanged regardless of growth rate (Table 2). These results indicate that a decrease in rrn gene dosage per se does not result in a deregulation of growth rate-dependent control.
TABLE 2.
Growth rate-dependent control still operates in strains with decreased rrn gene dosage
| Host | Lysogen | Promoter | β-Galactosidase activity
|
Slope (β-galactosidase vs μ)b | |
|---|---|---|---|---|---|
| Low μa | High μa | ||||
| W1485 | JV2978 | wt | 373 | 767 | 0.68 |
| JV2979 | C-1T | 755 | 777 | 0.02 | |
| ΔBAGH | JV2978 | wt | 774 | 1,210 | 0.59 |
| JV2979 | C-1T | 1,050 | 1,060 | 0.02 | |
Growth rates (in doublings per hour) in MOPS (morpholinepropanesulfonic acid)-glycerol media with 0.4% glycerol (low growth rate) were 0.55 for W1485 and 0.49 for ΔBAGH. Growth rates in LB media (high growth rate) were 1.6 for W1485 and 1.3 for ΔBAGH.
Plots were scaled to 1.0 at a μ of 1.0 as follows. Growth rate slopes were determined by linear regression analysis. β-Galactosidase activities at a μ of 1.0 were then scaled to a value of 1.0. Absolute activities were divided by the scaling factor and replotted. The final slope was then determined by linear regression on the scaled values.
Concentration of initiating NTPs in strains with altered rrn gene dosage.
Recent work by Gaal and coworkers (9) showed that the growth rate-dependent control of rRNA synthesis might be regulated by the cellular concentration of the initiating NTPs. Because there is a change in rRNA synthesis per operon in the strains with altered rrn gene dosage, we wished to determine if the initiating NTP concentrations had also changed in these strains. Extractions were performed essentially as described by Little and Bremer (13). M9 minimal medium containing uracil (50 μg/ml), thiamine (10 μg/ml), glucose (0.4%), and all amino acids (40 μg/ml) was inoculated with a single colony of each strain. Growth rates at 30°C were 1.25 doublings/h for W1485 and W1485(pBR322), 0.9 for ΔBAGH, and 0.95 for W1485(pNO1301). All cultures were grown to an optical density at 460 nm (OD460) of approximately 0.8, and then nucleotides were extracted following formaldehyde fixation by using NaOH, essentially as outlined by Little and Bremer (13). Nucleotides were separated and quantitated by fast protein liquid chromatography using a MonoQ HR 5/5 column (Pharmacia Biotech, Piscataway, N.J.). A NaCl gradient of 50 to 350 mM in 5 mM sodium phosphate (pH 7) was used for nucleotide separation. Nucleotide concentrations were calculated by using purified nucleotide standards. No significant change in the ATP concentration was detected. The average ATP concentrations (in picomoles per milliliter per OD460 unit, from at least two independent experiments) were as follows: W1485, 1,760; ΔBAGH, 1,780; W1485(pBR322), 1,930; and W1485(pNO1301), 1,720. We also found no change in GTP concentration (the initiating nucleotide for rrnD; data not presented). Our results suggest that the initiating NTP concentration is not the only effector of the feedback response.
Level of change of rrn expression in response to rrn gene dosage.
We found that the increase in expression from the rrn P1 promoter fragments in the ΔBAGH background was 1.3- to 1.5-fold higher than in the wt background. This increase was consistent with the increase in the number of RNA polymerase molecules on the rrn operons in ΔBAGH relative to the wt as found by Condon and coworkers (1.3) (4). In the strains containing the rrn plasmid, we found that expression from the rrn P1 promoter fragments was 0.65 to 0.8 that of the expression in the wt background. This decrease is in good agreement with one study (8) but was not as large as that found in other studies (10, 12). Our results are also in good agreement with the rrn plasmid-induced reduction in both the number of RNA polymerases on rrn operons (pNO1301/pBR322 = 0.66) as determined by electron microscopy and the level of tRNATrp (pNO1301/pBR322 = 0.69) as determined by RNA dot blot analysis (18). (The unique gene encoding tRNATrp is transcribed as part of the rrnC operon, so quantitation of cellular levels of tRNATrp provides a handy measure of cellular expression of rRNA genes.) The experiments by Voulgaris et al. (18) and Gaal and Gourse (8) and in the work reported here were performed in Luria-Bertani (LB) media or glucose minimal media supplemented with 0.5% Casamino Acids, while other published experiments showing a greater decrease in expression were performed in minimal media with a lower concentration of amino acids. The differences in media may explain the discrepancy in down-regulation observed. Another difference in our experiments is that we have used promoter fragments that contain at most the UP element and the FisI site. In other published experiments, promoter fragments which include the UP element and all three Fis sites have exhibited a greater decrease in expression than fragments containing only the core promoter (10).
The promoter fragment containing the double mutation C-1T,C-15G (JV901) responded normally to the presence of extra copies of rrn. This was an unexpected result, because Gaal and Gourse (8) had measured the expression from this doubly mutated promoter in the presence of the same rrn plasmid and found virtually no feedback sensitivity. These authors noted that this result was consistent with the model that feedback repression is the mechanism by which growth rate-dependent control is achieved. However, they used a different construct of this promoter mutation, RLG1019, while we used the construct JV901. JV901 contains the entire UP element and the FisI site, while RLG1019 does not. We have not ruled out a possible role for the UP element or Fis sites in the feedback response, and these contradictory results, obtained with mutations which are the same but are carried on different promoter fragments, suggest that a complex interaction may occur in which one control mechanism may compensate for another if one system is damaged or missing.
Relevance of NTP concentrations.
In the model for growth rate-dependent regulation of rRNA transcription proposed by Gaal et al. (9), the initiating NTP concentration in the cell is a regulator of the level of expression of the rrn operons and could explain the changes in rrn transcription as a function of gene dosage. Increased rrn gene dosage would lead to more ribosomes and thus increased consumption of the initiating nucleotides, which in turn would leave less available for further rRNA synthesis. The converse, for a reduction in rrn gene dosage, would also be true. We found, however, that the levels of ATP and GTP were essentially unchanged in the cells with altered gene dosage, suggesting that this model does not completely account for the feedback response. Also, the promoter fragment containing the C-1T mutation, which is insensitive to NTP concentration (9), responded normally to a decrease in rrn gene dosage. This mutation, however, did result in loss of the ability to respond to an increase in rrn gene dosage. Thus, our results do not rule out a role for NTP concentration in the feedback response.
Different promoter elements responsible for decreased and increased rrn gene dosage.
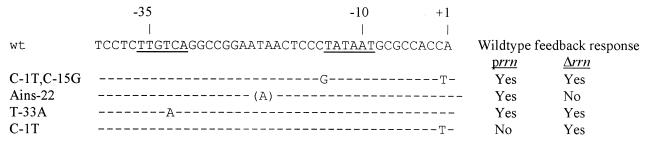
We found that in each of the two conditions of altered rrn gene dosage, there was one growth rate-independent control mutation that did destroy the feedback response (Fig. 1). Thus, it is clear that growth rate-dependent control and the feedback response do share some P1 promoter element requirements and presumably share some mechanistic properties as well (sensitivity to NTP concentration, for example). Surprisingly, however, the mutation that affected the feedback response to an increase in rrn gene dosage was not the same as the mutation that caused the loss of the ability to respond to a decrease in rrn gene dosage (Fig. 1). These results suggest that the mechanisms for responding to higher- and lower-than-normal numbers of rrn operons are not the same.
FIG. 1.
Summary of the effects of growth rate-independent control mutations on the rrn P1 feedback response. Dashes represent identity with the wt promoter; letters show the nucleotides which are substitutions (the letter in parentheses designates an insertion). prrn, strain with rrn-containing plasmid; Δrrn, strain with rrn deletions.
Except for the C-1T mutation, which has been shown to result in loss of sensitivity to NTP concentration (9), it is not known precisely how most of the mutations we have studied affect growth rate-dependent control. Because our results suggest that the feedback and growth rate controls can be separated, it may be that the feedback response operates at different steps in the initiation/promoter clearance pathway than does growth rate-dependent control and that most of the mutations that we have chosen to study may not affect the feedback-sensitive steps. Alternatively, different effector molecules may regulate the two processes.
Acknowledgments
We are grateful to Rick Gourse for providing the lysogenic strains containing the rrnB P1-lacZ fusions on lambda prophages and for plasmid pNO1301, to Ciarán Condon and Rick Gourse for their helpful feedback on the manuscript, and to Max Gottesman for generously hosting J.V. in his laboratory.
This work was supported by National Institutes of Health grants GM50747 to W.M.H. and GM24571 to C.L.S.
REFERENCES
- 1.Baracchini E, Bremer H. Control of rRNA synthesis in Escherichia coli at increased rrn gene dosage. J Biol Chem. 1991;266:11753–11760. [PubMed] [Google Scholar]
- 2.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole J R, Olsson C L, Hershey J W B, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli: requirement for initiation factor IF2. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- 4.Condon C, French S, Squires C, Squires C L. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickson R R, Gaal T, deBoer H A, deHaseth P L, Gourse R L. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J Bacteriol. 1989;171:4862–4870. doi: 10.1128/jb.171.9.4862-4870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaal T, Barkei J, Dickson R R, deBoer H A, deHaseth P L, Alavi H, Gourse R L. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J Bacteriol. 1989;171:4852–4861. doi: 10.1128/jb.171.9.4852-4861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaal T, Gourse R L. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 10.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 11.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 12.Jinks-Robertson S, Gourse R L, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- 13.Little R, Bremer H. Quantification of guanosine 5′,3′-bisdiphosphate in extracts from bacterial cells by ion-pair reverse phase high performance liquid chromatography. Anal Biochem. 1982;126:381–388. doi: 10.1016/0003-2697(82)90531-0. [DOI] [PubMed] [Google Scholar]
- 14.Maalöe O, Kjeldgaard N O. In control of macromolecular synthesis: a study of DNA, RNA, and protein synthesis in bacteria. New York, N.Y: Benjamin; 1966. [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Powell B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharrock R A, Gourse R L, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the musB5 mutant of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voulgaris J, French S, Gourse R L, Squires C, Squires C L. Increased rrn gene dosage causes intermittent transcription of rRNA in Escherichia coli. J Bacteriol. 1999;181:4170–4175. doi: 10.1128/jb.181.14.4170-4175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]