Abstract
Saccharomyces cerevisiae HTR1 mutants are severely impaired in the uptake of glucose. We have cloned dominant HTR1 mutant alleles and show that they encode mutant forms of the Mth1 protein. Mth1 is shown to be involved in carbon source-dependent regulation of its own, invertase and hexose transporter gene expression. The mutant forms block the transduction of the Snf3- and Rgt2-mediated glucose signals upstream of the Rgt1 transcriptional regulator.
HTR1 mutants show severely reduced glucose uptake rates (12). They were obtained by selecting for revertants of triosephosphate isomerase (tpi1Δ) mutants of Saccharomyces cerevisiae which had overcome the strong inhibitory effect of glucose on growth and carbon metabolism (12). The transcription of various known glucose transporter genes (HXT1, HXT3, and HXT4) is defective in HTR1 mutants. It was speculated that mutations in HTR1 affect a negative factor of hexose transporter gene expression.
(This work is part of the Ph.D. theses of F. Schulte and R. Wieczorke from Heinrich-Heine-Universität, Düsseldorf, Germany.)
To identify the dominant HTR1 allele, a DNA library was constructed from the HTR1-23 mutant strain (12). Genomic DNA was partially digested with Sau3A and cloned into the BamHI-digested vector YCp50. This library was transformed into the yeast wild-type strain MC971A (MATa ura3-52 his3-11,15 MAL2 SUC2 GAL MEL) and plated onto YNB medium (0.67% yeast nitrogen base, supplemented for auxotrophic demands) lacking uracil with 2% glucose. A total of about 10,000 transformants were replica plated onto YNB medium lacking uracil with 2% galactose additionally supplemented with 100-ppm 2-deoxyglucose (2-DOG), a toxic analogue of glucose (1), because HTR1 mutants are resistant to 2-DOG. The plasmids of two transformants which were able to grow in the presence of 2-DOG were isolated, amplified in Escherichia coli DH5αF′, and retransformed into yeast strain MC971A. One of the plasmids (pHTR1-23) conferred growth in the presence of 2-DOG to all of the transformants. A 5.9-kb HindIII/SalI fragment containing the complete insertion of plasmid pHTR1-23 was recloned into the integrative vector YIp5. This plasmid was linearized with XhoI within the inserted fragment and integrated into the genome of the original HTR1-23 mutant strain. The correct integration into the homologous genomic region was confirmed by Southern analysis. The strain was crossed with the isogenic wild-type strain MC996A (MATa ura3-52 his3-11,15 leu2-3,112 MAL2 SUC2 GAL MEL) and a tetrad analysis was performed. Eighteen tetrads were all parental ditype with regard to resistance to 2-DOG and uracil prototrophy, indicating that the cloned fragment is tightly linked to the dominant HTR1-23 allele.
The dominant HTR1-23 gene was localized on a 2.6-kb EcoRI/(Sau3A-BamHI) fragment, which contains the complete MTH1 gene. This gene originally had been isolated as a homologue of the STD1 (also known as MSN3) gene (4, 7). Both proteins have been shown to modulate glucose-regulated expression of SUC2 and HXT1-4 (7, 18, 19). The DNA sequence of the entire coding region of HTR1-23 and large parts of the promoter and terminator regions were determined and compared to the sequence of wild-type strain MC971A. Thymine, at position +254 of the coding region of MTH1, was replaced by an adenine in HTR1-23, leading to a substitution of asparagine in the mutant protein for isoleucine85 in the wild-type protein. Cloning and sequencing of additional dominant HTR1 mutant alleles revealed the same base pair change in HTR1-19. In HTR1-5, the thymine at position +254 was replaced by a guanine, resulting in a substitution of isoleucine85 for serine in the corresponding protein. Thus, substitution in Mth1 of isoleucine85 for either asparagine or serine was responsible for the dominant negative effect on yeast growth.
The region surrounding isoleucine85 is highly conserved between Mth1 and its homologue Std1 (Msn3). In order to see whether the replacement of the corresponding isoleucine94 in Std1 by serine has similar effects on yeast growth, the adenine and thymine residues at positions +280 and +281 of the STD1 coding region were replaced by thymine and cytosine, respectively, with the in vitro mutagenesis procedure described in reference 2 and the mutagenic primer 5′-GGCAAGAATAGAATCCATAAAAAGGTTATTGCC-3′. The mutagenized gene was cloned into YEplac181 (5) and transformed into the wild-type strain MC996A. However, in contrast to the HTR1-23 mutant strain, the transformants did not show any growth defect on glucose or raffinose medium, consistent with the recent observation that Mth1 and Std1, while homologous, are not functionally redundant (18).
To delete the MTH1 (HTR1) gene, a 0.9-kb AflII/EcoRI fragment containing 141 bp of the promoter and 804 bp of the coding region was replaced by a 1.5-kb AflII/EcoRI fragment containing the HIS3 gene and transformed into the diploid strain MC971. Histidine-prototrophic transformants were sporulated, and strain YFS3 (mth1Δ) was obtained. The correct replacement was confirmed by Southern analysis. The growth properties of the wild type, the dominant HTR1-23 mutant, and the mth1 deletion strain were compared. Whereas no growth differences between the three strains on yeast extract-peptone (YP)–2% maltose medium could be observed, after shifting the mth1 deletion mutant grew faster on YP–2% raffinose than the wild-type cells. The HTR1-23 mutant cells did not grow on raffinose medium. Moreover, in contrast to the wild-type cells and the mth1 mutant cells, the HTR1-23 mutants grew very slowly on YP–2% glucose medium and were resistant to 100-ppm 2-DOG on a YNB–2% galactose medium. These results show that the dominant phenotype of the HTR1 mutant allele is not a mere consequence of the loss of protein function.
It has been shown that the growth defect of HTR1-23 mutant cells is largely due to a decrease in glucose uptake activity, which is caused by a profound decrease in the transcription of HXT1, -3, and -4 genes (12). To compare the impact of the dominant mutant allele and the mth1 deletion allele on HXT gene expression, the E. coli lacZ gene was placed under control of the HXT7 promoter by cloning a 1.6-kb HindIII/PstI HXT7 promoter fragment into the multiple cloning site of vector YIp356 (10). The resulting plasmid and plasmid pBM2637 containing the lacZ gene under the control of the HXT1 promoter (a kind gift of S. Özcan) were linearized with StuI and integrated in single copy into the chromosomal ura3-52 alleles of the wild-type, the HTR1-23, and the mth1 deletion strains. Moreover, a 3.5-kb DNA fragment comprising the complete MTH1 promoter and parts of the MTH1 coding region was amplified by PCR, simultaneously adding a HindIII recognition site behind the second codon of MTH1. This fragment was cleaved with XhoI and HindIII, and a 2.7-kb fragment was inserted in front of the lacZ gene of vector YIp356 and integrated into the ura3 locus of all three strains. Determination of β-galactosidase and invertase (Suc2) activities in the different strains grown in the presence of different carbon sources (Table 1) revealed that transcription of the high-glucose-induced HXT1 gene was completely abolished in the dominant HTR1-23 mutant cells. Induction of HXT1 by high concentrations of glucose is dependent on the Rgt2-SCFGrr1-Rgt1 signaling pathway (8, 13). Moreover, the activity of the high-glucose-repressed and low-glucose-induced HXT7, SUC2, and MTH1 promoters was kept on a low and constitutive level in the HTR1-23 mutant cells. Raffinose was used as a carbon source to produce low concentrations of extracellular hexoses, as it is hydrolyzed by invertase and melibiase outside the cells. On raffinose and ethanol media, deletion of the MTH1 gene caused an increase in the promoter activity of HXT7, SUC2, and MTH1 itself (Table 1), demonstrating that Mth1 has a repressing function on the transcription of these genes. Moreover, the MTH1 gene seems to be subject to an autoregulatory mechanism. An HXT6 promoter-lacZ construct was regulated in ways very similar to that of HXT7 in the appropriate strains (data not shown). Glucose repression of HXT7, SUC2, and MTH1 was partially relieved in HTR1-23 cells (Table 1). To demonstrate that this was only a consequence of a lack of glucose transport (17), plasmid pFM-HXT1 containing the HXT1 gene under control of the strong ADH1 promoter (17) was transformed into the HTR1-23 mutant strain. Glucose repression of SUC2 expression was nearly completely restored (Table 1), indicating that the Htr1-23 protein has no direct function in the transduction of the glucose repression signal.
TABLE 1.
Expression of genes HXT1, HXT7, SUC2, and MTH1 in different yeast strainsa
| Gene | Strain genotype | Activity (nmol min−1 mg of protein−1)c on:
|
|||
|---|---|---|---|---|---|
| 2% Glucose | 2% Maltose | 2% Raffinose | 2% Ethanol | ||
| HXT1 | Wild type | 129 | 2 | 4 | <1 |
| HTR1-23 | <1 | <1 | <1 | <1 | |
| mth1Δ | 128 | 3 | 3 | 14 | |
| HXT7 | Wild type | 3 | 300 | 1,290 | 820 |
| HTR1-23 | 62 | 25 | 35 | 100 | |
| mth1Δ | 3 | 450 | 3,030 | 4,500 | |
| snf3Δ | —b | — | 86 | 166 | |
| grr1Δ | — | — | 38 | 67 | |
| snf3Δ rgt1Δ | — | — | 1,307 | — | |
| HTR1-23 rgt1Δ | 28 | 106 | 670 | 981 | |
| HTR1-23-YEpMTH1 | 230 | 65 | 200 | 190 | |
| Wild type-YEpMTH1 | 200 | 180 | 510 | 220 | |
| MTH1 | Wild type | 1 | 60 | 105 | 100 |
| HTR1-23 | 30 | 12 | 25 | 30 | |
| mth1Δ | 1 | 110 | 230 | 430 | |
| SUC2 | Wild type | 2 | 37 | 866 | — |
| HTR1-23 | 280 | 21 | 99 | — | |
| mth1Δ | 2 | 53 | 1,338 | — | |
| HTR1-23-pFM-HXT1 | 13 | — | — | — | |
| hxt1-7Δ | 980 | — | 1,240 | — | |
Cells were grown for about 15 h in YP medium or, in the case of plasmid-bearing cells, in YNB medium lacking uracil supplemented with different carbon sources to a density of 1 × 107 to 3 × 107 cells/ml. Mean values of data from at least two independent experiments are given. Standard deviations were less than 15%.
—, not determined.
For HXT1, HXT7, and MTH1, β-galactosidase activity; for SUC2, invertase activity.
On the other hand, Mth1 seems to be involved in the low-glucose-activated Snf3-SCFGrr1 signaling pathway (18), which has been shown to be responsible for the transcriptional induction of HXT2-4 and -6 and SUC2 genes (9, 11, 15, 20, 22). The SNF3 and GRR1 genes were deleted in the wild-type strain containing the HXT7 promoter-lacZ fusion, as described in reference 6. In both the snf3 and the grr1 mutants, HXT7 is not induced by low concentrations of hexoses (Table 1), indicating that HXT7 is regulated via the low-glucose-activated Snf3-SCFGrr1 signaling pathway. Surprisingly, the high-level expression of HXT7 on ethanol medium was reduced in the snf3 as well as in the grr1 mutant cells. RGT1, which encodes a transcriptional repressor that is inactivated by the Snf3-SCFGrr1 signaling pathway in response to low concentrations of glucose (11, 14), was deleted in the snf3 and HTR1-23 mutant strains, as described in reference 21. In both strains, deletion of RGT1 restored the high-level expression of HXT7 on raffinose and ethanol media (Table 1). Moreover, deletion of RGT1 in the HTR1-23 cells restored growth on glucose and partially on raffinose media. Thus, mutations in rgt1 are epistatic to the mutation in HTR1-23, suggesting that Mth1 acts upstream of Rgt1 in the glucose-signaling pathway. Lack of induction of glucose-induced genes in the HTR1-23 mutant strain was not a consequence merely of a lack of glucose transport, because the glucose transport-deficient yeast strain RE700A (hxt1-7Δ) (16) showed a very high expression of invertase (Suc2) activity on raffinose medium, in contrast to the HTR1-23 strain (Table 1).
The negative effect of the HTR1-23 allele on growth of the cells on glucose could be strengthened by integrating several copies of HTR1-23 into the genome of the wild-type strain. The negative effect of HTR1-23 on the transcription of HXT7 could be partially overcome by the overexpression of a 4.3-kb XhoI/Sau3A fragment containing the complete wild-type MTH1 gene cloned into YEplac181 (Table 1). These results suggest that both forms of the Mth1 protein compete for a common target. Overexpression of MTH1 partially relieved glucose repression of HXT7 even in the wild-type strain (Table 1), suggesting that Mth1 has an activating effect on the glucose derepression pathway (3, 7).
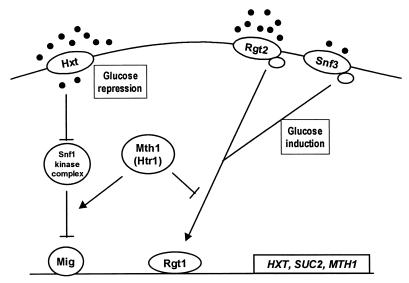
Our results, together with previous investigations (7, 18), demonstrate that Mth1 is a negative modulator of the Snf3-dependent glucose induction pathway and a positive modulator of the Snf1-dependent glucose derepression pathway, thus connecting the two glucose-responsive signaling pathways (Fig. 1). Substitution of isoleucine85 for serine or asparagine creates a mutant protein that is permanently locked in its inhibitory conformation. In this mutant form, Mth1 (Htr1) blocks the Snf3- and Rgt2-mediated glucose signals upstream of the Rgt1 transcriptional regulator.
FIG. 1.
Model for the involvement of Mth1 (Htr1) in glucose signal transduction in yeast. Arrows indicate activation and lines with perpendicular bars indicate inhibition. Filled circles represent glucose.
Acknowledgments
We acknowledge the work of Michael Ciriacy, who initiated the characterization of HTR1 in this laboratory.
This work was supported by grant BO 1517/1-1 from Deutsche Forschungsgemeinschaft and grant GR 97/9-02 from Internationale Brachet Stiftung to E.B.
REFERENCES
- 1.Biely P, Kratky Z, Bauer S. Metabolism of 2-deoxy-d-glucose by baker's yeast. IV. Incorporation of 2-deoxy-d-glucose into cell wall mannan. Biochim Biophys Acta. 1972;255:631–639. doi: 10.1016/0005-2736(72)90166-6. [DOI] [PubMed] [Google Scholar]
- 2.Boles E, Miosga T. A rapid and highly efficient method for PCR-based site-directed mutagenesis using only one new primer. Curr Genet. 1995;28:197–198. doi: 10.1007/BF00315788. [DOI] [PubMed] [Google Scholar]
- 3.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 4.Ganster R W, Shen W, Schmidt M C. Isolation of STD1, a high-copy-number suppressor of a dominant negative mutation in the yeast TATA-binding protein. Mol Cell Biol. 1993;13:3650–3659. doi: 10.1128/mcb.13.6.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 6.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard E J, Jiang R, Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 11.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Özcan S, Freidel K, Leuker A, Ciriacy M. Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J Bacteriol. 1993;175:5520–5528. doi: 10.1128/jb.175.17.5520-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Özcan S, Dover J, Rosenwald A G, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Özcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Özcan S, Vallier L G, Flick J S, Carlson M, Johnston M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast. 1997;13:127–137. doi: 10.1002/(SICI)1097-0061(199702)13:2<127::AID-YEA68>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- 17.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt M C, McCartney R R, Zhang X, Tillman T S, Solimeo H, Wölfl S, Almonte C, Watkins S C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillman T S, Ganster R W, Jiang R, Carlson M, Schmidt M C. STD1 (MSN3) interacts directly with the TATA-binding protein and modulates transcription of the SUC2 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3174–3180. doi: 10.1093/nar/23.16.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallier L G, Coons D, Bisson L F, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 22.Wendell D L, Bisson L F. Expression of high-affinity glucose transport protein Hxt2p of Saccharomyces cerevisiae is both repressed and induced by glucose and appears to be regulated posttranslationally. J Bacteriol. 1994;176:3730–3737. doi: 10.1128/jb.176.12.3730-3737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]